Summary
The establishment of a multi-cellular body plan requires coordinating changes in cell adhesion and the cytoskeleton to ensure proper cell shape and position within a tissue. Cell adhesion to the extracellular matrix (ECM) via integrins plays diverse, essential roles during animal embryogenesis and therefore must be precisely regulated [1]. Talin, a FERM-domain containing protein, forms a direct link between integrin adhesion receptors and the actin cytoskeleton, and is an important regulator of integrin function [2]. Similar to other FERM proteins, talin makes an intramolecular interaction that could autoinhibit its activity [3–6]. However, the functional consequence of such an interaction has not been previously explored in vivo. Here, we demonstrate that targeted disruption of talin autoinhibition gives rise to morphogenetic defects during fly development and specifically that dorsal closure (DC), a process that resembles wound healing, is delayed. Impairment of autoinhibition leads to reduced talin turnover at and increased talin and integrin recruitment to sites of integrin-ECM attachment. Finally, we present evidence that talin autoinhibition is regulated by Rap1-dependent signaling. Based on our data we propose that talin autoinhibition provides a switch for modulating adhesion turnover and adhesion stability that is essential for morphogenesis.
Results and Discussion
Integrins connect to the cytoskeleton through an intracellular adhesion complex (IAC); changes to the protein composition and interactions within the IAC have important implications for integrin-dependent cellular behaviors [1, 7–9]. Talin is an essential IAC component [2, 10] containing a conserved, integrin-binding FERM domain at its N-terminus, and an actin-binding domain at the C-terminus of its helical rod domain [2]. Structural studies identified residues in both the talin FERM and rod domains that mediate autoinhibition (Figure 1A) [4, 5]. The same region of the talin FERM domain that binds integrin also binds the rod to mediate autoinhibition [5]. It has been proposed that talin autoinhibition may provide a mechanism to down-regulate talin-dependent integrin activation and blocking talin autoinhibition leads to integrin activation [4,5]. The biological role of talin autoinhibition is currently not well defined but initial results in cell culture suggest that it plays an important role as expression of autoinhibition-impaired talin results in increased integrin activation and altered cell spreading [4, 11].
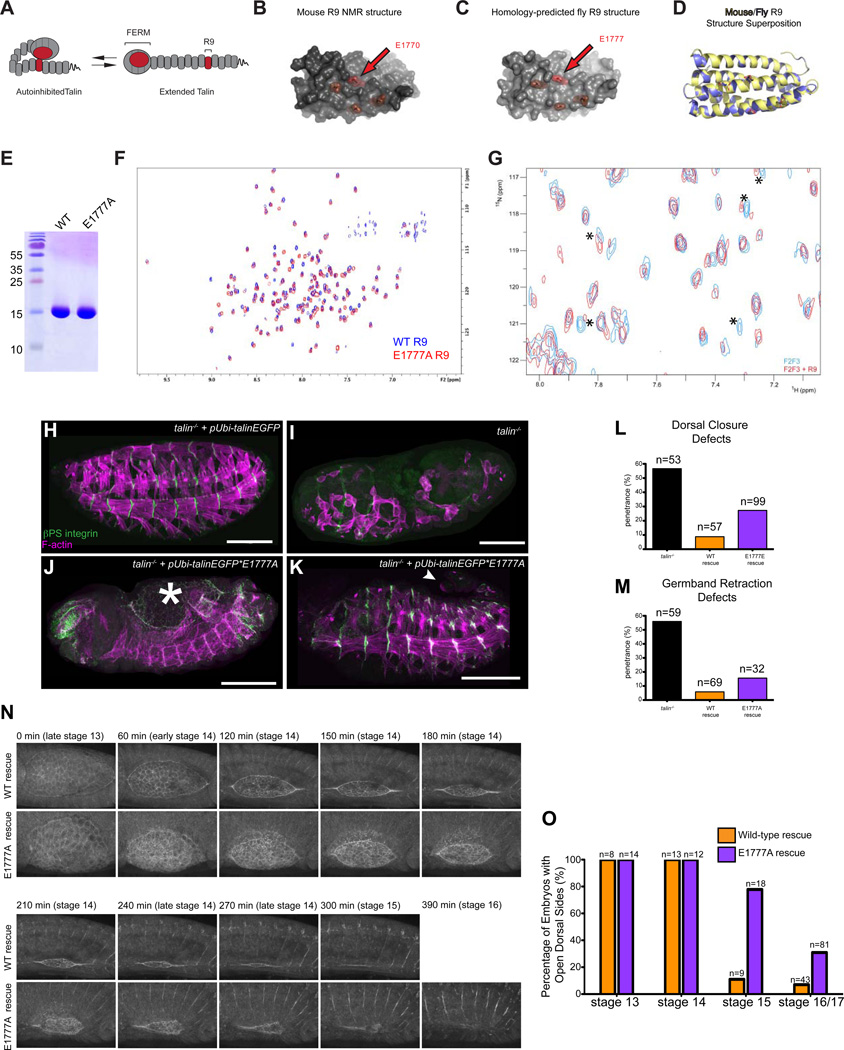
Figure 1. Disruption of a conserved autoinhibitory intramolecular interaction between the talin FERM and the talin rod leads to morphogenetic defects including delayed dorsal closure.
(A) Cartoon schematic of talin autoinhibition. (B-D) The NMR structure[5] of mouse R9 (B) and our homology-predicted model of fly talin R9 (C). Critical residues for F3-rod binding are highlighted in red. (D) Superposition of the mouse NMR structure (yellow) and the homology-modeled fly structure (blue). (E) Coomassie-stained SDS-PAGE gel showing that purified recombinant WT and E1777A fly R9 domains exhibit similar electrophoretic mobility at the expected molecular weight. (F) 1H,15N-TROSY-HSQC spectra of 150 µM 15N-labeled WT talin R9 (blue) and R9 E1777A (red). The R9 E1777A mutant shows a well dispersed NMR spectrum similar to that of the wildtype R9 indicating that the mutation does not affect the tertiary structure of the domain. (G) A 1H,15N-TROSY-HSQC spectra of 25 µM 15N-labeled fly talin F2F3 alone (blue) or in the presence of the talin rod R9 domain (red). In the presence of R9, some of the peaks have shifted and broadened (indicated by asterisks) compared to the spectra of the free F2F3 providing evidence of a direct interaction between fly F2F3 and R9. (H-M) Late stage talin-null embryos stained for integrin (green in H-K) and F-actin (magenta in H-K) were scored for phenotypes in the morphogenetic processes DC (J,L; asterisk in J demarcates open dorsal hole) and GBR (K,M; arrowhead in K shows un-retracted tail). Embryos were rescued with talinEGFP (H) construct or the talinEGFP*E1777A autoinhibition mutant construct (J-K). (N-O). Talin-null embryos rescued with either talinGFP or talinEGFP*E1777A were scored for dorsal holes at stage 13–17 (see Experimental Procedures). (O) Images from time-lapse movies of WT-rescued embryos (top) or E1777A mutant (bottom) embryos expressing talinGFP*E1777A and undergoing DC at the indicated time-points.
