Abstract
The zebrafish has become a commonly used model for studying hematopoiesis as a result of its unique attributes. Zebrafish are highly suitable for large-scale genetic and chemical screens compared to other vertebrate systems. It is now possible to analyze hematopoietic lineages in zebrafish and validate cell function via transplantation assays. Here, we review advancements over the past decade in forward genetic screens, chemical screens, fluorescence-activated cell sorting analysis, and transplantation assays. Integrating these approaches enables new chemical and genetic screens that assay cell function within the hematopoietic system. Studies in zebrafish will continue to contribute and expand our knowledge about hematopoiesis, and develop novel treatments for clinical applications.
Keywords: zebrafish, hematopoiesis, stem cell, screens, transplantation
Introduction
The zebrafish has unique advantages that make it an ideal model organism for studying hematopoiesis. The external fertilization of zebrafish allows for observation of and experimentation on embryos to commence as early as the one-cell stage. Their rapid early-stage development and transparency facilitates in vivo imaging of early embryonic processes, such as the onset of a beating heart and circulating blood cells. Moreover, one mating pair can produce hundreds of embryos at a time, making zebrafish powerful for large-scale genetic and chemical screens.
Zebrafish, like other vertebrate organisms, experience two distinct waves of hematopoiesis, primitive and definitive [1]. Primitive hematopoiesis occurs at two intraembryonic sites—the anterior lateral mesoderm (ALM) and the posterior lateral mesoderm (PLM), the latter of which gives rise to the intermediate cell mass (ICM). The ALM produces myeloid cells while the PLM predominately produces erythrocytes. The ventral mesoderm tissue of the ICM is the birthplace of primitive HSCs and the site of early erythropoiesis [2]. At 24 hours-post-fertilization (hpf), primitive blood cells move into circulation. In the definitive wave, multipotential HSCs, capable of producing all of the blood lineages, are produced in the ventral wall of the dorsal aorta [3], also known as the aorta-gonad-mesonephros (AGM). These HSCs then migrate to the caudal hematopoietic tissue (CHT) in the posterior tail region at 36 hpf [4]. Some HSCs travel directly from the AGM to the thymus where lymphopoiesis occurs at 3 dpf. By 4 dpf, HSCs from the AGM and CHT seed the kidney marrow, the equivalent of mammalian bone marrow (BM) and the site of larval and adult hematopoiesis.
Although the main hematopoietic sites in zebrafish differ from those in mammals, both zebrafish and mammals share all major blood cell types that arise from common hematopoietic lineages [5], making findings in zebrafish blood development translatable to mammalian systems. Moreover, many genes and signaling pathways involved in hematopoiesis are conserved between fish and mammals, notably transcription factors that mediate blood development and HSC differentiation. For example, scl is one of the first transcription factors expressed in early hematopoietic cells. During definitive hematopoiesis, runx1 marks the HSCs in both mouse and fish, whereas in the differentiated populations, gata1 is the erythroid lineage regulator, pu.1 and c/ebp are the myeloid lineage regulators, and ikaros marks the lymphoid population in accordance with the hematopoietic hierarchy in mammals [6].
During the past three decades, studies in zebrafish have provided insight into normal blood cell development and hematopoietic diseases. We highlight a few major advances from these studies using forward genetic screen and chemical screen approaches to illustrate the strength of this animal model. In addition to uncovering novel genetic and molecular pathways that govern hematopoiesis, previous studies have also made progress in developing new tools and assays that can be used to study blood development in zebrafish. Specifically, fluorescence-activated cell sorting (FACS) and hematopoietic cell transplantation, techniques commonly used to study hematopoiesis in mammals, have been implemented in zebrafish, permitting examination of zebrafish hematopoiesis in greater detail. We will also summarize techniques currently available or in progress in the field. In the future, genetic and chemical studies using transplantation and FACS as a readout will expand the capabilities of this area of research.
Forward Genetic Screens
Zebrafish embryos are amenable to a variety of genetic manipulations. The ex vivo development make them convenient for microinjections. mRNA and DNA-mediated overexpression and morpholino-mediated knockdown of genetic components provide rapid and effective ways to assess molecular functions of relevant genes in a live animal. Previously, targeted genome modification has been difficult in zebrafish. Several new strategies including the CRISPR-Cas system [7] and TALENs [8] have overcome this bottleneck and have provided efficient reverse genetic approaches for site-specific gene targeting. Zebrafish have entered a new age of genome manipulation; and, compared to other vertebrate organisms, they have a great advantage in forward genetic screens, which can be performed on a scale not feasible in any other vertebrate models [9; 10]. The mutations can be induced by different strategies, including chemical mutagenesis using ethylnitrosourea (ENU) and retroviral insertional mutagenesis [11].
Two decades ago, two large-scale ENU mutagenesis screens generated hundreds of mutants that affect different developmental processes, including hematopoiesis [12; 13; 14]. These mutants revealed the participation of several novel genes or novel functions during hematopoietic development. Subsequent studies elucidated the mechanism underlying these functions. Here, we use zebrafish moonshine (mon) mutant as an example to illustrate how zebrafish forward genetic studies permit us to understand the mechanism underlying the rapid and coordinated erythroid cell differentiation during development.
