Abstract
Enabled/VASP Homology-1 (EVH1) domains function primarily as interaction modules that link signaling proteins by binding to proline-rich sequences. EVH1 domains are ~115 residues in length and adopt the pleckstrin homology (PH) fold. Four different protein families contain EVH1 domains: Ena/VASP, Homer, WASP and SPRED. Except for the SPRED domains, for which no binding partners are known, EVH1 domains use a conserved hydrophobic cleft to bind a four-residue motif containing 2–4 prolines. Conserved aromatic residues, including an invariant tryptophan, create a wedge-shaped groove on the EVH1 surface that matches the triangular profile of a polyproline type II helix. Hydrophobic residues adjacent to the polyproline motif dock into complementary sites on the EVH1 domain to enhance ligand binding specificity. Pseudosymmetry in the polyproline type II helix allows peptide ligands to bind in either of two N-to-C terminal orientations, depending on interactions between sequences flanking the prolines and the EVH1 domain. EVH1 domains also recognize non-proline motifs, as illustrated by the structure of an EVH1:LIM3 complex and the extended EVH1 ligands of the verprolin family.
Keywords: EVH1 domain, polyproline, Wiskott-Aldrich-Syndrome, Pleckstrin homology domain, Review
2. INTRODUCTION
Protein-protein interactions regulate most cellular processes including gene expression, the cell cycle, protein trafficking, targeted proteolysis, and cytoskeletal reorganization. Cell surface receptors are often coupled to intracellular signaling pathways by recognition of modular protein interaction domains. Regulated signal transduction also depends on the ability to deactivate a pathway in response to changing conditions. Accordingly, protein interaction domains often bind with affinities in the low micromolar range to allow for rapid dissociation of active signaling complexes. Over 80 different protein interaction domain families have been characterized, many of which bind to short linear sequences containing specific amino acid patterns or modifications. Typically, these domains are self-contained modules consisting of 35–150 residues that can be expressed in isolation from their parent protein while retaining their ability to bind their physiological partners (1). This feature has allowed for the isolation and biochemical characterization of a number of protein interaction domains.
Protein interaction domains can be classified based on sequence homology, ligand-binding properties, or structural similarity (2). Thus, a typical class of ligand-binding proteins may contain a variety of protein interaction domains that recognize a common ligand whereas a family classified based on sequence homology contains a single fold that may recognize a variety of ligands. Some families function in a narrow cellular context, while others participate in a diverse range of processes. For example, PDZ domains typically help assemble protein complexes at the plasma membrane (3), but Src homology 2 (SH2) domains are found in a wide variety of signaling proteins (4). The paradigm originally established by the SH2 family, which binds phosphotyrosine-containing sequences, has expanded to include domains that recognize C-terminal sequences, acetyl-lysine residues, polyproline helices and other structural motifs. Four distinct families that recognize proline-rich sequences are now known: Src homology-3 (SH3), WW, glycine-tyrosine-phenylalanine (GYF), and Enabled/VASP homology-1 (EVH1) (5).
The EVH1 domain is a member of the Pleckstrin homology (PH) domain-like superfamily (Pfam ID – PF00169, CL0266). The PH fold consists of two perpendicular anti-parallel β-sheets followed by a C-terminal α-helix (Figure 1). Because sequence homology is low and loops regions are of variable length, sequence-based identification of PH domains can be difficult. Phosphotyrosine-interaction/phosphotyrosine-binding domains (PID/PTB), Ran-BP1, GRAM, FERM-C, and EVH1 domains all share the PH fold, and appear in a wide array of intracellular signaling proteins (Table 1) (6–8). These domains recognize a diverse set of ligands such as inositol lipids, phosphotyrosines, and proline-rich sequences, demonstrating that the PH fold is a stable scaffold that has been adapted to multiple functions. Uniquely in the PH domain superfamily, EVH1 domains interact with proline-rich sequences that bind a groove formed by β-strands 1, 2, 6 and 7. In contrast, PH and PTB domains contain an extra helix that occludes this binding groove shifting the binding sites for inositol lipids and phosphotyrosines to other regions of the PH superfold (9). Interestingly, a recent report shows that the EVH1 domain from Mena forms a stable complex with a Lim3 domain that lacks a polyproline motif (10). This interaction occludes the binding groove and competes with polyproline ligands. Lim binding reveals a new class of potential interactions for EVH1 domain containing proteins and suggests that EVH1 complexes may be dynamically regulated in the cell.
Figure 1.
The EVH1 fold is a member of the Pleckstrin Homlogy superfamily. Structural comparison of the EVH1 domain from Mena (PDB code: 1EVH) with the PH domain from phospholipase C δ (PDB code: 1MAI) and the phosphotyrosine binding domain (PTB) from tensin-1 (PDB code: 1WVH). The PH fold consists of two perpendicular anti-parallel β-sheets followed by a C-terminal α-helix that assemble to form a ligand binding groove between β-strands 1, 2, 6, and 7. In the PH and PTB domains, this groove is occupied by an additional α-helix that moves the recognition of inositol- (3,4,5) triphosphate and phosphotyrosines peptides by PH and PTB domains, respectively, to distinct regions of the EVH1 domain. Rotation by 90° about the vertical axis (lower panel) shows a side view of the PH fold.
Table 1.
EVH1 domain structures deposited in the protein data bank
| Structure | ||||
|---|---|---|---|---|
| EVH1 domain1 | Target | PDB ID | Ligand | Ref |
| Hs SPRED-2 | Unknown | 2jp2 | ||
| Xt SPRED-1 | Unknown | 1xod | ||
| Rn Homer | mGluRI5, IP3 receptor types 1 and 3, Shank 1 and 3, TRPCI, PIKE-L, DynaminIII, Oligophrenin-1, and DrebrinE | 1ddv (1ddw)2 | TPPSPF | (34, 69–78) |
| Rn Homer 1c | 1i2h | |||
| Mm Homer 2b | 2p8v | |||
| Hs Homer 3 | 1i7a | |||
| Mm EVL | ActA, Zyxin, LPP, Vinculin, Fyb/SLAP, Robo, Semaphorin 6A-1, Drk, FE65, and Profilin I and II | 1qc6 | FEFPPPPTDEE | (21–24, 26, 29, 30, 32, 79–93) |
| Mm Mena | 1evh | FPPPPT | ||
| Hs VASP | 1egx | |||
| Rn N-WASP | WIP, WIRE/WICH, and CR16 | 2ifs (1mke)3 | ESRFYFHPISDLPPPEPVQTTKSYPSKLARNESR | (43–49, 94) |
Species codes are as follows: Hs – Homo sapiens, Xt – Xenopus tropicalis, Rn – Rattus norvegicus, and Mm – Mus musculus
PDB code 1ddw is the structure of the rat Homer EVH1 domain in absence of ligand.
PDB code 1mke is the NMR structure of the rat N-WASP with the 25 C-terminal residues of the peptide listed in Table 1.
EVH1-containing proteins function in several cellular contexts including cytoskeletal dynamics, postsynaptic signal transduction, proliferation, and differentiation. Missense mutations in EVH1 domains that disrupt binding to their target proteins have been identified as the cause of inherited disease in humans (11–16). EVH1 domains recognize their target sequences in a different manner than other polyproline binding domains. This review aims to highlight the unique aspects of EVH1 complexes, including the enhancement of specificity and protein stability by extended binding motifs flanking the central proline motif.