We hypothesized that the mechanism of autoinhibition is conserved between flies and vertebrates. The autoinhibitory regions have been mapped to the F3 lobe of the FERM domain (residues 309–400 in Human Talin1; 318–409 in Drosophila Talin), and a region of the rod called R9, which forms an amphipathic helical bundle (residues 1655–1826 in mammals; 1662–1831 in fly) [5]. The F3 domain is highly conserved across species, with 85.7% protein sequence similarity and 74.7% identity between human Talin1 and fly talin (Figure S1A). The protein sequence of R9 is also highly conserved with 56.3% similarity and 33.5% identity (Fig. S1B). We used homology modeling to predict the structure of the rod R9 domain based on the NMR structure of mouse talin and found the fly structure closely resembles that of mouse (Fig. 1B,C). Notably, four negatively charged surface residues in the rod that are important for autoinhibitory interactions between the FERM and the rod domains are conserved in sequence and arrangement between flies and humans (Fig. 1B,C). To quantify differences in secondary structure between the mouse NMR structure and the predicted fly structure, we calculated the root-mean square deviation (RMSD) of the superposition of the two structures (Fig. 1D). We obtained a RMSD of 0.148Å for 635 aligned atoms suggesting the two structures are very similar. Homology modeling of the FERM domain also showed excellent conservation between fly and vertebrate (P.L. & F.V.P; data not shown). We also used NMR spectroscopy to show that the fly R9 domain adopts a stable globular conformation in vitro, similar to the mouse protein homolog (Fig. 1E,F). Altogether, our homology modeling and NMR data suggest that the domains of mammalian talin and fly talin involved in autoinhibition are likely structurally conserved. Importantly, NMR spectroscopy confirmed an interaction between F2-F3 and R9 of fly talin (Fig. 1G). This result further confirms the notion that this interaction, which mediates autoinhibition, is conserved between flies and vertebrates.
We sought to design a fly mutant that would specifically disrupt talin autoinhibition. In the R9 domain, we chose to introduce a mutation that was shown, in vitro, to completely abrogate binding with the FERM domain and thus block autoinhibition [5]. This mutation changes a conserved glutamate residue in R9 (E1777 in fly; E1770 in mammalian talin) to an alanine residue (E1777A). NMR analyses demonstrated that the spectra of the region of talin containing the E1777A strongly resembled the spectra of the wild-type region indicating that the mutation does not disrupt protein folding (Fig. 1F). It was not feasible to choose a mutation in the FERM domain to abrogate autoinhibition for two reasons. Firstly, there have only been two mutations in the FERM domain that have been described to disrupt autoinhibition: the role of the first, M319A (equivalent to M328 in flies), is the subject of an unresolved dispute [4, 5, 12]. Secondly, the other mutation described to disrupt autoinhibition K324D [5] (equivalent to K333D in flies) is adjacent to a residue that is critical for talin function (L325 in vertebrates; L334 in flies) [11, 13,14]. Moreover, this region of the FERM domain is packed with interaction sites for talin binding partners (Supplemental Fig. S1A). These factors would make it very difficult to interpret, in vivo, the phenotype of mutations in the FERM domain that disrupt talin autoinhibition.
To assess the role of talin autoinhibition, wild-type (WT) endogenous talin was replaced in developing Drosophila embryos with rescue transgenes containing the E1777A mutation (see Supplemental Experimental Procedures). Previous analysis has shown that a ubiquitously expressed WT talin rescue transgene (talinGFP) rescues the embryonic lethality that results when embryos lack both maternal and zygotic talin protein ([9]; Fig. 1H,I). In comparison TalinGFP*E1777A failed to rescue the lethality associated with loss of talin (Fig. 1J,K). The ability of talin transgenes to rescue talin mutants was assayed in the context of three different integrin-dependent processes. Two of these, DC and Germband Retraction (GBR), represent dynamic morphogenetic processes while the third, muscle attachment, represents stable long-term adhesion. While talinGFP fully rescued GBR and DC, TalinGFP*E1777A only gave a partial and inconsistent rescue (Fig. 2L,M). DC occurs late in fly embryogenesis and involves the migration of two epidermal sheets over an extra-embryonic epithelium called the amnioserosa (AS); the AS actively contributes to DC [15, 16]. The end result of DC is to create a continuous epidermis on the dorsal side of the embryo. Of embryos rescued with talinGFP*E1777A 27.3% (n=99) failed to complete DC compared with 49.2% (n=53) of talin null embryos and 8% (n=57) of talinGFP rescued embryos (Fig. 2L). However, closer examination of earlier stage embryos revealed a more penetrant phenotype (Fig. 2N-O); DC normally concludes at stage 15 in talinGFP-rescued embryos (89% completion rate/stage 15; n=9) but this was not the case for the majority of talinGFP*E1777A rescued embryos (22.2% completion rate/stage 15; n=18). Therefore, talin mutants rescued with talinE1777A exhibited delayed DC (Fig. 2O). We confirmed that DC was delayed in talinGFP*E1777A rescued talin mutants using live time-lapse imaging of rescue embryos (Fig. 2N, Movies S1–S2).