The moonshine mutant was identified in the original large-scale forward genetic screen because it results in defective erythropoiesis and severe anemia [14]. In moonshine mutant, the primitive and definitive erythroid progenitors are specified, but fail to express normal level of blood-specific genes and undergo cell death. Thus, mon is required for normal terminal differentiation and survival of erythroid progenitor cells [15]. Positional cloning identified the mon gene as the zebrafish ortholog of transcription intermediary factor1γ (TIF1γ), a nuclear factor expressed throughout the zebrafish embryo and enriched in hematopoietic tissue. TIF1γ belongs to the TIF1 family of transcriptional coactivators and corepressors that contain an N-terminal TRIM domain composed of a RING finger, two B boxes, and a coiled-coil domain, and a C-terminal PHD finger and bromodomain [15]. These domains are implicated in protein5 protein interaction and chromatin modification, which compounds the precise role mediated by TIF1γ. In human hematopoietic cells, it was found that TIF1γ interacts with phosphorylated Smad2/3 in competition with Smad4, and that Smad2/3-Tif1γ and Smad2/3-Smad4 act as complementary effector arms mediating different cellular responses by the transforming growth factor-beta (TGFβ) pathway [16]. In other studies primarily done in Xenopus embryos, Dupont et al showed that TIF1γ acts as a Smad4 monoubiquitin ligase and inhibits TGFβ/bone morphogenetic protein (BMP) signaling through the inhibition of Smad2/3 complex formation by ubiquintinated Smad4 [17; 18]. These studies suggest that the function of TIF1γcould be cell context-dependent.
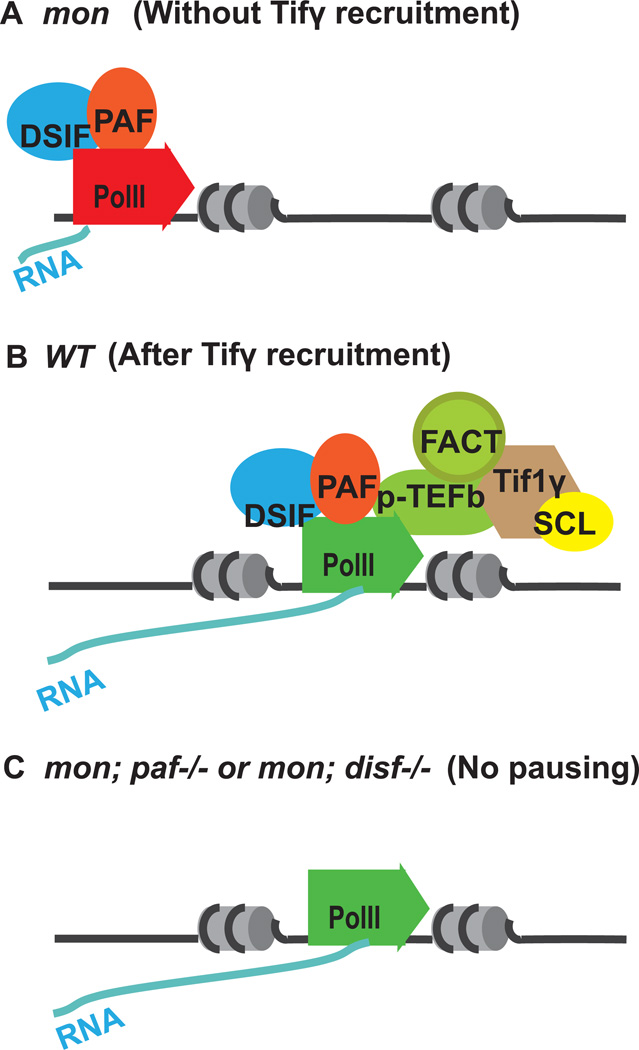
To understand the precise role of TIF1γ during developmental hematopoiesis, a genetic suppressor screen was performed to identify additional mutations that could bypass TIF1γ and restores erythropoiesis in mon [19]. This approach has been widely used in lower organisms as an unbiased way to isolate interacting components of an genetic pathway of interest. It has not been commonly used in vertebrates. The mon mutants are homozygous lethal, which makes the suppressor screen difficult. To overcome this problem, a viable mon homozygous line was created in which one copy of a bacterial artificial chromosome (BAC) transgene containing the entire TIF1γ gene locus, recombined with a GFP maker driven by an actin promoter was inserted into the genome. Haploid embryos from F1 progeny were used to for the suppressor screen, which greatly shortened the screen procedure [20]. From 800 ENU-mutagenized haploid genomes, two suppressor mutants were identified that rescue erythroid defects in mon mutants. One of the mutants has a mutation in cdc73 gene, which encodes a subunit of the Pol II Associated Factor (PAF) complex. PAF is a transcription elongation factor and functionally interacts with other elongation factors including DSIF, FACT and p-TEFb. Further genetic studies revealed that knockdown of DSIF subunits also restored erythropoiesis in mon embryos. In separate biochemical studies, TIF1γ was found to physically interact with blood-specific SCL transcription complex and the positive elongation factors p-TEFb and FACT. Together, these results suggest a model in which TIF1γ recruits positive elongation factors, through interacting with SCL transcription complex, to erythroid genes, thereby releasing paused PolII and turning on the erythroid program (Figure 1). TIF1γ controls the erythroid differentiation through regulating transcription elongation, which likely ensures a rapid and synchronized response in erythroid precursors to differentiation signals.