3. FUNCTIONAL ORGANIZATION OF THE EVH1 FAMILY
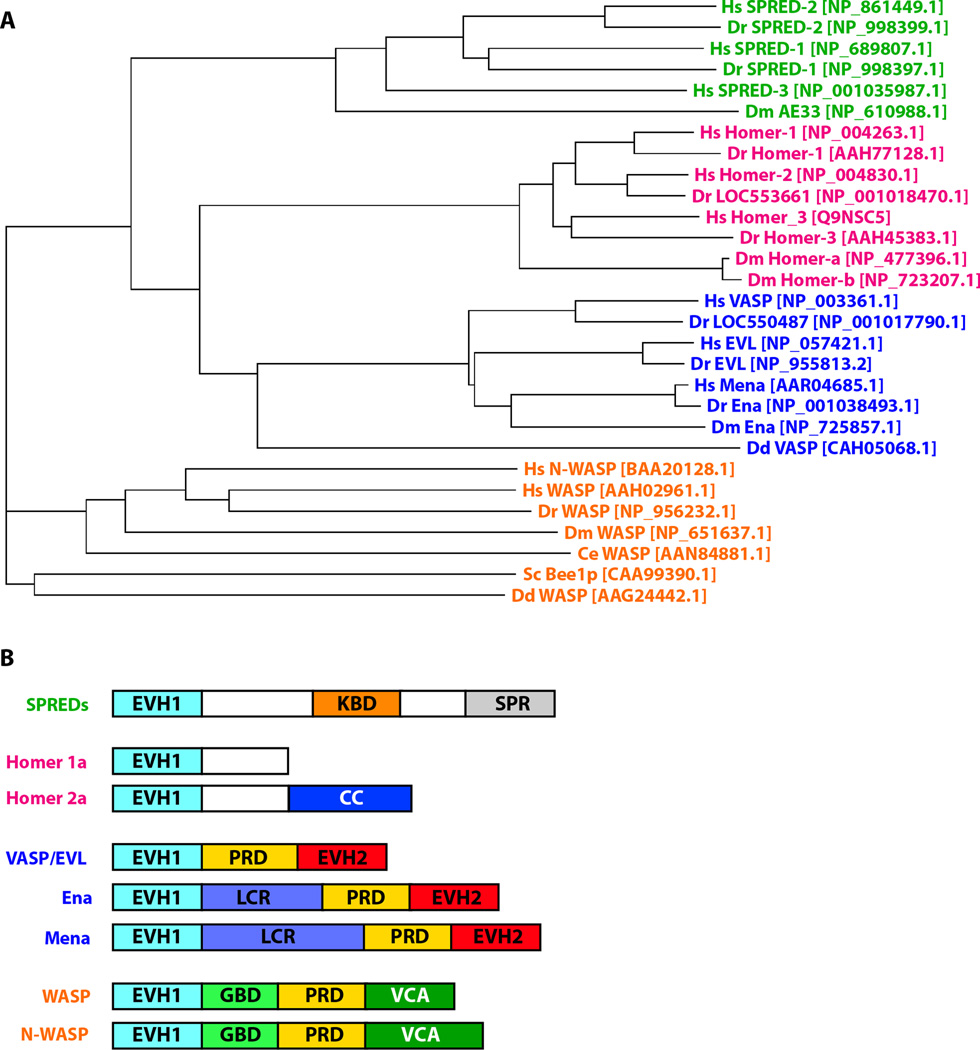
EVH1 domains have been found in ~630 human genes and are classified into four distinct protein families based on amino acid sequence analysis: Wiskott-Aldrich-Syndrome protein (WASP); Enabled/vasodilator-stimulated phosphoprotein (Ena/VASP); Homer/Vesl; and Sprouty-related proteins with an EVH1 domain (SPRED) (Figure 2A). In all instances the EVH1 domain is ~115 residues in length and found solely at the N-terminus. Regions C-terminal to the EVH1 domain are highly divergent in sequence and domain composition (Figure 2B). Each EVH1 subclass recognizes a distinct pattern of amino acids, but all of them bind a proline-rich sequence in the left-handed polyproline type II conformation. Residues flanking the polyproline motif contribute to the binding specificity of EVH1 complexes as discussed below in more detail.
Figure 2.
Domain organization and phylogenetic tree of EVH1 domain proteins. A) Protein sequences corresponding to the EVH1 domain boundaries identified by Pfam (95) for human (Homo sapiens, Hs), fly (Drosophila melanogaster, Dm), fish (Danio rerio, Dr), worm (Caenorhabditis elegans, Ce), slime mold (Dictyostelium discoideum, Dd) and yeast (Saccharomyces cerevisiae, Sc) were aligned. NCBI accession numbers for the proteins are indicated in the brackets. Multiple sequence alignments and the phylogenetic analysis were performed using ClustalW version 1.83 (96). EVH1 domains within a particular branch are more related to each other than to those within a particular species. B) Domain organization of representative members from each branch of the four EVH1 protein families. In all families the EVH1 domain is located at the N-terminus and is followed by a variety of domains including proline rich domains (PRD), coiled-coil (CC), low complexity regions (LCR), Ena-VASP homology 2 domains (EVH2), GTPase binding domains (GBD), verprolin-cofilin-acidic motifs (VCA), Sprouty-like cysteine-rich domains (SPR), and c-Kit binding domains (KBD).
Enabled-VASP Homology 1 domains were first identified in proteins of the Ena/VASP family that includes Ena, the product of the Drosphila enabled gene (17, 18), the mammalian protein vasodilator-stimulated phosphoprotein (VASP) (19, 20), mammalian Enabled (Mena), and Enabled/VASP-like (EVL) protein (21). Ena/VASP proteins also share two other common structural elements: a central proline-rich region and a C-terminal EVH2 domain, each of which varies in size among the family members. The EVH1 domains within this family recognize protein ligands that contain a core (F/L)PPPP motif (Figure 3) (22–24). A number of proteins such as zyxin (25), vinculin (25), roundabout (Robo) (26), Semaphorin 6A-1 (24), RIAM (27), lamellipodin (28), and the Fyn-binding and SLP-76 associated protein are ligands for Ena/VASP family members and play roles in focal adhesions (21, 29, 30), cell-cell adherens junctions (31), and cell motility. For example, zyxin and vinculin are focal adhesion proteins while Robo and Semaphorin 6A-1 participate in axon guidance. In contrast, RIAM and lamellipodin participate in cell motility and regulate the formation of lamellipodia. RIAM functions as an adaptor protein that provides a link between Rap1-GTP and profilin (27) while lamellipodin colocalizes with Ena/VASP proteins at the tips of lamellipodia and filopodia and plays a regulatory role (28). In addition, lamellipodin is recruited by Vaccinia virus and Enteropathogenic E. coli to facilitate their own motility (28). Similarly, the ActA protein from the intracellular bacterial pathogen Listeria monocytogenes utilizes both VASP and Mena to hijack the host’s actin nucleation and polymerization machinery to become mobile within the cell (32). ActA binding to the VASP or Mena EVH1 domain is mediated by the consensus sequence (D/E)FPPPPT (D/E) (D/E)EL, in which the core FPPPP is essential for the interaction (22, 33).
Figure 3.
Interaction of polyproline containing lignads with their EVH1 domains. A) Ribbon and surface diagrams depicting the interaction of Mena with ActA peptide (PDB code: 1EVH), of Homer-1 with the mGluR peptide (PDB code: 1DDV), of N-WASP with the WIP peptide (PDB code: 2IFS), and SPRED-1 (PDB code: 1XOD). No ligands for SPRED domains are known. The purple surface represents residues that are within 5Å of the bound peptide.
The Homer/Vesl family of adapter proteins consists of three members, Homer1, Homer2, and Homer3, each of which is subject to alternative pre-mRNA splicing to yield long and short isoforms (34–36). All Homer splice variants retain the N-terminal EVH1 domain but the long isoforms contain a C-terminal coiled coil that is absent in the short form. EVH1 domains from the Homer family interact with ligands that contain a core PPxxF motif, where×is any amino acid (Figure 3) (37). In the cell, members of the Homer family are primarily expressed in the nervous system and are localized to the post-synaptic density (PSD), an actin-rich structure found on the spines of neuronal dendrites. Homer proteins interact via their EVH1 domain with a number of receptors localized to the PSD including the metabotropic glutamate receptor (mGluR5), inositol 1,4,5-trisphosphate receptors, ryanodine receptors and Shank, an adaptor for the N-methyl-D-aspartate receptor complex. Within these complexes, Homer proteins can regulate the synaptic localization of target proteins or modulate cross talk between signaling proteins localized in the PSD. For example, overexpression of Homer1b (long form) retained the mGluR5 receptor in the endoplasmic reticulum preventing it from being localized to the cell-surface (38, 39). In contrast, overexpression of Homer1a (short form) induced cell-surface localization and clustering of the mGluR5 receptor (38). These results were confirmed in a number of cell types and suggest that the Homer proteins regulate cell-surface targeting and clustering of the mGluR5 receptor. Members of this family also participate in neuronal development and contribute to behavioral phenomena like drug addiction (40). While no clear role for has been established from Homer proteins in neurological disease, recent studies have suggested roles for certain family members in schizophrenia, X-linked mental retardation and Fragile X syndrome.
