Summary
A host of cancer types exhibit aberrant histone modifications. Recently, distinct and recurrent mutations in a specific histone variant, histone H3.3, have been implicated in a high proportion of malignant pediatric brain cancers. The presence of mutant H3.3 histone disrupts epigenetic post-translational modifications near genes involved in cancer processes and in brain function. Here, we propose several possible mechanisms by which mutant H3.3 histones may act to promote tumorigenesis. Furthermore, we discuss how perturbations in normal H3.3 chromatin-related and epigenetic functions may more broadly contribute to the formation of human cancers.
Introduction
Histones are linked to the genesis of a multitude of cancers primarily through alterations in their post-translational modification (PTM) and the epigenetic machinery controlling these modifications. Recurrent mutations in histone modifying enzymes and chromatin remodelers are apparent in various cancer types (Dawson and Kouzarides, 2012), and a number of studies have together provided growing insight into the interplay between histone modifying enzymes, specific histone PTMs, and tumorigenesis (Suva et al., 2013).
An emerging line of investigation is focused on cancer-related mutations in histones themselves as recent studies estimate that 30–40% of sequenced glioblastoma multiforme (GBM) tumors contain some disruption in epigenetic regulatory machinery, with roughly 11% of all samples bearing specific and reoccurring histone mutations (Sturm et al., 2012). When stratified by patient age, these estimates become much more striking. Approximately 70–80% of pediatric gliomas are characterized by precisely the same histone mutations manifested in the variant member of the histone H3 family, H3.3 (Attieh et al., 2013; Bjerke et al., 2013; Chan et al., 2013; Fontebasso et al., 2013a; Fontebasso et al., 2013b; Gessi et al., 2013; Je et al., 2013a; Je et al., 2013b; Jones et al., 2013; Khuong-Quang et al., 2012; Lewis et al., 2013; Schwartzentruber et al., 2012; Sturm et al., 2012; Venneti et al., 2013; Wiestler et al., 2013; Wu et al., 2012; Zhang et al., 2013). Together, these studies strongly implicate three separate, specific amino acid substitution mutations in H3.3 in the pathogenesis of several forms of human pediatric gliomas, including GBM and diffuse intrinsic pontine glioma (DIPG).
Unique qualities of histone variant H3.3
The human histone H3 family consists of a number of related proteins: H3.1 and H3.2 (commonly referred to as “canonical” H3), histone variant H3.3, the centromere-specific variant CENP-A/CenH3, the testes-specific H3t (Szenker et al., 2011), and the testes-specific H3.5 (Schenk et al., 2011). In eukaryotes there are two genes, known as H3f3a and H3f3b in mice and H3F3A and H3F3B in humans, that produce identical H3.3 proteins though each contains different mRNA untranslated regions and regulatory sequences (Akhmanova et al., 1995; Albig et al., 1995; Wells et al., 1987; Witt et al., 1997). While H3.3 can function much the same as canonical H3 as a core part of the nucleosome, H3.3 is also deposited into transcriptionally active regions to replace displaced nucleosomes throughout the cell cycle (Ahmad and Henikoff, 2002; Ray-Gallet et al., 2011; Tagami et al., 2004), in contrast to its canonical counterparts H3.1 and H3.2, which are deposited in a replication-dependent manner. H3.3 is found in genomic regions exhibiting “active” or “poised” transcription – domains commonly enriched for lysine 4 trimethylation of H3 (H3K4me3) or possessing both lysine 27 trimethylation (H3K27me3) and H3K4me3 (Delbarre et al., 2010) – in addition to pericentromeric and telomeric regions (Szenker et al., 2011). H3.3 comprises approximately 25% of the total pool of H3 histones in Drosophila melanogaster (Sakai et al., 2009) and is found at comparable levels in Mus musculus (Bush et al., 2013).
Insight gained from H3.3 pathway disruption
To date, two major histone chaperone complexes have been identified as responsible for H3.3 incorporation: HIRA, which incorporates H3.3 into genic, euchromatic regions in a replication-independent manner (Goldberg et al., 2010; Tagami et al., 2004), and the death associated protein (DAXX)/α-thalassemia X-linked mental retardation protein (ATRX) complex, which incorporates H3.3 into pericentromeric and telomeric heterochromatin regions (Delbarre et al., 2013; Drane et al., 2010; Goldberg et al., 2010) and in response to neuronal signaling (Michod et al., 2012). Loss of HIRA, the major chaperone responsible for H3.3 deposition, causes defects in early embryogenesis (Roberts et al., 2002; Szenker et al., 2012); loss of ATRX results in aneuploidy and defects in chromosomal segregation (Baumann et al., 2010). Both ATRX and DAXX have been reported as factors mutated in neuroblastoma (Cheung et al., 2012) and in other cancer types (Heaphy et al., 2011; Jiao et al., 2011). Thus, the interplay between ATRX/DAXX and H3.3 histones may represent a critical, yet unexplored, axis leading to pediatric gliomas.
Loss-of-function studies for genes encoding H3.3 have also proven insightful. Individual homozygous disruption of His3.3A and His3.3B in Drosophila (orthologs of human H3F3A and H3F3B) has little phenotypic effect on the overall organism. In contrast, combined disruption of both genes results in reduced viability and sterility (Hodl and Basler, 2009; Sakai et al., 2009). H3.3 may be necessary to sustain transcription of genes involved in differentiation, as knockdown of H3.3 by morpholino in Xenopus laevis leads to defects in late gastrulation developmental programs (Szenker et al., 2012), and the introduction of a dominant-negative form of H3.3 in zebrafish disrupts neural crest development (Cox et al., 2012). Loss-of-function studies of H3.3 in mammals have only disrupted one of the two H3.3-encoding genes successfully in mice (Bush et al., 2013; Couldrey et al., 1999; Tang et al., 2013), though disruption of either H3f3a or H3f3b imparts developmental defects, neonatal lethality, and reduced fertility.