Figure 1. Tifγ regulates transcription elongation of blood genes.
(A) In mon mutants, Pol II is paused by the DSIF and PAF and would not efficiently transcribe blood genes. (B) In wildtype cells, Tifγrecruits positive elongation factor p-TEFb and FACT to blood genes through the SCL complex and stimulates Pol II to proceed to finish transcription elongation. (C) In mon; cdc73−/− double mutants, the pausing factor PAF is lost, Pol II is no longer paused. Elongation continues without help from positive elongation factors.
The zebrafish studies prompted the study of TIF1γ in murine hematopoiesis. In mouse models with hematopoietic deletion of TIF1γ, erythroid maturation was blocked in the bone marrow and similar defects in transcription elongation were observed [21]. In addition, loss of TIF1γ in mice results in defects in other hematopoietic compartments including profound loss of B cells, decreased adult HSCs and expansion of granulocytes [21; 22]. A more recent study in zebrafish reveled similar expansion of definitive myeloid differentiation in mon mutants [23]. These studies support the evolutionarily conserved hematopoiesis between zebrafish and mammals.
Chemical screening in the Zebrafish
In addition to traditional genetic approaches, chemical screens, both genetic and phenotypic, have identified many unknown players in primitive and definitive hematopoiesis. Thousands of wild type embryos can be easily used in chemical screens to uncover modulators of blood development. Screening in zebrafish is more powerful when mutant strains are used to identify specific pathways of interest [1]. Zebrafish disease models present an excellent platform for screening chemical suppressors—the molecules identified will expand our understanding of pathways involved in disease development, such as leukemogenesis [24]. Notably, the small molecules identified can be translated into therapeutic studies and may have profound clinical applications. Screening of libraries that contain FDA approved drugs, in combination with the fact that thousands of molecules can be screened in a few weeks, makes zebrafish a powerful in vivo model for drug discovery.
Prostaglandin E2
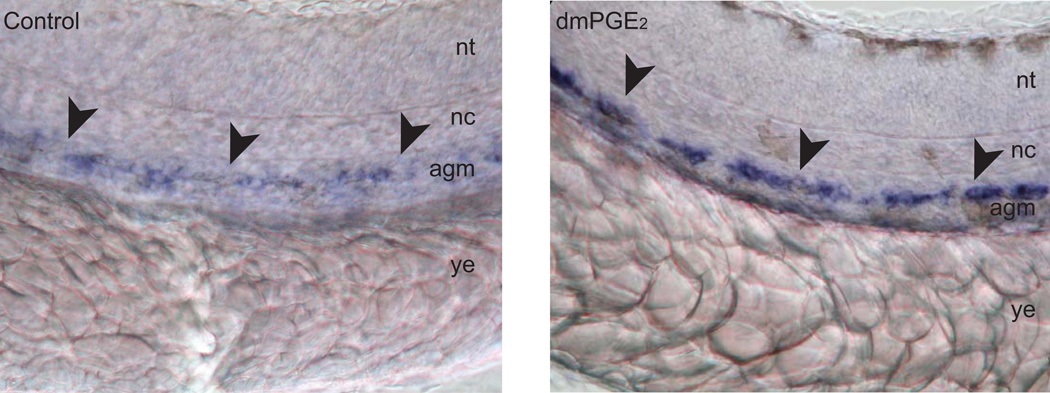
A chemical screen on a panel of small, biologically active molecules was carried out to identify compounds that altered the number of HSCs in the AGM during wild-type zebrafish embryogenesis. The screen revealed various molecules involved in the prostaglandin pathway, including prostaglandin (PG) E2, the main effector prostanoid in the zebrafish. Embryos treated with PGE2 and a long-acting derivative of PGE2, 16,16-dimethyl-PGE2 (dmPGE2), significantly increased HSC number (Figure 2), whereas treatment with chemical inhibitors of cyclooxygenase (Cox)-1 and Cox-2, modulators of the PGE2 synthesis, decreased the number of HSCs. Morpholino knockdown of Cox-1 and -2 expression in zebrafish embryos decreased the number of HSCs in the AGM. However, the inhibitory effect of Cox-1 and -2 knockdown on HSC development was rescued by treatment with dmPGE2, confirming that PGE2 signaling plays an important role in modulating HSC formation. When sublethally irradiated fish were treated with dmPGE2, the rate of recovery of the kidney marrow was augmented, showing increased rates of expansion in the precursor myeloid and lymphoid populations, revealing that the PGE2 pathway functions not only in the formation of HSCs during embryonic development but also in HSC homeostasis in the adult animal [25].
Figure 2.
In situ hybridization shows increased expression of HSC markers runx1 and cmyb in the AGM (indicated with arrowheads) of zebrafish embryos at 36hpf treated with dmPGE2. Lateral view with anterior to the left. nt = nueral tube, nc = notochord, agm = aorta-gonad-mesonephros, ys = yolk extension. Magnification = 40X. Figure panels are adapted from reference (Goessling et. al., 2007).