The WASP family consists of three members: WAS protein (WASP), expressed in cells of the hematopoietic lineage, its ubiquitously expressed homolog neuronal-WASP (N-WASP), and the yeast homolog Bee1p. WASP proteins are multidomain proteins that contain an N-terminal EVH1 domain, also referred to as a WASP homology 1 (WH1) domain, followed by a basic motif, a GTPase binding domain, a proline-rich region and verprolin-cofilin-acidic motif that binds directly to the actin-related protein (Arp)2/3 actin nucleating complex (41, 42). The functional importance of the EVH1 module is reflected in the high proportion of disease-causing missense mutations that occur within the domain (12–16).
In contrast to the Ena/VASP and Homer families, WASP proteins recognize an extended peptide motif that is far longer than the canonical EVH1 ligands (Figure 3) (43–45). Members of the mammalian verprolin family, including WASP interacting protein (WIP) (43–46), WIP-related protein (WIRE/WICH) (47, 48), and CR16 (49), contain a sequence near the carboxyl terminus that interacts with the WASP EVH1 domain. A polyproline sequence binds the conserved hydrophobic groove as in other EVH1 domains, but is flanked on both ends by additional sequences required for stable binding and biological WASP activity. The WASP protein is stabilized through its constitutive association with WIP (50, 51), and PKC θ–mediated phosphorylation of serine 488 near the WIP C-terminus reportedly weakens the interaction (52). To assess the structural impact of phosphorylation, we used 2D NMR to detect changes in an EVH1-WIP complex upon the introduction of mutations that mimic WIP phosphorylation (S488D and S488E). Our NMR analysis suggested that WIP phosphorylation does not disrupt the WIP-WASP complex (F. C. Peterson and B. F. Volkman, unpublished results). Consistent with our observations, a recent study by Dong et al. (53) showed that phosphorylation of serine 488 does not disrupt the WIP-WASP complex in cells. Thus, WIP-WASP interactions appear to be unaffected by phosphorylation at serine 488.
Disease causing mutations in the EVH1 domain are thought to destabilize the WASP/WIP complex leading to degradation of WASP (11, 51). Inherited mutations in the wasp gene lead to Wiskott-Aldrich Syndrome (WAS), an X-lined recessive disorder first identified in 1937 and characterize by immunodeficiency, eczema and thrombocytopenia (54). In lymphocytes, greater than 95% of WASP is normally bound to WIP (52). Most missense mutations identified in WAS families occur within the EVH1 domain and likely disrupt its interaction with WIP. This hypothesis is correlated with the observation that the most severe disease-causing WASP mutations result in the greatest reduction of WASP levels in circulating platelets, even though WASP mRNA levels remain unchanged (51). This observation lead Lutskiy et al. to conclude that WIP plays a WASP-protective role in leukocytes, and that disease-causing mutations lead to degradation of WASP protein and abnormal cytoskeletal regulation (51). These cytoskeletal defects result in impaired leukocyte chemotaxis and reduced T and natural killer cell engagement with targets cells leading to immune dysfunction in WAS patients (55).
Member of the SPRED family of proteins contain an N-terminal EVH1 domain that is followed by a central c-Kit binding domain, and a C-terminal Sprouty-like cysteine-rich domain (56, 57). First identified in Drosophila as inhibitors of the RAS/MAPK signaling pathway (56–58), homologs have now been identified in Xenopus, mice and humans. At present, the binding partners recognized by the SPRED EVH1 remain unknown and direct interactions with other proteins through the EVH1 domain have not been shown. Structural studies with the SPRED1 EVH1 domain from Xenopus tropicalis suggest that the conserved polyproline binding groove is narrower than in other EVH1 domains (59). Harmer et al. concluded that SPRED domains are likely to bind polyproline peptides that are less proline-rich than other EVH1 ligands (59). Potential roles for SPRED-1 and SPRED-2 in disease have been identified through the use of knock-out mouse models (58, 60–62). These models showed that SPRED-1 and SPRED-2 were not essential for fertility or development, but showed a dwarf phenotype similar to hypochondroplasia in young adult mice. Roles for SPRED-1 in mature late phase hematopoiesis and for SPRED-2 as a negative regulator of embryonic hematopoiesis has also been suggested (58, 62). Presently, no functional role has been demonstrated for SPRED-3.
The lack of known binding partners suggests that SPRED domains may utilize an interaction surface distinct from the conserved EVH1 polyproline docking site. Like many proteins, the conserved PH domain fold has been adapted to support multiple interaction surfaces (Figure 1). Some SH3 domains also have binding sites distinct from the conserved proline-binding surface (51). Recent structural studies show that some EVH1 domains are multifunctional, recognizing binding partners that are not restricted to polyproline sequences. For example, the N-WASP EVH1 domain employs three distinct epitopes to form a WIP binding surface that extends well beyond the conserved polyproline binding site. Likewise, the Mena EVH1 domain binds to the third LIM domain of testin, which lacks a polyproline motif altogether (10). Further experimental work is required to establish whether SPREDs are functionally distinct from EVH1 domains that bind proline-rich sequences.
4. POLYPROLINE RECOGNITION BY EVH1 DOMAINS
Proline-rich regions are among the most numerous sequence motifs in the fly and nematode genomes (5). Proline is unique among the 20 naturally occurring amino acids for its cyclic side chain, which creates a substituted amide nitrogen and restricts the range of available conformations. As a consequence, proline-rich sequences can adopt a left-handed helical structure called the polyproline type II (PPII) helix. Proline recognition domains from the SH3, WW, GYF and EVH1 families target distinct proline-rich sequences, but structural studies have shown that the polyproline motif adopts the PPII helix upon binding to domains of each type. With a periodicity of three residues per turn, the PPII structure is triangular in cross-section, and displays both hydrophobic surface (aliphatic side chains) and hydrogen bond acceptors (backbone carbonyls). Strikingly, amide substitution in the PPII helix results in a structure that is nearly superimposable on itself in either the N- to C-terminal or C- to N-terminal orientation. This twofold pseudosymmetry enables the bidirectional ligand binding modes observed for members of the SH3, WW and EVH1 families (44, 63–66).
It has been noted previously that SH3, WW, and GYF domains use a relatively flat surface that mates with one face of the prism-shaped polyproline helix (Figure 4). In contrast, a wedge-shaped hydrophobic groove in the EVH1 family recognizes an apex of the triangular PPII helix which is rotated by ~60° relative to the other proline binding domains (9). Aromatic side chains and other conserved residues at five sequence positions in the β1, β2 and β6 strands of the EVH1 domain line the sides of the binding cleft (Figure 5A). An invariant tryptophan in the β2-strand creates a ridge across the center of the hydrophobic groove. Ligand peptides dock over the indole ring of the Trp sidechain, defining the register and orientation of the bound polyproline motif. Depending on the EVH1 subfamily, the first position is always Phe (Homer), Tyr (WASP) or Met (Ena/VASP and SPRED), while position 2 requires Ile (Homer), Tyr (Ena/VASP) or Arg (SPRED). The third position is absolutely conserved as a Phe residue in all EVH1 sequences, and position 4 contains a Gln (Homer and Ena/VASP) or His (SPRED). Positions 2 and 4 in the WASP family are less strongly conserved, but typically occupied by Ala and Thr, respectively.
Figure 4.
Polyproline recognition by SH3, GYF, WW and EVH1 domains. The interaction of proline rich ligands is shown for the four distinct families that recognize proline rich sequences. A) The SH3 domain from Sem5 (PDB code: 1SEM). B) The GYF domain from CD2BP2 (PDB code: 1L27). C) The WW domain from dystrophin (PDB code: 1EG4). D) The EVH1 domain from N-WASP (PDB code: 2IFS).
Figure 5.
EVH1 family members use a conserved aromatic-rich binding site to recognize proline-rich ligands. A) Residues used in the recognition of the proline-rich ligands are conserved in the WASP, Homer, Ena/VASP and SPRED families. However, the orientation of the bound PPII helix is reversed when the Homer and Ena/VASP families are compared with the WASP proteins. B) Representative ligand peptide sequences for the Homer, Ena/VASP and WASP EVH1 domain families.