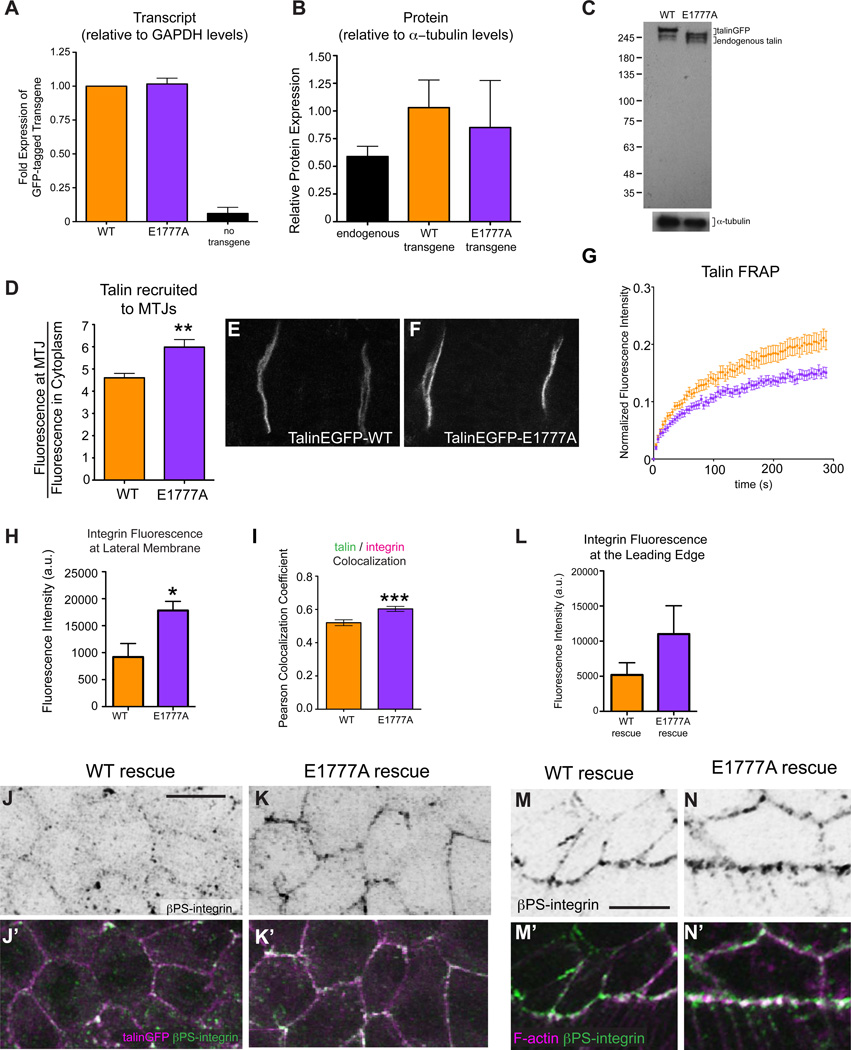
Figure 2. Talin stability and dynamics are compromised by the E1777A autoinhibition mutation.
(A-C) qPCR (A) and Western blot data (B-C) for talinEGFP (orange), autoinhibition mutant talinEGFP*E1777A (purple) and endogenous untagged talin (black). Talin was detected with a polyclonal antibody raised to the C-terminus (see [10]) and Westerns were done in a wild-type background. (D-F) The recruitment of talinGFP (D,E) and talinGFP*E1777A (D,F) at MTJs (C, p<0.01). (F) Fluorescence recovery curves of talinGFP (orange) and talinGFP*E1777A (purple) obtained from FRAP experiments on embryonic MTJs, *p<0.001. (G-N) βPS-integrin signal was quantified at the lateral membrane of AS cells (H) and the leading edge of the epidermis (L) and colocalization of talin (magenta in J’,K’) and βPS integrin (green in J’-K’,M’-N’; black in J’-K’,M’-N’) was measured at the lateral membrane of AS cells using Pearson correlation co-efficients (I; n>25 cells, *p<0.05, ***p<0.001). F-actin is shown in magenta in M’-N’ to highlight the leading edge.
A possible explanation for the delayed and incomplete DC observed in TalinGFP*E1777A-rescued embryos is insufficient expression of the mutant talin. Quantitative RT-PCR analysis revealed that transcript levels of talinGFP and the talinGFP*E1777A mutant were approximately equivalent (Fig. 2A). Western blot analysis showed that talinGFP*E1777A protein levels were slightly less than those of the talinGFP WT transgene (Fig. 2B). However, the mutant transgenic protein is still present at levels that are comparable to, and even slightly higher than, the levels of endogenous talin protein since the use of the ubi promoter results in slight over-expression of both talinGFP*E1777A and TalinGFP relative to endogenous protein (Fig 2B-C). Intriguingly, we observe a slight difference in size between talinGFP and talinGFP*E1777A, but we have no evidence to suggest that this has any functional consequence. Importantly, we could not detect a reduction in talin levels via antibody staining at myotendinous junctions (MTJs) suggesting that talinGFP*E1777A transgene expresses sufficiently (Fig. S2). We also quantified the recruitment of WT talinGFP and talinGFP*E1777A to the prominent integrin adhesions at the MTJs of embryonic muscles using our established protocol [9, 17]. TalinGFP*E1777A was recruited to sites of integrin-mediated adhesion at MTJs better than TalinGFP (Fig. 2D-F). This result is reminiscent of recent reports in cultured cells showing that mutating the talin rod to prevent autoinhibition results in increased talin localization in the membrane fraction [12]. Altogether, the defects we observe in talinGFP*E1777A mutant embryos are likely not caused by reduced expression and/or mislocalization of talin but by the specific effects of the mutation.
To investigate whether the TalinGFP*E1777A impairs the assembly of the IAC and/or its attachment to the ECM we analyzed the fly MTJs as they provide an established and quantitative model to study disruptions in IAC recruitment and ECM attachment [9, 17–19]. We did not find any defects in MTJ integrity, IAC recruitment or ECM attachment in talin mutant embryos rescued with TalinGFP*E1777A (Fig. S2A-E,I-O). Previous studies suggested that the ability of talin to autoinhibit might comprise a mechanism to modulate vinculin recruitment and actin association. However, we were unable to find any differences in either actin or vinculin recruitment (Fig. S2D-E,I-L). Additionally, vinculin was not expressed in the AS providing further evidence that a disruption in vinculin binding to talin was unlikely to underlie the dorsal closure defects we observed in the talinGFP*E1777A-rescued embryos (Fig. S2F). We also used gel filtration to confirm that the R9 of the rod domain does not bind vinculin in vitro (Fig. S1G,H).