After this initial chemical screen brought the important mediating effects of PGE2 on HSCs to light, the biochemical and genetic aspects of the pathway were further examined. Through the use of transgenic reporter embryos, PGE2 was directly linked to wnt signaling. Treatment of embryos with dmPGE2 caused an increase in wnt activity within HSCs and the hematopoietic niche during HSC development. Furthermore, inhibition of PGE2 abrogated the enhancing effect of wnt induction on HSC number, illustrating the requirement of PGE2 for wnt-mediated regulation of HSC and progenitor cell formation. The genetic interaction of PGE2 and wnt was then determined by exposing transgenic fish with inducible negative regulators of the wnt pathway to dmPEG2. This revealed that PGE2 acts via cAMP/PKA to modify wnt-mediated HSC proliferation and apoptosis at the level of b-catenin [26].
The effects of PGE2 on HSCs were recapitulated in the murine model, indicating the conserved function of PGE2 in zebrafish and mammalian hematopoiesis. Addition of dmPGE2 led to increased multipotential hematopoietic progenitors during mouse embryonic stem (ES) cell differentiation, enhanced bone marrow (BM) repopulation after injury, and competitive advantage and increased engraftment in competitive transplant assays [26; 27].
Exposure to dmPGe2 resulted in increased CXCR4 mRNA and surface expression on HSCs in mice and human cord blood (hCB), implicating the up-regulation of SDF-1α/CXCR4 signaling in the improved homing of dmPGE2 treated HSCs to the BM niche [28; 29]. Inhibition of endogenous PGE2 with non-steroidal anti-inflammatory drug treatment in mice resulted in a significant increase of HSC mobilization to the blood and decreased CXCR4 expression, reiterating the positive regulatory role of dmPGE2 on HSC CXCR4 expression and trafficking to the niche [28].
The extensive study of PGE2-mediated HSC regulation and the mechanistic aspects of the PGE2 pathway in both zebrafish and murine systems presented dmPGE2 as a potential therapeutic agent for enhancing hematopoietic engraftment in a clinical transplant setting. hCB transplants, which are more readily available and less immunologically stringent, have emerged as an efficacious alternative to marrow and mobilized blood stem cell transplants for patients without a matched donor. However, hCB contains significantly fewer HSCs than marrow and blood. Thus, the improved production, function, and multilineage hematopoietic recovery of HSCs after dmPGE2 exposure indicated PGE2 as a therapeutic option for improving hCB therapy. Indeed, both CD34+ enriched hCB cells and whole hCB units stimulated with dmPGE2 exhibited enhanced HSC number and engraftment capability in xenotransplants [29]. These findings led to an FDA approved clinical trial examining dmPGE2 in hCB transplantation. The phase I trial was completed and a phase II trial is underway. PGE2 was the first compound found in zebrafish to be translated to human patients and emphasized the therapeutic potential latent in zebrafish embryonic chemical screens. Since the onset of this clinical trial, research to optimize the effects of PGE2 on transplantation has continued.
Analyzing blood lineages by FACS
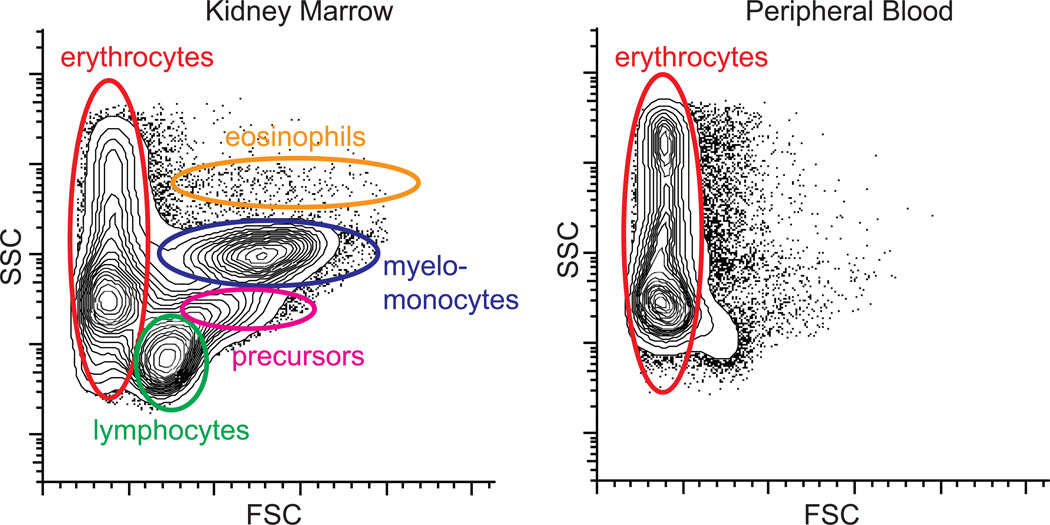
Flow cytometry measures light-scatter and fluorescence profiles of individual cells flowing in a stream of fluid. The scattering of incident light depends on cell volume and cell granularity or internal complexity, which can be quantified a forward scatter (FSC) and side scatter (SSC) parameters, respectively. In combination, FSC and SSC parameters can be used to resolve the five major zebrafish hematopoietic lineages found in whole kidney marrow (WKM) triturates: erythroid, lymphoid, precursors, myeloid, and eosinophil (Figure 3) [5; 30; 31]. Cytospins of cells purified only by light scatter profiles show high purity of the more abundant cell types such as mature erythrocytes, myelomonocytes, and lymphocytes. The mature cell types have conserved morphology when compared to mammalian counterparts, except that erythrocytes and thrombocytes are nucleated [5; 32]. WKM scatter profiles can be used to determine the missing hematopoietic lineages in mutants. For example, in homozygous mutant for the erythrocyte membrane protein band 4.1 (ebp41b −/−), erythrocytes are entirely missing and accumulation of precursors is observed [5]. In contrast, juvenile homozygous mutants for transcription factor c-myb are devoid of all lineages except primitive erythrocytes, consistent with blocked definitive hematopoiesis in these mutants [33]. Hematopoietic stem and progenitor cellss do not have a distinct light