At the chromatin level, loss of H3.3 in Drosophila results in a compensatory gap-filling mechanism, whereby HIRA and XNP (the Drosophila ATRX homolog) are bound to previously H3.3-associated regions (Schneiderman et al., 2012). In the mouse, H3.3 acts as a placeholder for CENP-A during cell division (Dunleavy et al., 2011), and partial loss-of-function of H3.3 via knockout of H3f3b causes ectopic CENP-A foci formation (Bush et al., 2013), with CENP-A acting in a possible compensatory gap-filling mechanism for lost H3.3. Partial loss of H3.3 in Mus also results in defects in cell cycling as well as chromosomal and karyotypic abnormalities (Bush et al., 2013). Similarly, knockdown of H3.3 leads to embryonic developmental arrest, chromosome missegregation, and chromatin condensation (Lin et al., 2013). Collectively, these data suggest that H3.3 may be necessary for proper chromosome segregation. These studies have provided insight into the normal function of endogenous H3.3 in the genome and have raised additional possibilities of what goes awry with H3.3 mutations in cancer.
H3.3 as an epigenetic memory and plasticity factor
How might H3.3 influence epigenetic states? Studies in Drosophila found that only specific residues in place of lysine 4 (a K4 arginine replacement, but not K4 alanine) could restore defects in fertility upon H3.3 removal (Hodl and Basler, 2009; Sakai et al., 2009), suggesting that the incorporation of a specific PTM, and not just the histone itself, is necessary for proper germ cell function. In Xenopus, H3.3 overexpression augmented an embryo’s memory of the transcriptional state of donor nuclei genes following l nuclear transfer to an enucleated egg (Ng and Gurdon, 2008). H3.3 was heavily incorporated into regions exhibiting this epigenetic memory. When K4 of H3.3 was replaced with glutamic acid [K4E], incorporation of K4E effectively silenced regions that were previously transcriptionally active (Ng and Gurdon, 2008; Yang et al., 2011). The presence of H3K4me3 marks is strongly associated with active transcription, and H3.3 protein is enriched for PTMs associated with active transcription when compared to its canonical H3 counterparts (Hake et al., 2006; McKittrick et al., 2004), further supporting the notion that modification of the N-terminal tail of H3.3 (H3.3K4 methylation, for example) and the pattern of H3.3 incorporation uniquely affect the transcriptional state of a cell (Ng and Gurdon, 2008).
In Mus, exploration of mutations on the N-terminal tail of H3.3 revealed that incorporation of a lysine 27 to arginine [K27R] mutant form of H3.3 (and not canonical H3.1) in zygotes altered embryonic stage-specific development, caused nuclear segregation abnormalities, and decreased H3K27me3 levels by approximately 65% (Santenard et al., 2010). Though not the same H3.3 mutation found in pediatric gliomas (K27R instead of a lysine 27 to methionine [K27M] substitution), the data nonetheless demonstrate the adverse consequences from continuous incorporation of K27-mutated H3.3 histone. One such consequence is a failure to establish or delineate proper heterochromatic regions. Finally, recent studies in Xenopus have established that H3.3 and its histone regulator A (HIRA) chaperone are necessary to reprogram a nucleus from one transcriptional state to another (Jullien et al., 2012). Thus, H3.3 appears to play a major role in transcriptional plasticity.
Reprogramming somatic cells to produce induced pluripotent stem cells has some parallels to tumorigenesis, particularly in regards to the transcriptional programs activated (Riggs et al., 2013; Suva et al., 2013). The transcriptional plasticity function of H3.3 may be needed to activate genes during oncogenic transformation as it is needed for maintaining transcriptional memory (Ng and Gurdon, 2008) or for switching transcriptional states (Jullien et al., 2012). It remains to be seen what role H3.3 may play in early oncogenic processes.
H3.3 mutations and gliomagenesis
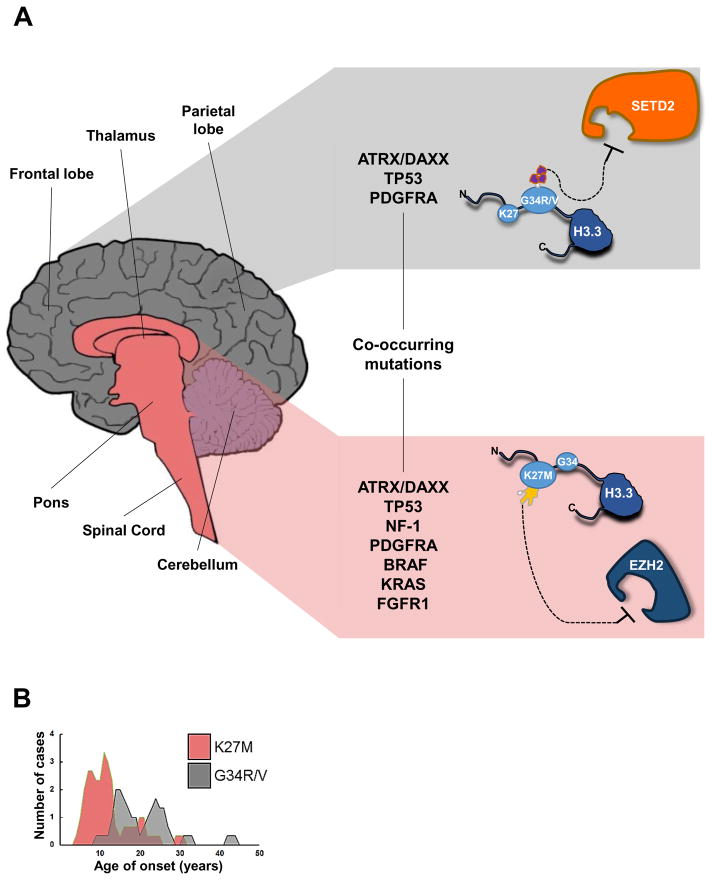
To date, all reported H3.3 mutations identified in human tumors have been in the H3F3A gene leading to single codon changes within the N-terminal tail of the H3.3 protein, a region enriched in PTMs. The first studies identified mutations encoding a K27M substitution, in addition to a smaller number of mutations encoding a glycine 34 to arginine or valine [G34R/V] substitution (Schwartzentruber et al., 2012; Wu et al., 2012). Relatively fewer gliomas exhibited K27M substitutions in HIST1H3B, one gene of several encoding canonical H3.1 histone; interestingly, H3.1 mutations appear to be restricted to DIPG and non-brainstem pediatric GBM in a younger range of patients (median = 4.75 years) (Wu et al., 2012). In tumor samples, H3.3K27M and H3.3G34R/V mutations are heterozygously expressed, with one allele of H3F3A being wildtype (Schwartzentruber et al., 2012). K27M and G34R/V mutations are mutually exclusive in tumors and show distinct gene expression profiles and DNA methylation patterns (Schwartzentruber et al., 2012; Sturm et al., 2012). Mutated tumors display different patterns of localization within the central nervous system that correlate to the normal patterns of brain expression in which the tumors are found. K27M tumors (both low- and high-grade) are primarily restricted to midline locations (spinal cord, thalamus, pons, brainstem) and G34R/V tumors to cerebral hemispheres (Fig. 1A) (Bjerke et al., 2013; Sturm et al., 2012). Additionally, both mutations correspond to different patient age ranges (Fig. 1B), with K27M mutations more prevalent in younger patients (range 5–29 years) and G34R/V mutations occurring in slightly older patients (range 9–42 years) (Schwartzentruber et al., 2012; Sturm et al., 2012). These data together suggest that K27M and G34R/V mutations may arise from independent cellular precursors and niches within the brain, and at different developmental time points.