Defective morphogenesis could result from improper regulation of stability and turnover of integrin-mediated adhesions. To test this, we studied the adhesion dynamics exhibited by the autoinhibition defective talinGFP*E1777A using our previously established Fluorescence Recovery After Photobleaching (FRAP) protocol to examine the turnover of integrin and IAC components at MTJs in living Drosophila embryos and larvae [18]. FRAP analysis revealed that talinGFP*E1777A is more stable at MTJs than WT talinGFP (Fig. 2G). This data suggests that talin autoinhibition can modulate the turnover of integrin-based adhesion and that, specifically, preventing talin autoinhibition stabilizes the adhesion complex. Further examination of integrin-mediated adhesions in the AS supported this idea. We found that embryos rescued with talinGFP*E1777A exhibited greater integrin recruitment to the membrane of AS cells (Fig. 2H, J-K) and also to the leading edge of the epidermal cells that crawl over the AS (Fig. 2L-N). We also observed increased co-localization of talin and integrin in the AS (Fig. 2I-K). These observations are in line with reports in culture that expression of the talinE1770A autoinhibition mutant resulted in increased focal adhesion assembly [11].
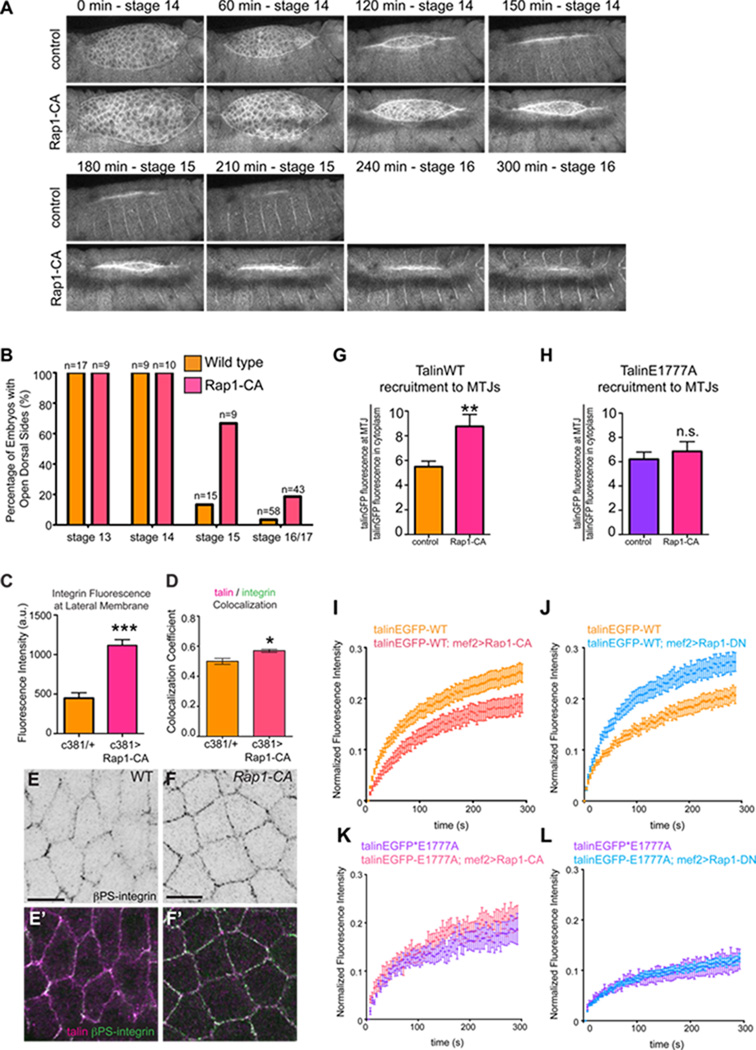
Our results indicated a link between autoinhibition and the regulation of the turnover and stability of integrin-based adhesions. The signaling molecules FAK and Rap1 have been implicated in such regulation [20–22] and we sought to see if either effector acts to regulate talin autoinhibition. Analysis of FAK failed to show any phenotypic parallels or genetic interactions with talin autoinhibition; loss of FAK does not lead to defects in embryogenesis or disrupt viability [23]. Moreover, modulation of FAK activity does not impinge on turnover of either WT talin or the talinE1777A at MTJs (Fig. S3). We also tested the small GTPase Rap1, which has been implicated as part of a putative complex that localizes talin from the cytoplasm to adhesion complexes at the plasma membrane [24, 25] where it has been speculated autoinhibition can be relieved [14, 26, 27]. Our hypothesis was that increasing Rap1 activity would give rise to similar phenotypes to those observed in talinEGFP*E1777A-rescued embryos. To test this, we expressed a constitutively-active form of Rap1 (Rap1-Q63E; Rap1-CA) in the AS using the tissue specific Gal4 driver, c381; we observed similar DC defects to those seen with the autoinhibition defective talin (Fig. 3). Specifically, more than 60% of the Rap1-CA expressing embryos had open dorsal holes at the end of stage 15 with about 20% of the embryos failing to complete DC altogether (Figure 3A-B). We confirmed this delay using time-lapse imaging (Fig. 3A, Movie S3–4). Furthermore, we found that colocalization of integrin and talin was increased in integrin-mediated adhesions of AS cells expressing Rap1-CA – this manifested itself as an increase in integrin signal at the membrane (Fig. 3C-F). We also tested the effect of expressing a dominant-negative form of Rap1 (Rap1-S17A; Rap1-DN) specifically in the AS and found that it also gave rise to DC defects. However, the Rap1-DN phenotype is different from that observed with the Rap1-CA in two ways: i) DC was not delayed but rather failed outright and ii) other morphogenetic problems, such as failed GBR, were observed (Fig. S4). Our data is consistent with previous work showing that expressing either Rap1-CA or Rap1-DN in the fly epidermis impairs DC, though the severity and range of phenotypes observed was different [28]. Altogether, these results indicated that Rap1 modulates integrin adhesion in the AS and is required for DC.