scatter profile, but cells from the lymphoid gate are capable of long-term transplantation [5]. The relative position and separation of the different cell populations by light scatter is instrument-dependent. On BD LSRII and FACSVantage instruments, erythroid and myeloid cells are separable by FSC, whereas on BD Aria and Miltenyi MACSquant the two populations are superimposed. Light scatter is also inherently noisy, making it incompatible with quantifying rare cell populations, or purifying cells from heterogenous cell mixtures such as embryonic cells. For example, the differential cell count on zebrafish blood is 99.2 +/− 0.8 % by manual counts and 97.6 +/− 1.5 % by FACS, with false positive cells in non-erythroid gates [5].
Figure 3.
FACS light scatter profiles of the zebrafish kidney marrow, the site of adult hematopoiesis, and peripheral blood. FSC = forward light scatter. SSC = side light scatter.
Fluorescence profiles can be used in addition to light scatter to purifying specific and rare cell populations. In zebrafish, this is typically done using transgenes with fluorescent proteins driven by tissue-specific regulatory elements instead of immunofluorescent staining, as it has been difficult to obtain specific monoclonal antibodies to teleost cell surface proteins. Transgenic lines have been developed that label all major zebrafish hematopoietic cells types. In some cases, different populations can be further resolved by using transgene intensity. For example, in CD41:eGFP transgenic zebrafish, the GFPhi cells are mature thrombocytes, whereas the GFPlo cells are mostly precursors, but also include transplantable HSCs [32; 34]. Combinations of transgenes can be used to further purify sub-populations. For example, Tg(gata1:dsRed;lmo2:eGFP) embryos were used to FACS-purify the double-transgenic definitive erythromyeloid progenitors from blood islands, as only those cells are dsRedhi and eGFPhi [35]. There are several caveats inherent to using transgenes to label-specific cell populations. Transgene fluorescence intensity can vary between homozygous and heterozygous lines and the distinction between “high” and “low” transgene levels needs to be determined experimentally in each case using pure reference populations or mutants, morphological phenotyping, and functional assays. Transgenes can also be stochastically silenced and label only a fraction of the population, complicating purification by negative selection.
Transplantation
Mammalian systems have been used in transplantation experiments to identify HSC populations and mechanisms of gene function. With the ability to distinguish transplanted donor cells from endogenous host cells, one can test self-renewal and differentiation abilities of distinct subpopulations of hematopoietic cells, further segregating populations of HSCs and purifying them from progenitors. Zebrafish transgenic cell subpopulations can be transplanted adult-to-adult, adult-to-embryo, and embryo-to-embryo. Hematopoietic cellular transplantation (HCT) is performed in recipients at the blastula stage (optimal between 1000-cell state and dome stage), gastrula stage (5.25 hpf to 10 hpf), at 48 hpf [36], and in adults between 2–3 months, often with donor subpopulations from adult kidney marrow (site of adult hematopoiesis).
In adult zebrafish, myeloablation using γ-irradiation is necessary to reduce endogenous leukocytes for successful engraftment. A minimum lethal dose of 40 Gy ablated the zebrafish blood-forming systems and nearly all leukocyte populations one week before death; however, HCT is able to rescue 75% of recipients with repopulation of kidney marrow, thymus, and spleen by 30 days [37]. A sublethal gamma irradiation dose of 20–25 Gy is optimal for robust engraftment, long-term repopulation, and high survival of transplant; 20 Gy dose γ—irradiation ablated lymphocytes and enabled the transplantation of a lethal T-cell leukemia from rag2-EGFP-cMyc transgenic zebrafish. Transplantation of double transgenic β-actin:GFP (leukocytes) and gata1:dsred (erythrocyte) donor cells enabled multi-lineage engraftment and repopulation by donorderived cells at this dose and visualization of engraftment via FACS analysis.
The casper mutant, a transparent adult fish that lacks melanocytes and iridophores (nacre−/−. Roy −/−) facilitates the adult zebrafish transplantation assay, allowing for direct visualization and tracking of exogenous donor cells in vivo [38]. Four week post-transplantation of β-actin:GFP WKM, in vivo visualization of transgenic donor cells can occur only in casper recipients, despite no significant difference in FACS analysis of survival and engraftment.