Figure 1. Distribution and characteristics of H3.3-mutated gliomas.
G34R/V and K27M mutations of H3F3A display distinct and independent characteristics from one another. (A) Characteristics of H3.3-mutated tumors. G34R/V mutations (gray, top) in H3F3A localize primarily to cerebral/cortical hemispheres, specifically in frontal, parietal, occipital, and temporal lobes. K27M mutations (pink, bottom) in H3F3A localize primarily to midline locations, including the spinal cord, thalamus, pons, and brainstem. G34R/V mutations significantly overlap with mutations in TP53 and ATRX/DAXX, but are also found simultaneously at low rates with mutations in PDGFRA. K27M mutations overlap with mutations in TP53 and ATRX/DAXX, although not at significantly different rates over control groups. In addition, K27M mutations are found simultaneously at low rates with mutations in NF-1, PDGFRA, BRAF, KRAS, and FGFR1. Mutations in H3.3 directly (K27M) or indirectly (G34R/V) alter post-translationally modified residues. G34R/V mutations appear to affect K36me3 levels, possibly through inhibition of the methyltransferase SETD2, while K27M mutations attenuate EZH2 methyltransferase function, decreasing global K27me3 levels. (B). Age of onset of H3.3-mutated tumors. K27M mutations are more prevalent in younger patients (median age 11 years) while G34R/V mutations are more prevalent in older patients (median age 20 years). The age distribution and tumor characteristics together suggest that H3.3 mutations arise at different developmental timepoints as well as from independent niches and cellular precursors within the brain.
Intriguingly, H3.3 mutations are also found to simultaneously overlap with other specific mutations within the same tumor (Fig. 1A). Approximately 30% of K27M mutations are associated with mutations in ATRX/DAXX and 60% with mutations in TP53 (Schwartzentruber et al., 2012). K27M mutations have also been found at much lower frequency alongside mutations in NF-1, PDGFRA, BRAF, KRAS, and FGFR1 in gliomas (Jones et al., 2013; Khuong-Quang et al., 2012; Schwartzentruber et al., 2012; Zhang et al., 2013). Meanwhile, G34R/V mutations completely overlap with tumors containing mutations in TP53 and ATRX/DAXX, but have also been found together with mutations in PDGFRA (Schwartzentruber et al., 2012). Mutations in IDH1 and IDH2, which are commonly found in adult GBM, were largely absent from mutant H3.3 tumors (Khuong-Quang et al., 2012; Schwartzentruber et al., 2012). Tumors with mutations in H3F3A, TP53, and ATRX/DAXX contain higher numbers of copy number alterations (CNAs) (Schwartzentruber et al., 2012) though there was not a clear association between increases in CNAs and H3F3A mutations alone. Given that H3F3A mutations are found in heterozygous form and that these gliomas exhibit similar rates of TP53 mutation with other glioma types, H3.3K27M and H3.3G34R/V may act as driver mutations with TP53 mutations occurring as a second hit (Khuong-Quang et al., 2012; Schwartzentruber et al., 2012), although it is important to note that G34R/V mutations display much higher association with mutations in ATRX/DAXX and TP53.
How might N-terminal tail mutations on H3.3 promote gliomagenesis? H3.3 normally incorporates into regions that are actively transcribed and exhibit nucleosome displacement, and is the major histone variant synthesized outside of S-phase including in differentiated brain cells that could give rise to glioblastoma (Meshorer, 2007; Pina and Suau, 1987; Wu et al., 1982). Mutations in H3F3A (H3.3) are expected to manifest far more than mutations in H3.1, as HIST1H3B is only one gene of many encoding H3.1 and is only expressed at one point in the cell cycle. In addition to directly modifying residues at, or near, critical epigenetically regulated sites, mutation of the N-terminal tail could alter recognition of H3.3 by chromatin remodeling enzymes or chaperones, leading to aberrant mutant H3.3 incorporation that may disrupt normal ATRX/DAXX-mediated incorporation of H3.3 in pericentromeric regions and furthermore negatively impact chromosomal segregation and genome integrity as was observed when H3.3 levels were reduced experimentally (Bush et al., 2013; Lin et al., 2013). Alternatively, these mutations could affect H3.3 turnover kinetics, induce conformational changes that disrupt normal chromatin architecture, or even disrupt the PTM of residues near K27 or G34. Strong evidence from the earliest studies (Schwartzentruber et al., 2012; Wu et al., 2012) and supported by more recent work (discussed further below) suggest that H3F3A mutations act in a dominant negative manner and disrupt normal post-translational modification of residues on or near K27 or G34 of H3.3 that may drive oncogenic processes. The overlapping association between TP53/H3F3A mutations in pediatric gliomas or TP53/IDH1 mutations in adult gliomas (Schwartzentruber et al., 2012; Sturm et al., 2012) suggests that IDH1 and H3F3A mutations at least in part affect similar pathways. In this regard, IDH1 mutations may lead to the production of an onco-metabolite that inhibits H3 and H3.3 K27 and K36 histone demethylases, potentially affecting the same residues mutated in H3.3 mutant gliomas (Fontebasso et al., 2013b; Schwartzentruber et al., 2012). Methylation of H3K27 is a marker of heterochromatic states typically associated with transcriptional repression, mediated through the Polycomb repressive complex (PRC) (Ringrose et al., 2004). Though G34 cannot be directly post-translationally modified, it is just two amino acid residues away from K36, a critical residue that is a marker of euchromatic states when methylated (Wagner and Carpenter, 2012).