Figure 3. Rap1 functions upstream of talin autoinhibition during morphogenesis.
(A) WT embryos and embryos expressing Rap1-CA in the AS were scored for openings in the dorsal epidermis at stage 13–17. (B) Images from time-lapse movies of control embryos (top) or embryos expressing Rap1-CA in the AS (bottom) undergoing DC at the indicated time-points. (C-F) β-integrin signal localized at the lateral membrane of AS cells was quantified (C) and colocalization of talin (magenta in E’,F’) and β-integrin (black in E,F; green in E’,F’) was measured at the membrane of AS cells using Pearson Correlation Co-efficients (D; n>25 cells, *p<0.05,***p<0.001). (G,H) The recruitment of talinGFP (D) and talinGFP*E1777A (E) to MTJs was measured in control embryos (orange in G; purple in H) and embryos expressing Rap1CA (pink; **p<0.01). (H-K) FRAP experiments were performed on talinGFP (I,J) and talinGFP*E1777A (K,L) to determine the effect of expressing either Rap1-CA (I,K) or Rap1-DN (J,L) on the mobility of talin at MTJs.
In addition to regulating integrin recruitment to the membrane in the AS, we also found that Rap1-CA increased the recruitment of talinGFP to MTJs (Fig. 3G). Therefore, we predicted that Rap1 might also regulate IAC turnover. FRAP analysis of talinGFP dynamics at MTJs revealed decreased turnover upon expression of Rap1-CA in the muscle (Fig. 3I). In comparison, expression of Rap1-DN elicited the opposite effect: turnover of talinGFP increased (Fig. 3J). To test whether Rap1 conferred its effect upstream of talin autoinhibition, either Rap1-DN or Rap1-CA were expressed in the presence of the talin autoinhibition mutant, talinGFP*E1777A. We found that Rap1-CA did not affect talinGFP*E1777A recruitment (Fig. 3H) and that neither Rap1-CA nor Rap1-DN modulated talinGFP*E1777A turnover (Fig. 3K,L). These results suggest that active Rap1 increases talin recruitment to and stabilization at cell-ECM adhesions, and that this effect occurs upstream of talin autoinibition.
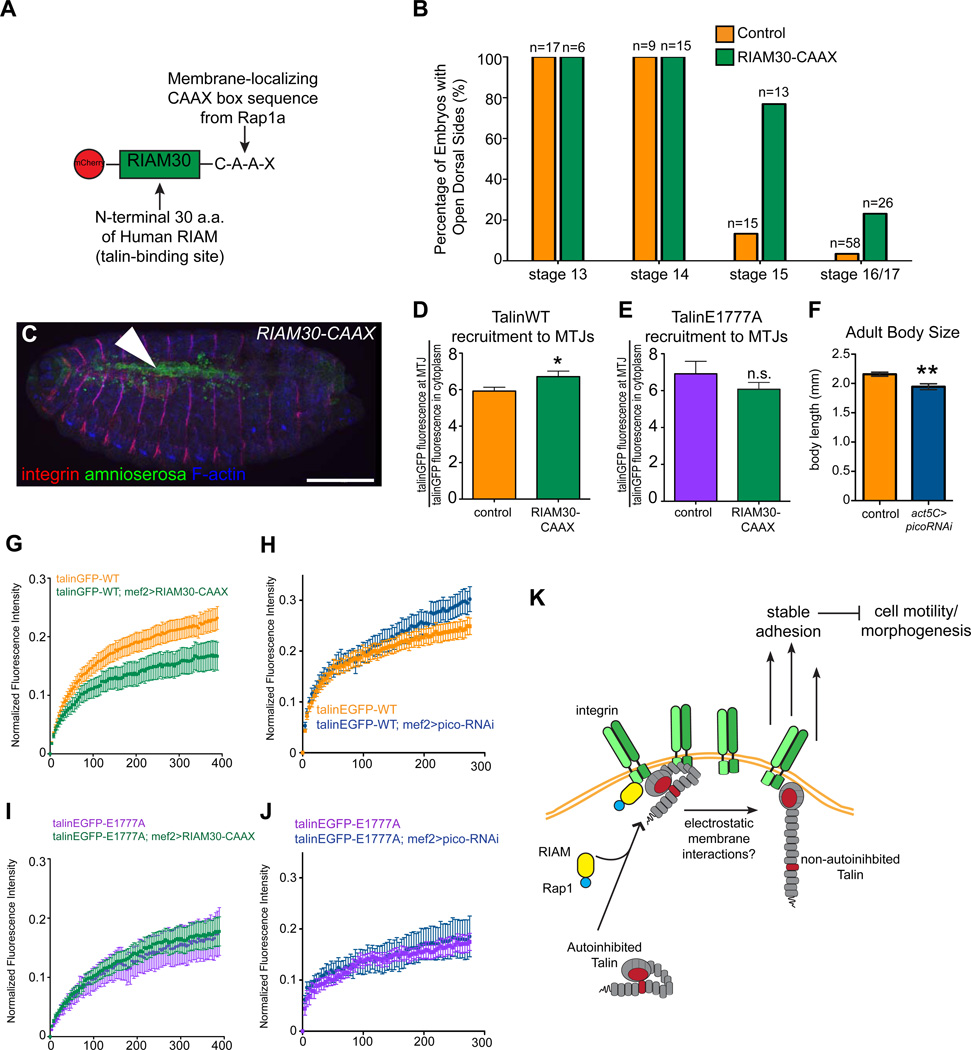
It has been shown that the MRL-family protein RIAM links membrane targeting sequences in Rap1 to talin, thereby recruiting talin to the plasma membrane which leads to activation of integrin and enhanced adhesion [24, 29]. In general, the functions assigned to RIAM, including recruiting talin to the membrane and promoting stable adhesions are similar to those obtained by the relief of autoinhibition [12, 25, 29, 30]. Comprehensive analyses of the embryonic role of the Drosophila RIAM homolog, pico, are precluded at this time because the original loss of function allele has been lost (D. Bennett, personal communication). To circumvent this problem and to test whether RIAM may also be involved in Rap1-dependent regulation of talin autoinhibition in the fly, we developed alternative approaches to modulate Pico/RIAM levels in the embryo. First, we used a minimal RIAM-Rap1 chimera (Fig. 4A; “RIAM30-CAAX”) comprised of the first 30 amino acids of human RIAM, which contains a talin binding site, and the membrane-targeting CAAX sequence of Rap1a, that was previously shown to be sufficient to activate integrins in CHO cells [24]. We found that expression of RIAM30-CAAX in the AS leads to delays in dorsal closure: approximately 80% of embryos exhibited open dorsal sides at the end of stage 15 (Fig. 4B-C). Furthermore, we found that RIAM30-CAAX induced increased recruitment of talinGFP to the membrane (Fig. 4D), and that the turnover dynamics of talinGFP decreased (Fig. 4G). The phenotypes conferred by increasing pico/RIAM via RIAM30-CAAX closely resembled those elicited by both the talinGFP*E1777A mutant and Rap1-CA suggesting that pico/RIAM could play a similar role in regulation of talin function. The ability of a human protein chimera to work as well as it does in flies illustrates the conservation of this system throughout evolution. Second, using an RNAi-induced knockdown of pico in the muscles, we found that the turnover of talinGFP increased (Fig. 4F,H), recapitulating the observed effect of expressing Rap1-DN. Importantly, neither the recruitment of talinGFP*E1777A to the membrane (Fig. 4E) nor the turnover dynamics of talinGFP*E1777A changed upon modulation of pico/RIAM (Fig. 4I-J), indicating that, like Rap1, Pico/RIAM modulates talin behavior via an autoinhibition-dependent mechanism. We propose that Rap1 and RIAM act upstream of talin to relieve autoinhibition; this promotes its recruitment to sites of adhesion where it forms a stabilizing link between integrins and the cytoskeleton (Fig. 4K). Our results also support the notion that a non-autoinhibited talin molecule can be recruited independent of Rap1/RIAM activity.
Figure 4. Pico/RIAM functions upstream of talin autoinhibition during morphogenesis.