Crucial to optimizing the HCT assay in adult zebrafish is immune-matching, which can prevent the rejection of transplanted tissue and graft-versus-host-disease (GVHD) [34]. Unlike human major histocompatibility complex genes (MHC) genes, Zebrafish MHC class I and class II genes are located on distinct chromosomes: chromosome 19 contains MHC class I genes; chromosome 8 contains MHC class II alpha and beta genes. Excitingly, MHC matched recipients to donors (locus on chromosome 19) dramatically increased the effectiveness of adult transplant; the matched setting had higher donor cell engraftment and greater percentage of multilineage (lymphoid and myeloid) donor chimerism compared to that of the unmatched setting. This improvement to the adult transplantation assay prevented immunologic rejection of transplanted tissue and decreased incidence of GVHD.
Results of large-scale mutagenesis screens have been a diverse set of zebrafish mutants with embryonic hematopoietic defects that can be used as recipients without primitive hematopoiesis, such as the moonshine mutant, vlad tepes mutant, gata1−/− mutant, and bloodless mutant. Transplantation of donor kidney marrow expression gata1:GFP (erythroid cells) into 48-hpf moonshine mutant embryos successfully demonstrated moonshine acts in cell-autonomous ways to generate erythrocytes. Although the mutation was still lethal and recipients did not out-survive their non-transplanted siblings, transplanted moonshine mutants showed robust reconstitution of donor-derived blood cells and marked increased in erythroid cells over the observation period [15]. Transplantation experiments in the vlad tepes mutant rescued both hematopoiesis and long-term survival [15]. HCT can also rescue multilineage hematopoiesis in embryo recipients with lethal gata1 −/− mutants: transgenic WKM gata1:dsred and β-actin:GFP led to long-term survival and hematopoiesis of gata1−/− mutants for at least 6 months, indicating that HSCs in donor WKM could engraft in the mutant and reconstitute red blood cells. These transplanted gata1−/− mutants showed robust β-actin:GFP cells to thymus and pronephros and expansion of gata1:dsred occurred after 7 days, ultimately leading to proliferation of donor derived cells into adulthood. Similarly, transplantation of bloodless mutant with donor-derived blood cells from adult β-actin: GFP and gata1: dsred resulted in circulating donor-derived cells after 8 weeks [5].
These mutant models help address whether early primitive blood precursors can generate definitive hematopoietic cells [39; 40]. For example, lmo2:eGFP expressing cells carrying gata1a:dsRed at the 8–12 somites stage transplanted into 1000 cell stage blastulae vlad tepes recipients are capable of generating populations of donor endothelium and circulating erythrocytes. However, the transplant did not display longterm repopulation of erythroid lineage cells of donor descent in circulation, despite the appearance of circulating donor-derived hematopoietic cells, blood, and blood vessels, indicating that lmo2 hematopoietic precursors could not give rise to definitive hematopoietic organs to repopulate host blood system.
Alternatively, mutant zebrafish homozygous for the c-mybt25127 allele (c-myb mutant) provide a niche suitable for the survival and self-renewal of donor HSCs without myoablative conditioning [33]. The c-myb mutant carries a missense mutation (I181N) in a highly conversed DNA binding domain: phenotypically, these fish are anemic, display developmental retardation, and lack sexual maturity. 6–9 week old c-myb mutants transplanted with wild-type donor kidney marrow transgenic for ikaros:eGFP demonstrate normal hematopoietic activity in the kidney, densely populated lymphoid cells in the thymus, and an indistinguishable phenotype from wild-type siblings five weeks post transplantation. These rescued mutants survived several months longer than non-transplanted siblings and did not display incidence of graft versus host disease that appears in unmatched MHC donor and recipient fish. The c-myb mutant provides an additional adult model for HCT that combines benefits of embryo and adult transplant.
Transplantation-based screening of chemicals in zebrafish has also been successful. Competitive transplantation of WKM treated with a pilot library of 480 bioactive chemicals resulted in the discovery of novel compounds with the ability to improve marrow cell engraftment (P. Li, E. Pugach, and L. Zon, unpublished findings). Additionally, transplanted fish with fluorescently labeled T-ALL can be screened using a high-throughput imaging system based on the LED fluorescence microscope [41]. These screening approaches are promising for expediting the discovery of the molecular and cellular mechanism of diseases and the development of novel therapies.
Conclusion
Zebrafish is a useful genetic model for studying hematopoiesis. The molecular and genetic studies can be coupled with high-throughput screens and in vivo visualization of fluorescently labeled cell populations. Previous forward genetic screens have generated hundreds of zebrafish mutants and identified many novel players in hematopoietic development. The modified/suppressor genetic screens have been successful in revealing biological mechanisms, such as the role of TIF1γ in the moonshine mutant. With the molecular cloning of many existing mutants, a genetic modifier screen will be a powerful approach to examine the underlying mechanism. Zebrafish is also cost effective for high-through chemical screens. Thousands of small molecules can be screened within weeks. As a whole organism, zebrafish offers great advantages over cell-line-based screens, which undoubtedly make the transition of the identified compounds into clinical applications more successful, such as PGE2 that in only two years entered into clinical trials.