The rotational difference of ~60° in how the PPII helix binds relative to other proline recognition domains may explain the less restrictive sequence requirements for EVH1 ligands. Proline is preferred but not absolutely conserved at each of five positions (-FPPPPP-), and few EVH1 ligands contain all five proline residues, as illustrated in Figure 5B. Thus it appears that while SH3, WW and GYF domains directly recognize the proline side chain at specific positions in the target sequence, EVH1 domains simply require that proline content be sufficient to stabilize a PPII helical conformation. However, since EVH1 domains recognize the general features of a proline-rich motif, but do not bind all proline-rich sequences, additional factors are required to achieve high peptide binding specificity as described in section 5.
Interestingly, while the polyproline motif is required for binding to most EVH1 domains, no peptide ligands have been identified for the SPRED family. Substitution of the positively charged Arg and His sidechains at positions 2 and 4 of the binding cleft may significantly alter the binding preferences for these domains, even though the invariant Phe and Trp residues are still present. A similar degree of sequence divergence within the aromatic binding cleft of WASP EVH1 domains probably explains why a proline-rich sequence is necessary but not sufficient for binding to WASP EVH1 domains (44, 45). However, low WASP EVH1 binding affinity to the PPII motif is compensated by extension of the peptide binding site relative to the Ena/VASP and Homer domains.
5. PEPTIDE BINDING SPECIFICITY OF EVH1 DOMAINS
Recognition of a secondary structure configuration like the PPII helix is, by itself, insufficient for selective binding of a short peptide ligand. As with other polyproline binding domains, EHV1 binding specificity is enhanced by flanking residues that dock in complementary sites adjacent to the conserved aromatic binding cleft. For example, a phenylalanine (or other hydrophobic) residue adjacent to the proline motif in ligands for Ena/VASP (FPxϕP) or Homer (PPxxF) family members binds a corresponding hydrophobic pocket in the EVH1 domain. Complementary electrostatic interactions between acidic residues of the peptide and basic residues on the EVH1 surface also contribute to ligand binding specificity in some cases (9).
Ena/VASP and Homer EVH1 domains recognize prototypical ligands that are analogous to the short sequence motifs recognized by SH3, WW, and GYF domains in that they combine a common proline recognition site with an adjacent binding pocket, typically for a hydrophobic side chain (5). Structures have been solved for relatively few EVH1-ligand complexes, so general rules for the binding preferences of all EVH1 domains are lacking. However, it is clear that EVH1 domains utilize interaction surfaces that extend well beyond the conserved polyproline-binding groove, as illustrated by novel complexes formed by N-WASP and Mena.
6. VERPROLINS CONTAIN AN EXTENDED EVH1 BINDING MOTIF
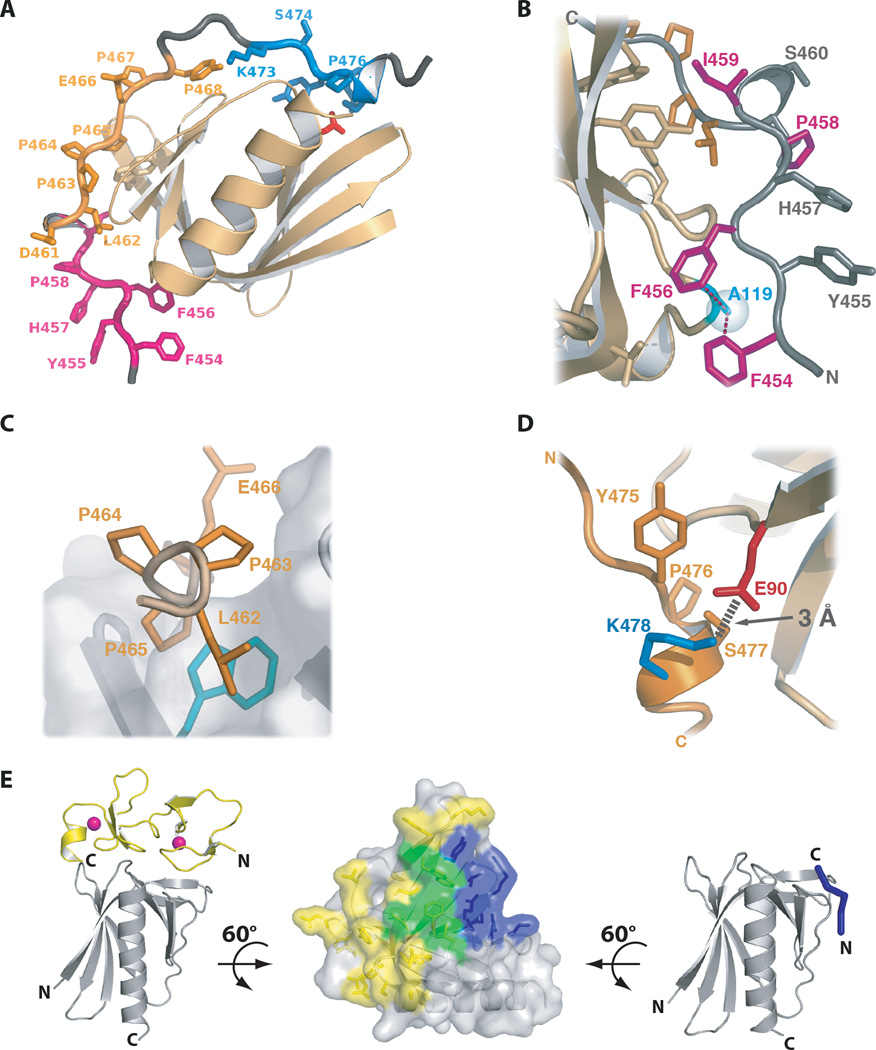
WASP interacting protein (WIP) forms a constitutive complex with an N-terminal EVH1 domain in WASP/WAVE proteins. WIP, WIP-related (WIRE/WICH), and CR16 comprise the mammalian verprolin family and function as regulators of the actin cytoskeleton (67). The verprolins are proline-rich proteins that bind profilin and various SH3-domain containing proteins, and contain an elongated EVH1 binding domain near the C-terminus. WIP residues 451–485 wrap more than halfway around the EVH1 domain, making specific contacts with three separate epitopes that correspond to regions of high sequence conservation in the verprolin family (Figure 6A). A central polyproline motif occupies the canonical binding site, but in a reversed orientation relative to other EVH1 complexes. Specific interactions involving WIP residues on either side of the polyproline motif specify the direction of peptide binding.
Figure 6.
Atypical EVH1 complexes. A) Three distinct WIP epitopes are required for N-WASP binding. Epitopes correspond to WIP amino acids 454–459 (magenta), 461–468 (gold), and 473–478 (blue). B) Hydrophobic contacts between the aromatic WIP residues Phe 454 and Phe 456 (magenta) and Ala 119 (cyan) from N-WASP. C) The WIP polyproline motif straddles the conserved Trp side chain of the EVH1 domain, but in the opposite orientation relative to peptide ligands for Mena and Homer. D) Lys 477 in WIP epitope 3 makes a conserved salt bridge to N-WASP residue Glu 90. E) Structures of the Mena EVH1 bound to the Tes LIM3 domain (left) and the FPPPP peptide (right) employ overlapping binding sites shown in green on the EVH1 domain surface (center).
Aromatic WIP residues in epitope 1 (454FYFHPIS460) bind a hydrophobic surface on the EVH1 domain (Figure 6B). Hydrophobic WIP residues Phe 454, Phe 456 and Ile 459 contribute ~20% of the total EVH1 contact surface. This N-terminal WIP epitope is linked to the polyproline sequence (epitope 2) by a well-defined structural element consisting of the His 457–Pro 458 cis peptide bond followed by a tight helical turn (residues 459–461).
WIP residues 461–468 contain the canonical polyproline motif and form the second binding epitope, which contributes just over 40% of the total buried surface of the WIP-EVH1 complex. WIP residues 462LPPP465 form a PPII helical turn that surrounds the conserved Trp side chain of the EVH1 binding surface (Figure 6C), similar to other EVH1-peptide complexes, but with the polypeptide chain running in the opposite direction (44).
The third WIP epitope, 473KSYPSK478, is separated from the polyproline motif by a flexible linker and contributes slightly less than 40% of the interface. While Lys 478 does not make extensive contact with the EVH1 surface, it is positioned by the WIP475–478 helical turn to form a salt bridge with Glu90 of N-WASP (Figure 6D), which corresponds to the site of a disease-causing mutation in the WASP sequence (11–16, 43).