The Polycomb connection
Although introduction of the H3.3K27M mutation into p53-null, nestin-expressing progenitors in the neonatal mouse brainstem was unable to generate gliomas, the expression of H3.1/H3.3K27M did lead to precancerous changes in the form of ectopic, proliferative cell clusters in 72% of mice (Lewis et al., 2013). H3.3K27M expression in cultured DIPG coincided with reduced global levels of K27me3 and increased acetylation of K27 (K27Ac) (Chan et al., 2013; Lewis et al., 2013), a mark that is mutually exclusive to K27me3 and is associated with active transcription. K27M nucleosomes not only inhibit K27me3 on the same and nearby nucleosomes, but also result in modest increases in K27Ac on H3/H3.3 (Lewis et al., 2013). Similar results were found in mouse embryonic fibroblast cells, human astrocytes, and 293T cells in which H3.3K27M was introduced (Chan et al., 2013; Lewis et al., 2013). K27M peptide allosterically inhibited methyltransferase activity of the Polycomb repressive complex group 2 (PRC2) member, enhancer of zeste homolog 2 (EZH2), on wild-type nucleosomes in a dose dependent manner (Lewis et al., 2013), and H3.3K27M histone protein strongly immunoprecipitates with EZH2 (Chan et al., 2013), suggesting an interaction between K27M and EZH2 active site. However, K27M peptides do not seem to interact with the PRC2 group member EED, which specifically recognizes H3K27me3 residues (Lewis et al., 2013; Margueron et al., 2009), suggesting that K27M may mimic K27me1/me2 residues in vivo. Finally, in a cohort of brain tumors 6/20 GBM tumors displayed lowered or absent levels of H3K27me3 also bore H3F3A K27M mutations (Venneti et al., 2013). Therefore, the K27M mutation likely acts to alter global H3K27me3 levels through inhibition of PRC2 in regions where mutant histones are deposited (Fig. 1A), although other components of PRC2 (i.e. SUZ12, EZH1) may also be involved (Shen et al., 2008; Xu et al., 2010).
De-regulated expression of both PRC members target genes is implicated in a variety of cancer types (Bracken and Helin, 2009; Hock, 2012; Sparmann and van Lohuizen, 2006). The methyltransferase activity of EZH2 may be an important contributor to GBM formation, as EZH2 is expressed at higher levels in GBM tumors when compared to other tumor types. However, a significant difference in EZH2 levels was not evident between GBM tumors bearing or lacking the K27M mutation (Chan et al., 2013; Lewis et al., 2013; Venneti et al., 2013). Aberrant levels of histone methylation such as H3K27me3 have also been reported in other cancers (Dawson and Kouzarides, 2012; Hock, 2012; Kondo et al., 2008; Yoo and Hennighausen, 2012).
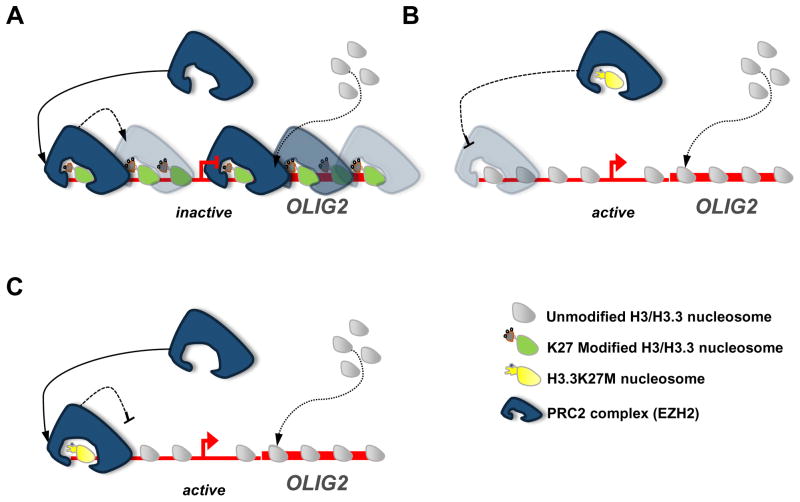
K27M mutations may affect transcriptional programs within the tumors. As EZH2 dysregulation may promote tumorigenesis depending on the cellular context (Hock, 2012), EZH2-regulated genes may become aberrantly activated or de-activated in the presence of mutant H3.3K27M histone, abrogating normal PRC-mediated repression (Fig. 2A). Data strongly suggest that K27M peptide and H3.3K27M histone allosterically bind and inhibit the active site of EZH2. Part of the cancer-related function of free H3.3K27M histones within the nucleus may be to sequester (Fig. 2B) or to trap (Fig. 2C) EZH2 at H3.3K27M chromatin-bound regions. Both scenarios would prevent PRC2 from binding and spreading K27me3 marks throughout genomic regions it normally targets, thereby causing de-repression of these elements (some of which may be oncogenic). In cultured H3.3K27M DIPG, chromatin immunoprecipitation sequencing (ChIP-seq) measured fewer H3K27me3 peaks compared to differentiation-matched neural stem cell (NSC) controls. Interestingly, the relatively lower total abundance of H3K27me3 peaks was not uniform across genic regions as some genes exhibited elevated H3K27me3 in DIPG. Genes in which H3K27me3 was depleted in H3.3K27M DIPG shared ontology with neurological processes (e.g. OLIG2, Fig. 2B, C), while genes in which H3K27me3 was enriched possessed ontology of cancer-related pathways (e.g. P16INK4A) (Chan et al., 2013). The presence of H3.3K27M inhibits EZH2 and H3K27me3 on mono and oligo-nucleosomes (Lewis et al., 2013), suggesting that H3.3K27M itself does not directly lead to hypermethylated regions. How PRC2 methylates these domains, such as tumor suppressor genes (e.g. P16INK4A), in K27M-mutated tumors is still unclear. OLIG2 expression in mutant gliomas was also found to be significantly higher in K27M samples (Sturm et al., 2012), and several studies have implicated OLIG2 in gliomagenesis, at least in part through p53 inactivation (Mehta et al., 2011; Sturm et al., 2012). In this way abnormal H3K27me3 levels could be altering specific transcriptional processes within a cell, contributing to oncogenesis.