The advances in genomic manipulation through techniques like CRISPR-Cas and TALENs for site-specific gene targeting have improved reverse genetic approaches in zebrafish. We are entering a new era in using zebrafish to dissect genetic components of hematopoiesis and model human hematopoietic disorders. Zebrafish disease models coupled with chemical screening will aid in the discovery of molecules that could improve various hematopoietic defects.
Ultimately, genetic and chemical screens may be performed by utilizing transplantation assays, not only in embryos but also in adult fish, assessing small molecules or genes that promote or stimulate HSC migration and engraftment during hematopoietic development and in adult hematopoiesis. Much effort has been devoted to improve the efficiency and reliability of the assay such as MHC matched recipients and donors, the c-myb mutants, or immunodeficient fish will propel the field closer to prevention of GVHD and graft rejections in transplantations.
As the transplantation assays will be optimized, purification of zebrafish HSCs will be improved. FACS analysis enables the purification of fluorescently labeled cells, which in turn can be transplanted into embryo or adult zebrafish to assess their repopulation capacity and therefore their “stem cell-ness”. Finally, the development of monoclonal antibodies to zebrafish cell surface markers will further accelerate the ability to characterize the pureness of HSC population in zebrafish.
Abbreviations
- AGM
aorta-gonad-mesonephros
- ALM
anterior lateral mesoderm
- BM
bone marrow
- CHT
caudal hematopoietic tissue
- ENU
ethylnitrosourea
- FACS
fluorescence-activated cell sorting
- FSC
forward light scatter
- GVHD
graft-versus-host-disease
- hCB
human cord blood
- HCT
hematopoietic cellular transplantation
- hpf
hours post fertilization
- ICM
intermediate cell mass
- MHC
major histocompatibility complex
- PLM
posterior lateral mesoderm
- PAF
Pol II Associated Factor
- PG
prostaglandin
- SSC
side light scatter
- TGFβ
transforming growth factor beta
- TIF1γ
transcription intermediary factor1γ
- WKM
whole kidney marrow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Paik EJ, de Jong JL, Pugach E, Opara P, Zon LI. A chemical genetic screen in zebrafish for pathways interacting with cdx4 in primitive hematopoiesis. Zebrafish. 2010;7:61–68. doi: 10.1089/zeb.2009.0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Detrich HW, 3rd, Kieran MW, Chan FY, et al. Intraembryonic hematopoietic cell migration during vertebrate development. Proc Natl Acad Sci U S A. 1995;92:10713–10717. doi: 10.1073/pnas.92.23.10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson MA, Ransom DG, Pratt SJ, et al. The cloche and spadetail genes differentially affect hematopoiesis and vasculogenesis. Dev Biol. 1998;197:248–269. doi: 10.1006/dbio.1998.8887. [DOI] [PubMed] [Google Scholar]
- 4.Murayama E, Kissa K, Zapata A, et al. Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity. 2006;25:963–975. doi: 10.1016/j.immuni.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 5.Traver D, Paw BH, Poss KD, et al. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat Immunol. 2003;4:1238–1246. doi: 10.1038/ni1007. [DOI] [PubMed] [Google Scholar]
- 6.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang WY, Fu Y, Reyon D, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sander JD, Cade L, Khayter C, et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jing L, Zon LI. Zebrafish as a model for normal and malignant hematopoiesis. Dis Model Mech. 2011;4:433–438. doi: 10.1242/dmm.006791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patton EE, Zon LI. The art and design of genetic screens: zebrafish. Nat Rev Genet. 2001;2:956–966. doi: 10.1038/35103567. [DOI] [PubMed] [Google Scholar]
- 11.Jao LE, Maddison L, Chen W, Burgess SM. Using retroviruses as a mutagenesis tool to explore the zebrafish genome. Brief Funct Genomic Proteomic. 2008;7:427–443. doi: 10.1093/bfgp/eln038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Driever W, Solnica-Krezel L, Schier AF, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- 13.Haffter P, Granato M, Brand M, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- 14.Ransom DG, Haffter P, Odenthal J, et al. Characterization of zebrafish mutants with defects in embryonic hematopoiesis. Development. 1996;123:311–319. doi: 10.1242/dev.123.1.311. [DOI] [PubMed] [Google Scholar]
- 15.Ransom DG, Bahary N, Niss K, et al. The zebrafish moonshine gene encodes transcriptional intermediary factor 1gamma, an essential regulator of hematopoiesis. PLoS Biol. 2004;2:E237. doi: 10.1371/journal.pbio.0020237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He W, Dorn DC, Erdjument-Bromage H, et al. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell. 2006;125:929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 17.Dupont S, Mamidi A, Cordenonsi M, et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009;136:123–135. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 18.Dupont S, Zacchigna L, Cordenonsi M, et al. Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell. 2005;121:87–99. doi: 10.1016/j.cell.2005.01.033. [DOI] [PubMed] [Google Scholar]