These three WIP epitopes comprise a ~30 residue EVH1 binding domain that is conserved throughout the verprolin family of actin binding proteins (Figure 5B). Disruption of any of the three WIP epitopes reduces WASP or N-WASP binding in cells, demonstrating a functional requirement for a significantly longer sequence than the polyproline ligands recognized by other EVH1 domains. The WIP/N-WASP structure shows how semi-independent linear recognition motifs (68) can be used to make composite recognition motifs that are recognized in an extended conformation with enhanced specificity.
7. LIM DOMAIN RECOGNITION BY THE MENA EVH1 DOMAIN
While the verprolins contain unusually long EVH1 binding sequences, they still use the canonical PPII helix to recognize the conserved aromatic-rich binding groove in the same manner as all other known EVH1 ligands. Recently, however, the first non-polyproline binding partner was identified for an EVH1 domain. The EVH1 domain from Mena forms a specific complex with a small zinc-binding domain that lacks a proline-rich motif (10). The EVH1 domains of Mena, VASP and EVL all bind the FPPPP motif using the conserved proline recognition surface (9), but only Mena binds to the LIM3 domain of Tes, a putative tumor suppressor protein. The LIM3 and FPPPP binding surfaces overlap significantly (Figure 6D), consistent with the observation that they compete for binding to the EVH1 domain of Mena. Characterization of the Tes:Mena interaction raises the possibility of EVH1 complexes with other LIM domain proteins. By using a single site for multiple interaction partners, Mena also expands the potential range of EVH1 functions to include regulation of complex formation through competitive binding to overlapping recognition surfaces.
8. CONCLUSIONS
Structural studies of numerous EVH1-ligand complexes have defined a conserved aromatic-rich groove that recognizes a single turn of polyproline type II helix. Amino acids flanking the proline motif enhance ligand specificity and define the N-to-C orientation of peptide binding. Short (6–10 amino acids) peptide ligands of the Ena/VASP and Homer families bind in one orientation, but extended sequences (~30 amino acids) from the verprolin family wrap around EVH1 domains from the WASP family in the opposite orientation. The versatility of the EVH1 structure is further illustrated by the novel complex formed by the Mena EVH1 domain and the LIM3 domain from Tes, the first example of an EVH1 binding partner that lacks a polyproline motif. The unorthodox binding modes employed by N-WASP and Mena demonstrate that EVH1 interactions are more diverse than originally thought, and that competition between multiple EVH1 binding partners may be used to regulate the formation of signaling complexes in the cell.
Abbreviations
- EVH1
Enabled/VASP Homology-1
- PH
pleckstrin homology
- SH2
Src homology 2
- SH3
Src homology 3
- GYF
glycine-tyrosine-phenylalanine
- PID/PTB
Phosphotyrosine-interaction/phosphotyrosine-binding domains
- GRAM
(Glucosyltransferases, Rab-like GTPase activators-Myotubularins) domain
- FERM-C
(Four-point one, Ezrin, Merlin, Radixin) C-terminal domain
- SPED
Sprouty-related proteins with an EVH1 domain
- WASP
Wiskott-Aldrich-Syndrome protein
- N-WASP
neuronal-Wiskott-Aldrich-Syndrome protein
- Mena
mammalian Enabled
- EVL
Enabled/VASP-like protein
- RIAM
Rap1-GTP interacting adapter molecule
- WH1
WASP homology 1
- WIP
WASP interacting protein
- WIRE/WICH
WIP-related protein
- PPII
polyproline type II.
REFERENCES
- 1.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 2.Copley RR, Doerks T, Letunic I, Bork P. Protein domain analysis in the era of complete genomes. FEBS Lett. 2002;513:129–134. doi: 10.1016/s0014-5793(01)03289-6. [DOI] [PubMed] [Google Scholar]
- 3.Harris BZ, Lim WA. Mechanism and role of PDZ domains in signaling complex assembly. J Cell Sci. 2001;114:3219–3231. doi: 10.1242/jcs.114.18.3219. [DOI] [PubMed] [Google Scholar]
- 4.Pawson T, Gish GD, Nash P. SH2 domains, interaction modules and cellular wiring. Trends Cell Biol. 2001;11:504–511. doi: 10.1016/s0962-8924(01)02154-7. [DOI] [PubMed] [Google Scholar]
- 5.Zarrinpar A, Bhattacharyya RP, Lim WA. The structure and function of proline recognition domains. Sci STKE. 2003;2003:RE8. doi: 10.1126/stke.2003.179.re8. [DOI] [PubMed] [Google Scholar]
- 6.Begley MJ, Taylor GS, Brock MA, Ghosh P, Woods VL, Dixon JE. Molecular basis for substrate recognition by MTMR2, a myotubularin family phosphoinositide phosphatase. Proc Natl Acad Sci U S A. 2006;103:927–932. doi: 10.1073/pnas.0510006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearson MA, Reczek D, Bretscher A, Karplus PA. Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell. 2000;101:259–270. doi: 10.1016/s0092-8674(00)80836-3. [DOI] [PubMed] [Google Scholar]
- 8.Blomberg N, Baraldi E, Nilges M, Saraste M. The PH superfold: a structural scaffold for multiple functions. Trends Biochem Sci. 1999;24:441–445. doi: 10.1016/s0968-0004(99)01472-3. [DOI] [PubMed] [Google Scholar]
- 9.Prehoda KE, Lee DJ, Lim WA. Structure of the enabled/VASP homology 1 domain-peptide complex: a key component in the spatial control of actin assembly. Cell. 1999;97:471–480. doi: 10.1016/s0092-8674(00)80757-6. [DOI] [PubMed] [Google Scholar]
- 10.Boeda B, Briggs DC, Higgins T, Garvalov BK, Fadden AJ, McDonald NQ, Way M. Tes, a specific Mena interacting partner, breaks the rules for EVH1 binding. Mol Cell. 2007;28:1071–1082. doi: 10.1016/j.molcel.2007.10.033. [DOI] [PubMed] [Google Scholar]
- 11.Derry JM, Kerns JA, Weinberg KI, Ochs HD, Volpini V, Estivill X, Walker AP, Francke U. WASP gene mutations in Wiskott-Aldrich syndrome and X-linked thrombocytopenia. Hum Mol Genet. 1995;4:1127–1135. doi: 10.1093/hmg/4.7.1127. [DOI] [PubMed] [Google Scholar]
- 12.Greer WL, Shehabeldin A, Schulman J, Junker A, Siminovitch KA. Identification of WASP mutations, mutation hotspots and genotype-phenotype disparities in 24 patients with the Wiskott-Aldrich syndrome. Hum Genet. 1996;98:685–690. doi: 10.1007/s004390050285. [DOI] [PubMed] [Google Scholar]
- 13.Kolluri R, Shehabeldin A, Peacocke M, Lamhonwah AM, Teichert-Kuliszewska K, Weissman SM, Siminovitch KA. Identification of WASP mutations in patients with Wiskott-Aldrich syndrome and isolated thrombocytopenia reveals allelic heterogeneity at the WAS locus. Hum Mol Genet. 1995;4:1119–1126. doi: 10.1093/hmg/4.7.1119. [DOI] [PubMed] [Google Scholar]
- 14.Zhu Q, Watanabe C, Liu T, Hollenbaugh D, Blaese RM, Kanner SB, Aruffo A, Ochs HD. Wiskott-Aldrich syndrome/X-linked thrombocytopenia: WASP gene mutations, protein expression, and phenotype. Blood. 1997;90:2680–2689. [PubMed] [Google Scholar]
- 15.El-Hakeh J, Rosenzweig S, Oleastro M, Basack N, Berozdnik L, Molina F, Rivas EM, Zelazko M, Danielian S. Wiskott-Aldrich syndrome in Argentina: 17 unique, including nine novel, mutations. Hum Mutat. 2002;19:186–187. doi: 10.1002/humu.9013. [DOI] [PubMed] [Google Scholar]