Figure 2. The Polycomb connection.
Mutant H3.3K27M can interact with PRC2 to alter transcription in a number of possible scenarios using a representative gene that is dysregulated in K27M gliomas (OLIG2). (A). “Wild type.” In this context, H3.3K27M has not been incorporated into the chromatin of OLIG2 (no mutant protein present) and thus does not inhibit PRC2 function. PRC2 is able to trimethylate H3K27, spreading H3K27me3 marks throughout the promoter region and gene body, effectively repressing transcription. (B). “Sequestered.” H3.3K27M peptide mimics K27 methylation and allosterically inhibits EZH2 methyltransferase function outside of chromatin. Free H3.3K27M in the nucleoplasm may bind to EZH2 through interaction with the active site, sequestering it in the nucleoplasm. Such binding lessens the likelihood of EZH2 binding and silencing its genomic targets. (C). “Trapped.” In this scenario, H3.3K27M has been incorporated into the promoter/transcriptional start site region. However, binding of EZH2 to H3.3K27M blocks the active site and interferes with methyltransferase activity of EZH2 on the same and nearby nucleosomes. Thus, this region is not properly silenced and does not recruit additional silencing factors to spread H3K27me3.
Oncogenic mechanisms linked to H3.3G34R/V
The mechanisms by which H3.3G34R/V mutations contribute to gliomagenesis are relatively less clear than those proposed for H3.3K27M. While one study reported an increase in H3K36me3 levels in a GBM line harboring an H3.3G34V mutation (Schwartzentruber et al., 2012), H3K36me3 levels do not appear to be changed in 293T lines expressing H3.3G34R/V mutations (Lewis et al., 2013), nor in a H3.3G34V GBM line (Bjerke et al., 2013). However, introduction of G34R/V mutant histone results in substantial decreases in K36me3 on the same, and nearby, nucleosomes (Chan et al., 2013; Lewis et al., 2013). In humans, SETD2 is the only methyltransferase that catalyzes K36me3 of H3/H3.3 (Edmunds et al., 2008), indicating that the decreases in K36me3 are likely the result of G34R/V mutant histone blocking SETD2 function (Fig. 1A). H3K36me2/me3 and H3K27me3 rarely co-exist in chromatin, and H3K36 methylation antagonizes the methylation of H3K27 by PRC2 (Yuan et al., 2011). Likewise, H3.3G34R/V mutations do not appear to affect the levels of H3K27me3 and H3.3K27M mutations do not appear to affect the levels of H3K36me3 (Chan et al., 2013; Lewis et al., 2013). These data suggest that H3.3K27M and G34R/V-mediated gliomagenesis occur through independent mechanisms.
Methylation of H3/H3.3K36, particularly K36me3, plays some role in high-grade gliomas as mutations in SETD2 were found in 15% of exome-sequenced pediatric high-grade gliomas (Fontebasso et al., 2013b). Along with H3.3G34R/V mutated tumors, SETD2-mutant high-grade gliomas also localize to cerebral hemispheres, though SETD2-mutant tumors frequently contain IDH1 mutations, suggesting that H3.3G34R/V and SETD2/IDH1 mutations act to disrupt K36me3 in tumors (Fontebasso et al., 2013b). ChIP-seq analysis of H3K36me3 from a H3.3G34V pediatric GBM cell line identified 156 genes with differentially enriched H3K36me3 peaks when compared to a wildtype H3F3A pediatric GBM line, indicating that G34V may cause distinct changes in K36 methylation at specific regions if not globally (Bjerke et al., 2013). These genes were highly enriched for ontological processes involving brain development and cell proliferation. Intriguingly, one of the most highly enriched genes was MYCN, a known oncogene that can lead to context-dependent gliomagenesis in Mus (Swartling et al., 2012). Transduction of H3.3G34V into normal human astrocytes and transformed human fetal glial cells resulted in a 2- to 3-fold increase in MYCN transcript (Bjerke et al., 2013), further supporting the hypothesis that H3.3G34V induces MYCN upregulation in pediatric gliomas. The potential link between H3.3 and MYCN is notable given the association of H3.3 with actively transcribed genes and the role of MYCN in maintaining global euchromatin in neural stem cells (Knoepfler et al., 2006) and neuroblastoma (Cotterman et al., 2008).
Nearly all tumors bearing G34R/V mutations in H3F3A also exhibit mutations in ATRX/DAXX and display alternate lengthening of telomeres (ALT), a classical phenotype of cancerous cells (Khuong-Quang et al., 2012; Schwartzentruber et al., 2012; Sturm et al., 2012). ATRX/DAXX mediate deposition of H3.3 into pericentromeric and telomeric regions, and chromosome ends were found to have demethylated DNA in G34R/V mutant groups, further implicating a correlation between G34R/V mutations and ALT. Thus, G34R/V mutations may not only act to disrupt K36me3 levels and activate potential oncogenes, but mutations in H3.3 or ATRX/DAXX may potentially disrupt their proper interaction, leading to aberrant deposition of H3.3 near telomeric regions and resulting in ALT.