- 19.Bai X, Kim J, Yang Z, et al. TIF1gamma controls erythroid cell fate by regulating transcription elongation. Cell. 2010;142:133–143. doi: 10.1016/j.cell.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai X, Yang Z, Jiang H, Lin S, Zon LI. Genetic suppressor screens in haploids. Methods Cell Biol. 2011;104:129–136. doi: 10.1016/B978-0-12-374814-0.00007-0. [DOI] [PubMed] [Google Scholar]
- 21.Bai X, Trowbridge JJ, Riley E, et al. TiF1-gamma plays an essential role in murine hematopoiesis and regulates transcriptional elongation of erythroid genes. Dev Biol. 2013;373:422–430. doi: 10.1016/j.ydbio.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kusy S, Gault N, Ferri F, et al. Adult hematopoiesis is regulated by TIF1gamma, a repressor of TAL1 and PU.1 transcriptional activity. Cell Stem Cell. 2011;8:412–425. doi: 10.1016/j.stem.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 23.Monteiro R, Pouget C, Patient R. The gata1/pu.1 lineage fate paradigm varies between blood populations and is modulated by tif1gamma. EMBO J. 2011;30:1093–1103. doi: 10.1038/emboj.2011.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Wang J, Wheat J, et al. AML1-ETO mediates hematopoietic self-renewal and leukemogenesis through a COX/beta-catenin signaling pathway. Blood. 2013 doi: 10.1182/blood-2012-08-447763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.North TE, Goessling W, Walkley CR, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007–1111. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goessling W, North TE, Loewer S, et al. Genetic interaction of PGE2 and Wnt signaling regulates developmental specification of stem cells and regeneration. Cell. 2009;136:136–147. doi: 10.1016/j.cell.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lord AM, North TE, Zon LI. Prostaglandin E2: making more of your marrow. Cell Cycle. 2007;6:3054–3057. doi: 10.4161/cc.6.24.5129. [DOI] [PubMed] [Google Scholar]
- 28.Hoggatt J, Singh P, Sampath J, Pelus LM. Prostaglandin E2 enhances hematopoietic stem cell homing, survival, and proliferation. Blood. 2009;113:5444–5455. doi: 10.1182/blood-2009-01-201335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goessling W, Allen RS, Guan X, et al. Prostaglandin E2 enhances human cord blood stem cell xenotransplants and shows long-term safety in preclinical nonhuman primate transplant models. Cell Stem Cell. 2011;8:445–458. doi: 10.1016/j.stem.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balla KM, Lugo-Villarino G, Spitsbergen JM, et al. Eosinophils in the zebrafish: prospective isolation, characterization, and eosinophilia induction by helminth determinants. Blood. 2010;116:3944–3954. doi: 10.1182/blood-2010-03-267419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haining WN, Davies J, Kanzler H, et al. CpG oligodeoxynucleotides alter lymphocyte and dendritic cell trafficking in humans. Clin Cancer Res. 2008;14:5626–5634. doi: 10.1158/1078-0432.CCR-08-0526. [DOI] [PubMed] [Google Scholar]
- 32.Lin HF, Traver D, Zhu H, et al. Analysis of thrombocyte development in CD41-GFP transgenic zebrafish. Blood. 2005;106:3803–3810. doi: 10.1182/blood-2005-01-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hess I, Iwanami N, Schorpp M, Boehm T. Zebrafish model for allogeneic hematopoietic cell transplantation not requiring preconditioning. Proc Natl Acad Sci U S A. 2013;110:4327–4332. doi: 10.1073/pnas.1219847110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Jong JL, Burns CE, Chen AT, et al. Characterization of immune-matched hematopoietic transplantation in zebrafish. Blood. 2011;117:4234–4242. doi: 10.1182/blood-2010-09-307488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bertrand JY, Kim AD, Teng S, Traver D. CD41+ cmyb+ precursors colonize the zebrafish pronephros by a novel migration route to initiate adult hematopoiesis. Development. 2008;135:1853–1862. doi: 10.1242/dev.015297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li P, White RM, Zon LI. Transplantation in zebrafish. Methods Cell Biol. 2011;105:403–417. doi: 10.1016/B978-0-12-381320-6.00017-5. [DOI] [PubMed] [Google Scholar]
- 37.Traver D, Winzeler A, Stern HM, et al. Effects of lethal irradiation in zebrafish and rescue by hematopoietic cell transplantation. Blood. 2004;104:1298–1305. doi: 10.1182/blood-2004-01-0100. [DOI] [PubMed] [Google Scholar]
- 38.White RM, Sessa A, Burke C, et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008;2:183–189. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stachura DL, Traver AJ. Cellular dissection of zebrafish hematopoiesis. Methods Cell Biol. 2011;101:75–110. doi: 10.1016/B978-0-12-387036-0.00004-9. [DOI] [PubMed] [Google Scholar]
- 40.Zhu H, Traver D, Davidson AJ, et al. Regulation of the lmo2 promoter during hematopoietic and vascular development in zebrafish. Dev Biol. 2005;281:256–269. doi: 10.1016/j.ydbio.2005.01.034. [DOI] [PubMed] [Google Scholar]
- 41.Smith AC, Raimondi AR, Salthouse CD, et al. High-throughput cell transplantation establishes that tumor-initiating cells are abundant in zebrafish T-cell acute lymphoblastic leukemia. Blood. 2010;115:3296–3293. doi: 10.1182/blood-2009-10-246488. [DOI] [PMC free article] [PubMed] [Google Scholar]