- 16.Jin Y, Mazza C, Christie JR, Giliani S, Fiorini M, Mella P, Gandellini F, Stewart DM, Zhu Q, Nelson DL, Notarangelo LD, Ochs HD. Mutations of the Wiskott-Aldrich Syndrome Protein (WASP): hotspots, effect on transcription, and translation and phenotype/genotype correlation. Blood. 2004;104:4010–4019. doi: 10.1182/blood-2003-05-1592. [DOI] [PubMed] [Google Scholar]
- 17.Gertler FB, Comer AR, Juang JL, Ahern SM, Clark MJ, Liebl EC, Hoffmann FM. enabled, a dosage-sensitive suppressor of mutations in the Drosophila Abl tyrosine kinase, encodes an Abl substrate with SH3 domain-binding properties. Genes Dev. 1995;9:521–533. doi: 10.1101/gad.9.5.521. [DOI] [PubMed] [Google Scholar]
- 18.Gertler FB, Doctor JS, Hoffmann FM. Genetic suppression of mutations in the Drosophila abl proto-oncogene homolog. Science. 1990;248:857–860. doi: 10.1126/science.2188361. [DOI] [PubMed] [Google Scholar]
- 19.Reinhard M, Halbrugge M, Scheer U, Wiegand C, Jockusch BM, Walter U. The 46/50 kDa phosphoprotein VASP purified from human platelets is a novel protein associated with actin filaments and focal contacts. Embo J. 1992;11:2063–2070. doi: 10.1002/j.1460-2075.1992.tb05264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reinhard M, Jouvenal K, Tripier D, Walter U. Identification, purification, and characterization of a zyxin-related protein that binds the focal adhesion and microfilament protein VASP (vasodilator-stimulated phosphoprotein) Proc Natl Acad Sci U S A. 1995;92:7956–7960. doi: 10.1073/pnas.92.17.7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gertler FB, Niebuhr K, Reinhard M, Wehland J, Soriano P. Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell. 1996;87:227–239. doi: 10.1016/s0092-8674(00)81341-0. [DOI] [PubMed] [Google Scholar]
- 22.Ball LJ, Kuhne R, Hoffmann B, Hafner A, Schmieder P, Volkmer-Engert R, Hof M, Wahl M, Schneider-Mergener J, Walter U, Oschkinat H, Jarchau T. Dual epitope recognition by the VASP EVH1 domain modulates polyproline ligand specificity and binding affinity. Embo J. 2000;19:4903–4914. doi: 10.1093/emboj/19.18.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niebuhr K, Ebel F, Frank R, Reinhard M, Domann E, Carl UD, Walter U, Gertler FB, Wehland J, Chakraborty T. A novel proline-rich motif present in ActA of Listeria monocytogenes and cytoskeletal proteins is the ligand for the EVH1 domain, a protein module present in the Ena/VASP family. Embo J. 1997;16:5433–5444. doi: 10.1093/emboj/16.17.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klostermann A, Lutz B, Gertler F, Behl C. The orthologous human and murine semaphorin 6A-1 proteins (SEMA6A-1/Sema6A-1) bind to the enabled/vasodilator-stimulated phosphoprotein-like protein (EVL) via a novel carboxyl-terminal zyxin-like domain. J Biol Chem. 2000;275:39647–39653. doi: 10.1074/jbc.M006316200. [DOI] [PubMed] [Google Scholar]
- 25.Beckerle MC. Spatial control of actin filament assembly: lessons from Listeria. Cell. 1998;95:741–748. doi: 10.1016/s0092-8674(00)81697-9. [DOI] [PubMed] [Google Scholar]
- 26.Bashaw GJ, Kidd T, Murray D, Pawson T, Goodman CS. Repulsive axon guidance: Abelson and Enabled play opposing roles downstream of the roundabout receptor. Cell. 2000;101:703–715. doi: 10.1016/s0092-8674(00)80883-1. [DOI] [PubMed] [Google Scholar]
- 27.Lafuente EM, van Puijenbroek AA, Krause M, Carman CV, Freeman GJ, Berezovskaya A, Constantine E, Springer TA, Gertler FB, Boussiotis VA. RIAM, an Ena/VASP and Profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Dev Cell. 2004;7:585–595. doi: 10.1016/j.devcel.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 28.Krause M, Leslie JD, Stewart M, Lafuente EM, Valderrama F, Jagannathan R, Strasser GA, Rubinson DA, Liu H, Way M, Yaffe MB, Boussiotis VA, Gertler FB. Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev Cell. 2004;7:571–583. doi: 10.1016/j.devcel.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 29.Lambrechts A, Kwiatkowski AV, Lanier LM, Bear JE, Vandekerckhove J, Ampe C, Gertler FB. cAMP-dependent protein kinase phosphorylation of EVL, a Mena/VASP relative, regulates its interaction with actin and SH3 domains. J Biol Chem. 2000;275:36143–36151. doi: 10.1074/jbc.M006274200. [DOI] [PubMed] [Google Scholar]
- 30.Ahern-Djamali SM, Comer AR, Bachmann C, Kastenmeier AS, Reddy SK, Beckerle MC, Walter U, Hoffmann FM. Mutations in Drosophila enabled and rescue by human vasodilator-stimulated phosphoprotein (VASP) indicate important functional roles for Ena/VASP homology domain 1 (EVH1) and EVH2 domains. Mol Biol Cell. 1998;9:2157–2171. doi: 10.1091/mbc.9.8.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vasioukhin V, Bauer C, Yin M, Fuchs E. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell. 2000;100:209–219. doi: 10.1016/s0092-8674(00)81559-7. [DOI] [PubMed] [Google Scholar]
- 32.Cameron LA, Giardini PA, Soo FS, Theriot JA. Secrets of actin-based motility revealed by a bacterial pathogen. Nat Rev Mol Cell Biol. 2000;1:110–119. doi: 10.1038/35040061. [DOI] [PubMed] [Google Scholar]
- 33.Purich DL, Southwick FS. ABM-1 and ABM-2 homology sequences: consensus docking sites for actin-based motility defined by oligoproline regions in Listeria ActA surface protein and human VASP. Biochem Biophys Res Commun. 1997;231:686–691. doi: 10.1006/bbrc.1997.6158. [DOI] [PubMed] [Google Scholar]
- 34.Brakeman PR, Lanahan AA, O'Brien R, Roche K, Barnes CA, Huganir RL, Worley PF. Homer: a protein that selectively binds metabotropic glutamate receptors. Nature. 1997;386:284–288. doi: 10.1038/386284a0. [DOI] [PubMed] [Google Scholar]
- 35.Kato A, Ozawa F, Saitoh Y, Hirai K, Inokuchi K. vesl, a gene encoding VASP/Ena family related protein, is upregulated during seizure, long-term potentiation and synaptogenesis. FEBS Lett. 1997;412:183–189. doi: 10.1016/s0014-5793(97)00775-8. [DOI] [PubMed] [Google Scholar]
- 36.Berke JD, Paletzki RF, Aronson GJ, Hyman SE, Gerfen CR. A complex program of striatal gene expression induced by dopaminergic stimulation. J Neurosci. 1998;18:5301–5310. doi: 10.1523/JNEUROSCI.18-14-05301.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shiraishi-Yamaguchi Y, Furuichi T. The Homer family proteins. Genome Biol. 2007;8:206. doi: 10.1186/gb-2007-8-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ango F, Robbe D, Tu JC, Xiao B, Worley PF, Pin JP, Bockaert J, Fagni L. Homer-dependent cell surface expression of metabotropic glutamate receptor type 5 in neurons. Mol Cell Neurosci. 2002;20:323–329. doi: 10.1006/mcne.2002.1100. [DOI] [PubMed] [Google Scholar]
- 39.Roche KW, Tu JC, Petralia RS, Xiao B, Wenthold RJ, Worley PF. Homer 1b regulates the trafficking of group I metabotropic glutamate receptors. J Biol Chem. 1999;274:25953–25957. doi: 10.1074/jbc.274.36.25953. [DOI] [PubMed] [Google Scholar]
- 40.Szumlinski KK, Kalivas PW, Worley PF. Homer proteins: implications for neuropsychiatric disorders. Curr Opin Neurobiol. 2006;16:251–257. doi: 10.1016/j.conb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 41.Machesky LM, Insall RH. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- 42.Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner MW. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 43.Peterson FC, Deng Q, Zettl M, Prehoda KE, Lim WA, Way M, Volkman BF. Multiple WIP recognition motifs are required for a functional interaction with N-WASP. J Biol Chem. 2007 doi: 10.1074/jbc.M609902200. [DOI] [PubMed] [Google Scholar]