Future directions and clinical implications
A number of open questions remain, including why mutations specifically in H3F3A are so prominent in cases of pediatric glioma. In addition, where are H3.3K27M and H3.3G34R/V deposited in the genome? Equally unclear is what causes the buildup of H3K27me3 in certain genomic regions of tumors carrying H3.3K27M mutations, particularly at tumor suppressor genes. Also, do G34R/V mutations act to produce structural or conformational changes in the N-terminal tail of H3.3, thereby exerting effects on K36 and other residues that can be PTM? While H3.3G34V mutations appear to coincide with increased expression of MYCN, future studies will need to address the cellular context of H3K36me3 and H3.3G34V and how common the MYCN phenotype is in clinically-derived tumor samples.
There is a startling degree of overlap between mutations in H3F3A and ATRX/DAXX (Schwartzentruber et al., 2012). ATRX/DAXX are responsible for H3.3 deposition in pericentromeric/telomeric regions, and loss of ATRX/DAXX is associated with some degree of genetic instability (Baumann et al., 2010). As H3.3 mutant tumors contain CNAs and ALT (Schwartzentruber et al., 2012; Sturm et al., 2012), it is interesting to speculate whether altered H3.3 deposition through ATRX/DAXX or H3.3 mutations themselves lead to a loss in genomic integrity. Partial H3.3 loss-of-function in the mouse resulted in genomic instability (Bush et al., 2013), and H3.3 has been associated with machinery involved in double strand break repair and homologous recombination (Jones et al., 2011; Yang et al., 2013) and somatic hypermutation (Aida et al., 2013).
While some initial work has investigated targeted therapies in a few H3.3-mutated pediatric tumor types (Bjerke et al., 2013), a better understanding of these brain tumors will facilitate the development of novel targeted treatments. The prevalence of H3.3 mutations in a pediatric setting, and the fact that patients bearing K27M mutations have lower overall survival than patients bearing G34R/V mutations (Khuong-Quang et al., 2012; Sturm et al., 2012), highlights the importance of this. From a wider perspective, it is also possible that the H3.3 pathway is involved in a wider range of cancers through mutations in chaperones, histone modifying enzymes, or other molecules that ultimately feed into the H3.3 functional pathway. Future studies should address the role of both wildtype and mutant H3.3 in the genome and epigenome in order to offer better insight into how mutation of this intriguing histone variant causes cancer.
Acknowledgments
We thank Dr. Bonnie Barrilleaux, Kelly Bush, and Po-Yuan Tung for feedback on the manuscript. This work was supported by NIH Grant 1R01GM100782 and CIRM Grant RN2-00922-1 to PK, and in part by awards from the UC Davis HHMI: Integrating Medicine into Basic Science Program (Grant 56006769) and the CIRM Stem Cell Training Program (CIRM Grant TG2-01163) to BY.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- Aida M, Hamad N, Stanlie A, Begum NA, Honjo T. Accumulation of the FACT complex, as well as histone H3.3, serves as a target marker for somatic hypermutation. Proc Natl Acad Sci U S A. 2013;110:7784–7789. doi: 10.1073/pnas.1305859110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhmanova AS, Bindels PC, Xu J, Miedema K, Kremer H, Hennig W. Structure and expression of histone H3.3 genes in Drosophila melanogaster and Drosophila hydei. Genome. 1995;38:586–600. doi: 10.1139/g95-075. [DOI] [PubMed] [Google Scholar]
- Albig W, Bramlage B, Gruber K, Klobeck HG, Kunz J, Doenecke D. The human replacement histone H3.3B gene (H3F3B) Genomics. 1995;30:264–272. doi: 10.1006/geno.1995.9878. [DOI] [PubMed] [Google Scholar]
- Attieh Y, Geng QR, Dinardo CD, Zheng H, Jia Y, Fang ZH, Ganan-Gomez I, Yang H, Wei Y, Kantarjian H, et al. Low frequency of H3.3 mutations and upregulated DAXX expression in MDS. Blood. 2013;121:4009–4011. doi: 10.1182/blood-2012-11-466714. [DOI] [PubMed] [Google Scholar]
- Baumann C, Viveiros MM, De La Fuente R. Loss of maternal ATRX results in centromere instability and aneuploidy in the mammalian oocyte and pre-implantation embryo. PLoS Genet. 2010;6:e1001137. doi: 10.1371/journal.pgen.1001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerke L, Mackay A, Nandhabalan M, Burford A, Jury A, Popov S, Bax DA, Carvalho D, Taylor KR, Vinci M, et al. Histone H3.3 Mutations Drive Pediatric Glioblastoma through Upregulation of MYCN. Cancer Discov. 2013 doi: 10.1158/2159-8290.CD-12-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- Bush KM, Yuen BT, Barrilleaux BL, Riggs JW, O’Geen H, Cotterman RF, Knoepfler PS. Endogenous mammalian histone H3.3 exhibits chromatin-related functions during development. Epigenetics Chromatin. 2013;6:7. doi: 10.1186/1756-8935-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KM, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, Gupta N, Mueller S, James CD, Jenkins R, et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 2013;27:985–990. doi: 10.1101/gad.217778.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung NK, Zhang J, Lu C, Parker M, Bahrami A, Tickoo SK, Heguy A, Pappo AS, Federico S, Dalton J, et al. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA. 2012;307:1062–1071. doi: 10.1001/jama.2012.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotterman R, Jin VX, Krig SR, Lemen JM, Wey A, Farnham PJ, Knoepfler PS. N-Myc regulates a widespread euchromatic program in the human genome partially independent of its role as a classical transcription factor. Cancer Res. 2008;68:9654–9662. doi: 10.1158/0008-5472.CAN-08-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couldrey C, Carlton MB, Nolan PM, Colledge WH, Evans MJ. A retroviral gene trap insertion into the histone 3.3A gene causes partial neonatal lethality, stunted growth, neuromuscular deficits and male sub-fertility in transgenic mice. Hum Mol Genet. 1999;8:2489–2495. doi: 10.1093/hmg/8.13.2489. [DOI] [PubMed] [Google Scholar]