(A) Schematic diagram of RIAM30-CAAX (B) WT embryos and embryos expressing RIAM30-CAAX in the AS were scored for openings in the dorsal epidermis at stage 13–17. (C) Stage 15 embryo with an open dorsal hole (arrowhead) stained for amnioserosa (green), integrin (red), and F-actin (blue). (D-E) The recruitment of talinGFP (D) and talinGFP*E1777A (E) to MTJs in control embryos and embryos expressing RIAM30-CAAX. (F) To provide evidence of pico knockdown based on the previously described pico phenotype [34], we measured adult body size of control embryos and embryos expressing picoRNAi under the control of a ubiquitous driver. (G-J) FRAP experiments were performed on talinGFP (G,H) and talinGFP*E1777A (I,J) to determine the effect of expressing either RIAM30-CAAX (G,I) or picoRNAi (H,J) on the mobility of talin at MTJs. (M) Model for the role and regulation of talin autoinhibition. RIAM-Rap1 acts to localize autoinhibited talin to integrin-mediated adhesions where autoinhibition can be relieved by electrostatic membrane interactions. This mechanism promotes stable adhesion, thus down-regulating cell motility required for morphogenesis.
Overall, this study identifies an important role for the regulation of talin function through autoinhibition. Failure to autoinhibit talin impairs morphogenetic processes but this is not due to defects in integrin-mediated attachment to the ECM or in the assembly of the adhesion complex. Thus it is unlikely that the E1777A mutation blocks integrin-mediated Cell-ECM attachment in a dominant negative fashion. An alternative explanation for the phenotype is that the E1777A mutant behaves like a gain of function allele of talin, and that the morphogenetic defects we observe are due to too much rather than too little adhesion. This would not be the first time such a phenomenon has been observed, for example overexpression of integrins in either the wing or the muscle gives rise to phenotypes identical to those found in integrin null mutants [17, 31]. How could the E1777A mutation give rise to stronger adhesion? We show that this mutation enhances the recruitment and co-localization of talin and integrin at sites of adhesion. Importantly, we show that the E1777A mutation effectively reduces talin turnover at sites of adhesion. Indeed, our data fits with a gain-of-function model: blocking talin autoinhibition leads to increased integrin-mediated adhesion, and this impairs morphogenetic processes that require cyclic adhesion assembly and disassembly. Further consistent with this model is the observation that adhesion at MTJs, a non-morphogenetic context, is not perturbed upon blocking autoinhibition of talin. We cannot exclude the possibility that E1777A may confer its effect on talin function through a means other than disruption of autoinhibition. Encouragingly, however, our homology modeling and NMR analyses strongly suggest that the fly protein behaves much as the mammalian homolog does.
How does preventing autoinhibition stabilize integrin-mediated adhesion? We show that autoinhibition regulates talin recruitment to adhesions through a RIAM-Rap1 dependent mechanism. Interestingly, the E1777A autoinhibition mutant talin is more strongly recruited to adhesions than WT talin; this enhanced recruitment occurs independent of RIAM-Rap1 activity. Thus, it is possible that constitutively relieving autoinhibition works to stabilize and promote adhesion by enhancing recruitment of the talin molecule to adhesions, thus bypassing the need of the RIAM-Rap1 pathway for recruitment. At the membrane, adhesion strengthening may occur via talin’s scaffolding function, as talin can interact with multiple components of the IAC, and these interactions may increase and/or change when talin assumes a more extended conformation. Another possibility, consistent with structural studies, is that relieving autoinhibition frees up the FERM/IBS-1 domain of talin such that it can activate integrins [4, 5]. We would predict that mutations in talin that block IBS-1-mediated integrin activation would lead to more dynamic adhesions, and this is indeed what was observed [9]. According to the model we envision, talin recruitment is determined by the sum of interactions that a single molecule can make with other IAC components at any one time. For example, the autoinhibited form of talin relies on Rap1/RIAM for efficient recruitment, even though it may still bind integrin through its free IBS-2 domain [9]; both mechanisms may contribute to targetting of talin to adhesions. We speculate that relieving autoinhibition makes the IBS-1 available, as well as the many other binding sites for IAC components that are found in the talin rod domain (e.g. vinculin binding sites), thereby substantially increasing the number of possible interactions that can lead to talin recruitment to the IAC.