- 44.Volkman BF, Prehoda KE, Scott JA, Peterson FC, Lim WA. Structure of the N-WASP EVH1 domain-WIP complex: insight into the molecular basis of Wiskott-Aldrich Syndrome. Cell. 2002;111:565–576. doi: 10.1016/s0092-8674(02)01076-0. [DOI] [PubMed] [Google Scholar]
- 45.Zettl M, Way M. The WH1 and EVH1 domains of WASP and Ena/VASP family members bind distinct sequence motifs. Curr Biol. 2002;12:1617–1622. doi: 10.1016/s0960-9822(02)01112-0. [DOI] [PubMed] [Google Scholar]
- 46.Ramesh N, Anton IM, Hartwig JH, Geha RS. WIP, a protein associated with wiskott-aldrich syndrome protein, induces actin polymerization and redistribution in lymphoid cells. Proc Natl Acad Sci U S A. 1997;94:14671–14676. doi: 10.1073/pnas.94.26.14671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aspenstrom P. The WASP-binding protein WIRE has a role in the regulation of the actin filament system downstream of the platelet-derived growth factor receptor. Exp Cell Res. 2002;279:21–33. doi: 10.1006/excr.2002.5576. [DOI] [PubMed] [Google Scholar]
- 48.Kato M, Miki H, Kurita S, Endo T, Nakagawa H, Miyamoto S, Takenawa T. WICH, a novel verprolin homology domain-containing protein that functions cooperatively with N-WASP in actin-microspike formation. Biochem Biophys Res Commun. 2002;291:41–47. doi: 10.1006/bbrc.2002.6406. [DOI] [PubMed] [Google Scholar]
- 49.Ho HY, Rohatgi R, Ma L, Kirschner MW. CR16 forms a complex with N-WASP in brain and is a novel member of a conserved proline-rich actin-binding protein family. Proc Natl Acad Sci U S A. 2001;98:11306–11311. doi: 10.1073/pnas.211420498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gallego MD, de la Fuente MA, Anton IM, Snapper S, Fuhlbrigge R, Geha RS. WIP and WASP play complementary roles in T cell homing and chemotaxis to SDF-1alpha. Int Immunol. 2006;18:221–232. doi: 10.1093/intimm/dxh310. [DOI] [PubMed] [Google Scholar]
- 51.Lutskiy MI, Rosen FS, Remold-O'Donnell E. Genotype-proteotype linkage in the Wiskott-Aldrich syndrome. J Immunol. 2005;175:1329–1336. doi: 10.4049/jimmunol.175.2.1329. [DOI] [PubMed] [Google Scholar]
- 52.Sasahara Y, Rachid R, Byrne MJ, de la Fuente MA, Abraham RT, Ramesh N, Geha RS. Mechanism of recruitment of WASP to the immunological synapse and of its activation following TCR ligation. Mol Cell. 2002;10:1269–1281. doi: 10.1016/s1097-2765(02)00728-1. [DOI] [PubMed] [Google Scholar]
- 53.Dong X, Patino-Lopez G, Candotti F, Shaw S. Structure-function analysis of the WIP role in T cell receptor-stimulated NFAT activation: evidence that WIP-WASP dissociation is not required and that the WIP NH2 terminus is inhibitory. J Biol Chem. 2007;282:30303–30310. doi: 10.1074/jbc.M704972200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Derry JM, Ochs HD, Francke U. Isolation of a novel gene mutated in Wiskott-Aldrich syndrome. Cell. 1994;78:635–644. doi: 10.1016/0092-8674(94)90528-2. [DOI] [PubMed] [Google Scholar]
- 55.Notarangelo LD, Mori L. Wiskott-Aldrich syndrome: another piece in the puzzle. Clin Exp Immunol. 2005;139:173–175. doi: 10.1111/j.1365-2249.2005.02707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kato R, Nonami A, Taketomi T, Wakioka T, Kuroiwa A, Matsuda Y, Yoshimura A. Molecular cloning of mammalian Spred-3 which suppresses tyrosine kinase-mediated Erk activation. Biochem Biophys Res Commun. 2003;302:767–772. doi: 10.1016/s0006-291x(03)00259-6. [DOI] [PubMed] [Google Scholar]
- 57.Wakioka T, Sasaki A, Kato R, Shouda T, Matsumoto A, Miyoshi K, Tsuneoka M, Komiya S, Baron R, Yoshimura A. Spred is a Sprouty-related suppressor of Ras signalling. Nature. 2001;412:647–651. doi: 10.1038/35088082. [DOI] [PubMed] [Google Scholar]
- 58.Nobuhisa I, Kato R, Inoue H, Takizawa M, Okita K, Yoshimura A, Taga T. Spred-2 suppresses aorta-gonad-mesonephros hematopoiesis by inhibiting MAP kinase activation. J Exp Med. 2004;199:737–742. doi: 10.1084/jem.20030830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harmer NJ, Sivak JM, Amaya E, Blundell TL. 1.15 A crystal structure of the X. tropicalis Spred1 EVH1 domain suggests a fourth distinct peptide-binding mechanism within the EVH1 family. FEBS Lett. 2005;579:1161–1166. doi: 10.1016/j.febslet.2004.11.114. [DOI] [PubMed] [Google Scholar]
- 60.Bundschu K, Knobeloch KP, Ullrich M, Schinke T, Amling M, Engelhardt CM, Renne T, Walter U, Schuh K. Gene disruption of Spred-2 causes dwarfism. J Biol Chem. 2005;280:28572–28580. doi: 10.1074/jbc.M503640200. [DOI] [PubMed] [Google Scholar]
- 61.Inoue H, Kato R, Fukuyama S, Nonami A, Taniguchi K, Matsumoto K, Nakano T, Tsuda M, Matsumura M, Kubo M, Ishikawa F, Moon BG, Takatsu K, Nakanishi Y, Yoshimura A. Spred-1 negatively regulates allergen-induced airway eosinophilia and hyperresponsiveness. J Exp Med. 2005;201:73–82. doi: 10.1084/jem.20040616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nonami A, Kato R, Taniguchi K, Yoshiga D, Taketomi T, Fukuyama S, Harada M, Sasaki A, Yoshimura A. Spred-1 negatively regulates interleukin-3-mediated ERK/mitogen-activated protein (MAP) kinase activation in hematopoietic cells. J Biol Chem. 2004;279:52543–52551. doi: 10.1074/jbc.M405189200. [DOI] [PubMed] [Google Scholar]
- 63.Feng S, Chen JK, Yu H, Simon JA, Schreiber SL. Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions. Science. 1994;266:1241–1247. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- 64.Lim WA, Richards FM. Critical residues in an SH3 domain from Sem-5 suggest a mechanism for proline-rich peptide recognition. Nat Struct Biol. 1994;1:221–225. doi: 10.1038/nsb0494-221. [DOI] [PubMed] [Google Scholar]
- 65.Lim WA, Richards FM, Fox RO. Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Nature. 1994;372:375–379. doi: 10.1038/372375a0. [DOI] [PubMed] [Google Scholar]
- 66.Yu H, Chen JK, Feng S, Dalgarno DC, Brauer AW, Schreiber SL. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- 67.Aspenstrom P. The verprolin family of proteins: regulators of cell morphogenesis and endocytosis. FEBS Lett. 2005;579:5253–5259. doi: 10.1016/j.febslet.2005.08.053. [DOI] [PubMed] [Google Scholar]
- 68.Puntervoll P, Linding R, Gemund C, Chabanis-Davidson S, Mattingsdal M, Cameron S, Martin DM, Ausiello G, Brannetti B, Costantini A, Ferre F, Maselli V, Via A, Cesareni G, Diella F, Superti-Furga G, Wyrwicz L, Ramu C, McGuigan C, Gudavalli R, Letunic I, Bork P, Rychlewski L, Kuster B, Helmer-Citterich M, Hunter WN, Aasland R, Gibson TJ. ELM server: A new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kato A, Ozawa F, Saitoh Y, Fukazawa Y, Sugiyama H, Inokuchi K. Novel members of the Vesl/Homer family of PDZ proteins that bind metabotropic glutamate receptors. J Biol Chem. 1998;273:23969–23975. doi: 10.1074/jbc.273.37.23969. [DOI] [PubMed] [Google Scholar]