- Cox SG, Kim H, Garnett AT, Medeiros DM, An W, Crump JG. An essential role of variant histone H3.3 for ectomesenchyme potential of the cranial neural crest. PLoS Genet. 2012;8:e1002938. doi: 10.1371/journal.pgen.1002938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Delbarre E, Ivanauskiene K, Kuntziger T, Collas P. DAXX-dependent supply of soluble (H3.3-H4) dimers to PML bodies pending deposition into chromatin. Genome Res. 2013;23:440–451. doi: 10.1101/gr.142703.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbarre E, Jacobsen BM, Reiner AH, Sorensen AL, Kuntziger T, Collas P. Chromatin environment of histone variant H3.3 revealed by quantitative imaging and genome-scale chromatin and DNA immunoprecipitation. Mol Biol Cell. 2010;21:1872–1884. doi: 10.1091/mbc.E09-09-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Almouzni G, Karpen GH. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G(1) phase. Nucleus. 2011;2:146–157. doi: 10.4161/nucl.2.2.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontebasso AM, Liu XY, Sturm D, Jabado N. Chromatin remodeling defects in pediatric and young adult glioblastoma: a tale of a variant histone 3 tail. Brain Pathol. 2013a;23:210–216. doi: 10.1111/bpa.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontebasso AM, Schwartzentruber J, Khuong-Quang DA, Liu XY, Sturm D, Korshunov A, Jones DT, Witt H, Kool M, Albrecht S, et al. Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol. 2013b;125:659–669. doi: 10.1007/s00401-013-1095-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessi M, Gielen GH, Hammes J, Dorner E, Muhlen AZ, Waha A, Pietsch T. H3.3 G34R mutations in pediatric primitive neuroectodermal tumors of central nervous system (CNS-PNET) and pediatric glioblastomas: possible diagnostic and therapeutic implications? J Neurooncol. 2013;112:67–72. doi: 10.1007/s11060-012-1040-z. [DOI] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, Bazett-Jones DP, Allis CD, Hunt DF. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J Biol Chem. 2006;281:559–568. doi: 10.1074/jbc.M509266200. [DOI] [PubMed] [Google Scholar]
- Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C, Rodriguez FJ, Eberhart CG, Hebbar S, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock H. A complex Polycomb issue: the two faces of EZH2 in cancer. Genes Dev. 2012;26:751–755. doi: 10.1101/gad.191163.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodl M, Basler K. Transcription in the absence of histone H3.3. Curr Biol. 2009;19:1221–1226. doi: 10.1016/j.cub.2009.05.048. [DOI] [PubMed] [Google Scholar]
- Je EM, Yoo NJ, Kim YJ, Kim MS, Lee SH. Somatic mutation of H3F3A, a chromatin remodeling gene, is rare in acute leukemias and non-Hodgkin lymphoma. Eur J Haematol. 2013a;90:169–170. doi: 10.1111/ejh.12039. [DOI] [PubMed] [Google Scholar]
- Je EM, Yoo NJ, Lee SH. Mutational analysis of H3F3A, a chromatin remodeling gene in common solid tumors. APMIS. 2013b doi: 10.1111/apm.12124. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Hutter B, Jager N, Korshunov A, Kool M, Warnatz HJ, Zichner T, Lambert SR, Ryzhova M, Quang DA, et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45:927–932. doi: 10.1038/ng.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JM, Bhattacharyya A, Simkus C, Vallieres B, Veenstra TD, Zhou M. The RAG1 V(D)J recombinase/ubiquitin ligase promotes ubiquitylation of acetylated, phosphorylated histone 3.3. Immunol Lett. 2011;136:156–162. doi: 10.1016/j.imlet.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Jullien J, Astrand C, Szenker E, Garrett N, Almouzni G, Gurdon J. HIRA dependent H3.3 deposition is required for transcriptional reprogramming following nuclear transfer to Xenopus oocytes. Epigenetics Chromatin. 2012;5:17. doi: 10.1186/1756-8935-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khuong-Quang DA, Buczkowicz P, Rakopoulos P, Liu XY, Fontebasso AM, Bouffet E, Bartels U, Albrecht S, Schwartzentruber J, Letourneau L, et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 2012;124:439–447. doi: 10.1007/s00401-012-0998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, Yamochi T, Urano T, Furukawa K, Kwabi-Addo B, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CJ, Conti M, Ramalho-Santos M. Histone variant H3.3 maintains a decondensed chromatin state essential for mouse preimplantation development. Development. 2013;140:3624–3634. doi: 10.1242/dev.095513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKittrick E, Gafken PR, Ahmad K, Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc Natl Acad Sci U S A. 2004;101:1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S, Huillard E, Kesari S, Maire CL, Golebiowski D, Harrington EP, Alberta JA, Kane MF, Theisen M, Ligon KL, et al. The central nervous system-restricted transcription factor Olig2 opposes p53 responses to genotoxic damage in neural progenitors and malignant glioma. Cancer Cell. 2011;19:359–371. doi: 10.1016/j.ccr.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E. Chromatin in embryonic stem cell neuronal differentiation. Histol Histopathol. 2007;22:311–319. doi: 10.14670/HH-22.311. [DOI] [PubMed] [Google Scholar]
- Michod D, Bartesaghi S, Khelifi A, Bellodi C, Berliocchi L, Nicotera P, Salomoni P. Calcium-dependent dephosphorylation of the histone chaperone DAXX regulates H3.3 loading and transcription upon neuronal activation. Neuron. 2012;74:122–135. doi: 10.1016/j.neuron.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol. 2008;10:102–109. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- Pina B, Suau P. Changes in histones H2A and H3 variant composition in differentiating and mature rat brain cortical neurons. Dev Biol. 1987;123:51–58. doi: 10.1016/0012-1606(87)90426-x. [DOI] [PubMed] [Google Scholar]