There are likely multiple avenues leading to relief of talin autoinhibition. Recent super-resolution studies provided elegant evidence that autoinhibition is primarily relieved within adhesion complexes [27], implicating the need for a mechanism to specifically recruit autoinhibited talin to adhesions. Here we show that forcing talin to remain in an open, non-autoinhibited conformation gives rise to very similar phenotypes as activating the RIAM-Rap1 pathway. Based on the results obtained by us and other groups [6, 14, 25, 27, 29, 32, 33], we propose that RIAM-Rap1 brings autoinhibited talin to the membrane where autoinhibition can subsequently be relieved, possibly through electrostatic interactions with the membrane/PIP2. RIAM-Rap1 has a previously established role in mediating the recruitment of talin to sites of adhesion, but recently, it has been demonstrated that the requirement for RIAM-Rap1 is context-dependent. Structural and biochemical studies have revealed that the binding of talin to either RIAM or vinculin is mutually exclusive and likely dependent on force [32, 33]. Moreover, in cell culture, vinculin-stimulated integrin activation is RIAM-Rap1 independent raising the possibility that more mature adhesions might not need RIAM-Rap1 to promote talin activation in this case [32]. Along similar lines, we demonstrate that RIAM-Rap1 activity is dispensable for recruitment of a non-autoinhibited talin molecule.
In summary, our results suggest that talin autoinhibition confers a switch through which fine control of integrin-mediated adhesion can be exerted in vivo. Our findings also reveal RIAM-Rap1-mediated regulation of integrin adhesion to be an important modulator of morphogenesis and provide evidence for an autoinhibition-based pathway for control of talin function through RIAM-Rap1. Furthermore, this study exemplifies how subtle tuning of adhesion complex composition and stability elicits different adhesive functions and cellular behaviors during development.
Supplementary Material
Highlights.
Talin autoinhibition modulates cell-ECM adhesion stability during morphogenesis
Failure to regulate talin autoinhibition results in delayed morphogenesis
Defect arises as a result of cell-ECM adhesion that is too stable
Rap1/RIAM signalling acts upstream of talin autoinhibition
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bulgakova NA, Klapholz B, Brown NH. Cell adhesion in Drosophila: versatility of cadherin and integrin complexes during development. Current opinion in cell biology. 2012;24:702–712. doi: 10.1016/j.ceb.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Critchley DR. Biochemical and structural properties of the integrinassociated cytoskeletal protein talin. Annual review of biophysics. 2009;38:235–254. doi: 10.1146/annurev.biophys.050708.133744. [DOI] [PubMed] [Google Scholar]
- 3.Tepass U. FERM proteins in animal morphogenesis. Current opinion in genetics & development. 2009;19:357–367. doi: 10.1016/j.gde.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 4.Goksoy E, Ma YQ, Wang X, Kong X, Perera D, Plow EF, Qin J. Structural basis for the autoinhibition of talin in regulating integrin activation. Molecular cell. 2008;31:124–133. doi: 10.1016/j.molcel.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goult BT, Bate N, Anthis NJ, Wegener KL, Gingras AR, Patel B, Barsukov IL, Campbell ID, Roberts GC, Critchley DR. The structure of an interdomain complex that regulates talin activity. The Journal of biological chemistry. 2009;284:15097–15106. doi: 10.1074/jbc.M900078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song X, Yang J, Hirbawi J, Ye S, Perera HD, Goksoy E, Dwivedi P, Plow EF, Zhang R, Qin J. A novel membrane-dependent on/off switch mechanism of talin FERM domain at sites of cell adhesion. Cell research. 2012 doi: 10.1038/cr.2012.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Askari JA, Buckley PA, Mould AP, Humphries MJ. Linking integrin conformation to function. Journal of cell science. 2009;122:165–170. doi: 10.1242/jcs.018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geiger B, Yamada KM. Molecular architecture and function of matrix adhesions. Cold Spring Harbor perspectives in biology. 2011;3 doi: 10.1101/cshperspect.a005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis SJ, Pines M, Fairchild MJ, Tanentzapf G. In vivo functional analysis reveals specific roles for the integrin-binding sites of talin. Journal of cell science. 2011;124:1844–1856. doi: 10.1242/jcs.083337. [DOI] [PubMed] [Google Scholar]
- 10.Brown NH, Gregory SL, Rickoll WL, Fessler LI, Prout M, White RA, Fristrom JW. Talin is essential for integrin function in Drosophila. Developmental cell. 2002;3:569–579. doi: 10.1016/s1534-5807(02)00290-3. [DOI] [PubMed] [Google Scholar]
- 11.Kopp PM, Bate N, Hansen TM, Brindle NP, Praekelt U, Debrand E, Coleman S, Mazzeo D, Goult BT, Gingras AR, et al. Studies on the morphology and spreading of human endothelial cells define key inter- and intramolecular interactions for talin1. European journal of cell biology. 2010;89:661–673. doi: 10.1016/j.ejcb.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banno A, Goult BT, Lee H, Bate N, Critchley DR, Ginsberg MH. Subcellular localization of talin is regulated by inter-domain interactions. The Journal of biological chemistry. 2012;287:13799–13812. doi: 10.1074/jbc.M112.341214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wegener KL, Partridge AW, Han J, Pickford AR, Liddington RC, Ginsberg MH, Campbell ID. Structural basis of integrin activation by talin. Cell. 2007;128:171–182. doi: 10.1016/j.cell.2006.10.048. [DOI] [PubMed] [Google Scholar]