- 70.Shiraishi Y, Mizutani A, Mikoshiba K, Furuichi T. Coincidence in dendritic clustering and synaptic targeting of homer proteins and NMDA receptor complex proteins NR2B and PSD95 during development of cultured hippocampal neurons. Mol Cell Neurosci. 2003;22:188–201. doi: 10.1016/s1044-7431(03)00037-x. [DOI] [PubMed] [Google Scholar]
- 71.Xiao B, Tu JC, Petralia RS, Yuan JP, Doan A, Breder CD, Ruggiero A, Lanahan AA, Wenthold RJ, Worley PF. Homer regulates the association of group 1 metabotropic glutamate receptors with multivalent complexes of homer-related, synaptic proteins. Neuron. 1998;21:707–716. doi: 10.1016/s0896-6273(00)80588-7. [DOI] [PubMed] [Google Scholar]
- 72.Govek EE, Newey SE, Akerman CJ, Cross JR, Van der Veken L, Van Aelst L. The X-linked mental retardation protein oligophrenin-1 is required for dendritic spine morphogenesis. Nat Neurosci. 2004;7:364–372. doi: 10.1038/nn1210. [DOI] [PubMed] [Google Scholar]
- 73.Gray NW, Fourgeaud L, Huang B, Chen J, Cao H, Oswald BJ, Hemar A, McNiven MA. Dynamin 3 is a component of the postsynapse, where it interacts with mGluR5 and Homer. Curr Biol. 2003;13:510–515. doi: 10.1016/s0960-9822(03)00136-2. [DOI] [PubMed] [Google Scholar]
- 74.Inaba Y, Tian QB, Okano A, Zhang JP, Sakagami H, Miyazawa S, Li W, Komiyama A, Inokuchi K, Kondo H, Suzuki T. Brain-specific potential guanine nucleotide exchange factor for Arf, synArfGEF (Po), is localized to postsynaptic density. J Neurochem. 2004;89:1347–1357. doi: 10.1111/j.1471-4159.2004.02440.x. [DOI] [PubMed] [Google Scholar]
- 75.Rong R, Ahn JY, Huang H, Nagata E, Kalman D, Kapp JA, Tu J, Worley PF, Snyder SH, Ye K. PI3 kinase enhancer-Homer complex couples mGluRI to PI3 kinase, preventing neuronal apoptosis. Nat Neurosci. 2003;6:1153–1161. doi: 10.1038/nn1134. [DOI] [PubMed] [Google Scholar]
- 76.Tu JC, Xiao B, Naisbitt S, Yuan JP, Petralia RS, Brakeman P, Doan A, Aakalu VK, Lanahan AA, Sheng M, Worley PF. Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron. 1999;23:583–592. doi: 10.1016/s0896-6273(00)80810-7. [DOI] [PubMed] [Google Scholar]
- 77.Tu JC, Xiao B, Yuan JP, Lanahan AA, Leoffert K, Li M, Linden DJ, Worley PF. Homer binds a novel proline-rich motif and links group 1 metabotropic glutamate receptors with IP3 receptors. Neuron. 1998;21:717–726. doi: 10.1016/s0896-6273(00)80589-9. [DOI] [PubMed] [Google Scholar]
- 78.Yuan JP, Kiselyov K, Shin DM, Chen J, Shcheynikov N, Kang SH, Dehoff MH, Schwarz MK, Seeburg PH, Muallem S, Worley PF. Homer binds TRPC family channels and is required for gating of TRPC1 by IP3 receptors. Cell. 2003;114:777–789. doi: 10.1016/s0092-8674(03)00716-5. [DOI] [PubMed] [Google Scholar]
- 79.Krause M, Sechi AS, Konradt M, Monner D, Gertler FB, Wehland J. Fyn-binding protein (Fyb)/SLP-76-associated protein (SLAP), Ena/vasodilator-stimulated phosphoprotein (VASP) proteins and the Arp2/3 complex link T cell receptor (TCR) signaling to the actin cytoskeleton. J Cell Biol. 2000;149:181–194. doi: 10.1083/jcb.149.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ahern-Djamali SM, Bachmann C, Hua P, Reddy SK, Kastenmeier AS, Walter U, Hoffmann FM. Identification of profilin and src homology 3 domains as binding partners for Drosophila enabled. Proc Natl Acad Sci U S A. 1999;96:4977–4982. doi: 10.1073/pnas.96.9.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ermekova KS, Zambrano N, Linn H, Minopoli G, Gertler F, Russo T, Sudol M. The WW domain of neural protein FE65 interacts with proline-rich motifs in Mena, the mammalian homolog of Drosophila enabled. J Biol Chem. 1997;272:32869–32877. doi: 10.1074/jbc.272.52.32869. [DOI] [PubMed] [Google Scholar]
- 82.Lanier LM, Gates MA, Witke W, Menzies AS, Wehman AM, Macklis JD, Kwiatkowski D, Soriano P, Gertler FB. Mena is required for neurulation and commissure formation. Neuron. 1999;22:313–325. doi: 10.1016/s0896-6273(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 83.Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V, Jockusch BM, Walter U. The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. Embo J. 1995;14:1583–1589. doi: 10.1002/j.1460-2075.1995.tb07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Drees B, Friederich E, Fradelizi J, Louvard D, Beckerle MC, Golsteyn RM. Characterization of the interaction between zyxin and members of the Ena/vasodilator-stimulated phosphoprotein family of proteins. J Biol Chem. 2000;275:22503–22511. doi: 10.1074/jbc.M001698200. [DOI] [PubMed] [Google Scholar]
- 85.Pistor S, Chakraborty T, Walter U, Wehland J. The bacterial actin nucleator protein ActA of Listeria monocytogenes contains multiple binding sites for host microfilament proteins. Curr Biol. 1995;5:517–525. doi: 10.1016/s0960-9822(95)00104-7. [DOI] [PubMed] [Google Scholar]
- 86.Carl UD, Pollmann M, Orr E, Gertlere FB, Chakraborty T, Wehland J. Aromatic and basic residues within the EVH1 domain of VASP specify its interaction with proline-rich ligands. Curr Biol. 1999;9:715–718. doi: 10.1016/s0960-9822(99)80315-7. [DOI] [PubMed] [Google Scholar]
- 87.Harbeck B, Huttelmaier S, Schluter K, Jockusch BM, Illenberger S. Phosphorylation of the vasodilator-stimulated phosphoprotein regulates its interaction with actin. J Biol Chem. 2000;275:30817–30825. doi: 10.1074/jbc.M005066200. [DOI] [PubMed] [Google Scholar]
- 88.Huttelmaier S, Mayboroda O, Harbeck B, Jarchau T, Jockusch BM, Rudiger M. The interaction of the cell-contact proteins VASP and vinculin is regulated by phosphatidylinositol-4,5-bisphosphate. Curr Biol. 1998;8:479–488. doi: 10.1016/s0960-9822(98)70199-x. [DOI] [PubMed] [Google Scholar]
- 89.Jonckheere V, Lambrechts A, Vandekerckhove J, Ampe C. Dimerization of profilin II upon binding the (GP5)3 peptide from VASP overcomes the inhibition of actin nucleation by profilin II and thymosin beta4. FEBS Lett. 1999;447:257–263. doi: 10.1016/s0014-5793(99)00293-8. [DOI] [PubMed] [Google Scholar]
- 90.Lambrechts A, Verschelde JL, Jonckheere V, Goethals M, Vandekerckhove J, Ampe C. The mammalian profilin isoforms display complementary affinities for PIP2 and proline-rich sequences. Embo J. 1997;16:484–494. doi: 10.1093/emboj/16.3.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Laurent V, Loisel TP, Harbeck B, Wehman A, Grobe L, Jockusch BM, Wehland J, Gertler FB, Carlier MF. Role of proteins of the Ena/VASP family in actin-based motility of Listeria monocytogenes. J Cell Biol. 1999;144:1245–1258. doi: 10.1083/jcb.144.6.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- 93.Petit MM, Fradelizi J, Golsteyn RM, Ayoubi TA, Menichi B, Louvard D, Van de Ven WJ, Friederich E. LPP, an actin cytoskeleton protein related to zyxin, harbors a nuclear export signal and transcriptional activation capacity. Mol Biol Cell. 2000;11:117–129. doi: 10.1091/mbc.11.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rohatgi R, Nollau P, Ho HY, Kirschner MW, Mayer BJ. Nck and phosphatidylinositol 4,5-bisphosphate synergistically activate actin polymerization through the N-WASP-Arp2/3 pathway. J Biol Chem. 2001;276:26448–26452. doi: 10.1074/jbc.M103856200. [DOI] [PubMed] [Google Scholar]
- 95.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]