- Ray-Gallet D, Woolfe A, Vassias I, Pellentz C, Lacoste N, Puri A, Schultz DC, Pchelintsev NA, Adams PD, Jansen LE, et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol Cell. 2011;44:928–941. doi: 10.1016/j.molcel.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Riggs JW, Barrilleaux BL, Varlakhanova N, Bush KM, Chan V, Knoepfler PS. Induced pluripotency and oncogenic transformation are related processes. Stem Cells Dev. 2013;22:37–50. doi: 10.1089/scd.2012.0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Ehret H, Paro R. Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol Cell. 2004;16:641–653. doi: 10.1016/j.molcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Roberts C, Sutherland HF, Farmer H, Kimber W, Halford S, Carey A, Brickman JM, Wynshaw-Boris A, Scambler PJ. Targeted mutagenesis of the Hira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryonic lethality. Mol Cell Biol. 2002;22:2318–2328. doi: 10.1128/MCB.22.7.2318-2328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A, Schwartz BE, Goldstein S, Ahmad K. Transcriptional and developmental functions of the H3.3 histone variant in Drosophila. Curr Biol. 2009;19:1816–1820. doi: 10.1016/j.cub.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santenard A, Ziegler-Birling C, Koch M, Tora L, Bannister AJ, Torres-Padilla ME. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat Cell Biol. 2010;12:853–862. doi: 10.1038/ncb2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk R, Jenke A, Zilbauer M, Wirth S, Postberg J. H3.5 is a novel hominid-specific histone H3 variant that is specifically expressed in the seminiferous tubules of human testes. Chromosoma. 2011;120:275–285. doi: 10.1007/s00412-011-0310-4. [DOI] [PubMed] [Google Scholar]
- Schneiderman JI, Orsi GA, Hughes KT, Loppin B, Ahmad K. Nucleosome-depleted chromatin gaps recruit assembly factors for the H3.3 histone variant. Proc Natl Acad Sci U S A. 2012;109:19721–19726. doi: 10.1073/pnas.1206629109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, Pfaff E, Tonjes M, Sill M, Bender S, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- Suva ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartling FJ, Savov V, Persson AI, Chen J, Hackett CS, Northcott PA, Grimmer MR, Lau J, Chesler L, Perry A, et al. Distinct neural stem cell populations give rise to disparate brain tumors in response to N-MYC. Cancer Cell. 2012;21:601–613. doi: 10.1016/j.ccr.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szenker E, Lacoste N, Almouzni G. A developmental requirement for HIRA-dependent H3.3 deposition revealed at gastrulation in Xenopus. Cell Rep. 2012;1:730–740. doi: 10.1016/j.celrep.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Szenker E, Ray-Gallet D, Almouzni G. The double face of the histone variant H3.3. Cell Res. 2011;21:421–434. doi: 10.1038/cr.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Tang MC, Jacobs SA, Wong LH, Mann JR. Conditional allelic replacement applied to genes encoding the histone variant H3.3 in the mouse. Genesis. 2013;51:142–146. doi: 10.1002/dvg.22366. [DOI] [PubMed] [Google Scholar]
- Venneti S, Garimella MT, Sullivan LM, Martinez D, Huse JT, Heguy A, Santi M, Thompson CB, Judkins AR. Evaluation of Histone 3 Lysine 27 Trimethylation (H3K27me3) and Enhancer of Zest 2 (EZH2) in Pediatric Glial and Glioneuronal Tumors Shows Decreased H3K27me3 in H3F3A K27M Mutant Glioblastomas. Brain Pathol. 2013 doi: 10.1111/bpa.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells D, Hoffman D, Kedes L. Unusual structure, evolutionary conservation of non-coding sequences and numerous pseudogenes characterize the human H3.3 histone multigene family. Nucleic Acids Res. 1987;15:2871–2889. doi: 10.1093/nar/15.7.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiestler B, Claus R, Hartlieb SA, Schliesser MG, Weiss EK, Hielscher T, Platten M, Dittmann LM, Meisner C, Felsberg J, et al. Malignant astrocytomas of elderly patients lack favorable molecular markers: an analysis of the NOA-08 study collective. Neuro Oncol. 2013;15:1017–1026. doi: 10.1093/neuonc/not043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt O, Albig W, Doenecke D. Transcriptional regulation of the human replacement histone gene H3.3B. FEBS Lett. 1997;408:255–260. doi: 10.1016/s0014-5793(97)00436-5. [DOI] [PubMed] [Google Scholar]
- Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RS, Tsai S, Bonner WM. Patterns of histone variant synthesis can distinguish G0 from G1 cells. Cell. 1982;31:367–374. doi: 10.1016/0092-8674(82)90130-1. [DOI] [PubMed] [Google Scholar]
- Xu C, Bian C, Yang W, Galka M, Ouyang H, Chen C, Qiu W, Liu H, Jones AE, MacKenzie F, et al. Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2) Proc Natl Acad Sci U S A. 2010;107:19266–19271. doi: 10.1073/pnas.1008937107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, Song Y, Seol JH, Park JY, Yang YJ, Han JW, Youn HD, Cho EJ. Myogenic transcriptional activation of MyoD mediated by replication-independent histone deposition. Proc Natl Acad Sci U S A. 2011;108:85–90. doi: 10.1073/pnas.1009830108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Li L, Liang J, Shi L, Yang J, Yi X, Zhang D, Han X, Yu N, Shang Y. Histone Acetyltransferase 1 Promotes Homologous Recombination in DNA Repair by Facilitating Histone Turnover. J Biol Chem. 2013;288:18271–18282. doi: 10.1074/jbc.M113.473199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo KH, Hennighausen L. EZH2 methyltransferase and H3K27 methylation in breast cancer. Int J Biol Sci. 2012;8:59–65. doi: 10.7150/ijbs.8.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem. 2011;286:7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wu G, Miller CP, Tatevossian RG, Dalton JD, Tang B, Orisme W, Punchihewa C, Parker M, Qaddoumi I, et al. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013;45:602–612. doi: 10.1038/ng.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]