- 14.Saltel F, Mortier E, Hytonen VP, Jacquier MC, Zimmermann P, Vogel V, Liu W, Wehrle-Haller B. New PI(4,5)P2- and membrane proximal integrin-binding motifs in the talin head control beta3-integrin clustering. The Journal of cell biology. 2009;187:715–731. doi: 10.1083/jcb.200908134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frank LH, Rushlow C. A group of genes required for maintenance of the amnioserosa tissue in Drosophila. Development. 1996;122:1343–1352. doi: 10.1242/dev.122.5.1343. [DOI] [PubMed] [Google Scholar]
- 16.Solon J, Kaya-Copur A, Colombelli J, Brunner D. Pulsed forces timed by a ratchet-like mechanism drive directed tissue movement during dorsal closure. Cell. 2009;137:1331–1342. doi: 10.1016/j.cell.2009.03.050. [DOI] [PubMed] [Google Scholar]
- 17.Tanentzapf G, Martin-Bermudo MD, Hicks MS, Brown NH. Multiple factors contribute to integrin-talin interactions in vivo. Journal of cell science. 2006;119:1632–1644. doi: 10.1242/jcs.02859. [DOI] [PubMed] [Google Scholar]
- 18.Yuan L, Fairchild MJ, Perkins AD, Tanentzapf G. Analysis of integrin turnover in fly myotendinous junctions. Journal of cell science. 2010;123:939–946. doi: 10.1242/jcs.063040. [DOI] [PubMed] [Google Scholar]
- 19.Tanentzapf G, Brown NH. An interaction between integrin and the talin FERM domain mediates integrin activation but not linkage to the cytoskeleton. Nature cell biology. 2006;8:601–606. doi: 10.1038/ncb1411. [DOI] [PubMed] [Google Scholar]
- 20.Ren XD, Kiosses WB, Sieg DJ, Otey CA, Schlaepfer DD, Schwartz MA. Focal adhesion kinase suppresses Rho activity to promote focal adhesion turnover. Journal of cell science. 2000;113(Pt 20):3673–3678. doi: 10.1242/jcs.113.20.3673. [DOI] [PubMed] [Google Scholar]
- 21.Lawson C, Lim ST, Uryu S, Chen XL, Calderwood DA, Schlaepfer DD. FAK promotes recruitment of talin to nascent adhesions to control cell motility. The Journal of cell biology. 2012;196:223–232. doi: 10.1083/jcb.201108078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freeman SA, McLeod SJ, Dukowski J, Austin P, Lee CC, Millen-Martin B, Kubes P, McCafferty DM, Gold MR, Roskelley CD. Preventing the activation or cycling of the Rap1 GTPase alters adhesion and cytoskeletal dynamics and blocks metastatic melanoma cell extravasation into the lungs. Cancer research. 2010;70:4590–4601. doi: 10.1158/0008-5472.CAN-09-3414. [DOI] [PubMed] [Google Scholar]
- 23.Grabbe C, Zervas CG, Hunter T, Brown NH, Palmer RH. Focal adhesion kinase is not required for integrin function or viability in Drosophila. Development. 2004;131:5795–5805. doi: 10.1242/dev.01462. [DOI] [PubMed] [Google Scholar]
- 24.Lee HS, Lim CJ, Puzon-McLaughlin W, Shattil SJ, Ginsberg MH. RIAM activates integrins by linking talin to ras GTPase membrane-targeting sequences. The Journal of biological chemistry. 2009;284:5119–5127. doi: 10.1074/jbc.M807117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han J, Lim CJ, Watanabe N, Soriani A, Ratnikov B, Calderwood DA, Puzon-McLaughlin W, Lafuente EM, Boussiotis VA, Shattil SJ, et al. Reconstructing and deconstructing agonist-induced activation of integrin alphaIIbbeta3. Current biology : CB. 2006;16:1796–1806. doi: 10.1016/j.cub.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 26.Martel V, Racaud-Sultan C, Dupe S, Marie C, Paulhe F, Galmiche A, Block MR, Albiges-Rizo C. Conformation, localization, and integrin binding of talin depend on its interaction with phosphoinositides. The Journal of biological chemistry. 2001;276:21217–21227. doi: 10.1074/jbc.M102373200. [DOI] [PubMed] [Google Scholar]
- 27.Rossier O, Octeau V, Sibarita JB, Leduc C, Tessier B, Nair D, Gatterdam V, Destaing O, Albiges-Rizo C, Tampe R, et al. Integrins beta(1) and beta(3) exhibit distinct dynamic nanoscale organizations inside focal adhesions. Nature cell biology. 2012;14:1057–1067. doi: 10.1038/ncb2588. [DOI] [PubMed] [Google Scholar]
- 28.Boettner B, Harjes P, Ishimaru S, Heke M, Fan HQ, Qin Y, Van Aelst L, Gaul U. The AF-6 homolog canoe acts as a Rap1 effector during dorsal closure of the Drosophila embryo. Genetics. 2003;165:159–169. doi: 10.1093/genetics/165.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Worth DC, Hodivala-Dilke K, Robinson SD, King SJ, Morton PE, Gertler FB, Humphries MJ, Parsons M. Alpha v beta3 integrin spatially regulates VASP and RIAM to control adhesion dynamics and migration. The Journal of cell biology. 2010;189:369–383. doi: 10.1083/jcb.200912014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lafuente EM, van Puijenbroek AA, Krause M, Carman CV, Freeman GJ, Berezovskaya A, Constantine E, Springer TA, Gertler FB, Boussiotis VA. RIAM, an Ena/VASP and Profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Developmental cell. 2004;7:585–595. doi: 10.1016/j.devcel.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 31.Brabant MC, Fristrom D, Bunch TA, Brower DL. Distinct spatial and temporal functions for PS integrins during Drosophila wing morphogenesis. Development. 1996;122:3307–3317. doi: 10.1242/dev.122.10.3307. [DOI] [PubMed] [Google Scholar]
- 32.Lee HS, Anekal P, Lim CJ, Liu CC, Ginsberg MH. Two modes of integrin activation form a binary molecular switch in adhesion maturation. Molecular biology of the cell. 2013;24:1354–1362. doi: 10.1091/mbc.E12-09-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goult BT, Zacharchenko T, Bate N, Tsang R, Hey F, Gingras AR, Elliott PR, Roberts GC, Ballestrem C, Critchley DR, et al. RIAM and vinculin binding to talin are mutually exclusive and regulate adhesion assembly and turnover. The Journal of biological chemistry. 2013;288:8238–8249. doi: 10.1074/jbc.M112.438119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyulcheva E, Taylor E, Michael M, Vehlow A, Tan S, Fletcher A, Krause M, Bennett D. Drosophila pico and its mammalian ortholog lamellipodin activate serum response factor and promote cell proliferation. Developmental cell. 2008;15:680–690. doi: 10.1016/j.devcel.2008.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.