Abstract
The bean pathogen Pseudomonas syringae pv. syringae B728a expresses homologs of the type III effectors AvrPto and AvrPtoB, either of which can trigger resistance in tomato cultivars expressing Pto and Prf genes. We found that strain B728a also elicits nonhost resistance in tomato cultivars VFNT Cherry and Moneymaker that lack Pto but express other members of the Pto family (e.g., SlFen and SlPtoC). Here, we show that the AvrPtoB homolog from B728a, termed AvrPtoBB728a (also known as HopAB1), is recognized by ‘VFNT Cherry’ and ‘Moneymaker’ when the effector is expressed in P. syringae pv. syringae 61, a strain lacking the avrPto or avrPtoB homolog. Using a gene-silencing approach, this recognition was shown to involve one or more Pto family members and Prf. AvrPtoBB728a interacted with SlFen, SlPtoC, and SlPtoD, in addition to Pto, in a yeast two-hybrid assay. In P. syringae pv. tomato DC3000, the C-terminal domain of AvrPtoB is an E3 ubiquitin ligase that ubiquitinates Fen, causing its degradation and leading to disease susceptibility. Although the C-terminal domain of AvrPtoBB728a shares 69% amino acid identity with that of AvrPtoB, we found that it has greatly reduced E3 ligase activity and is unable to ubiquitinate Fen in an in vitro ubiquitination assay. Thus, the nonhost resistance of ‘VFNT Cherry’ and ‘Moneymaker’ to B728a appears to be due to recognition of AvrPtoBB728 as a result of the effector’s reduced E3 ligase activity, which prevents it from facilitating degradation of a Pto family member. We speculate that the primary plant host of B728a lacks a Fen-like protein and that, therefore, the E3 ligase of AvrPtoBB728 was unnecessary for pathogenicity and has diverged and become ineffective.
Continuously encountering attempted pathogen attack, plants have evolved different weapons, including preexisting and induced biochemical or structural defense mechanisms, to block or inhibit pathogen infection (Agrios 2005). Pathogens that manage to surmount physical barriers and enter plant tissues may be recognized extracellularly when conserved microbial molecules, termed pathogen-associated molecular patterns (PAMPs) (Medzhitov and Janeway 1997), are perceived by transmembrane pattern recognition receptors. This perception results in activation of mitogen-activated protein kinase signaling pathways, induction of the expression of pathogenesis-related genes, production of reactive oxygen species and nitric oxide, and callose deposition to reinforce cell wall structure. These responses, termed PAMP-triggered immunity (PTI), act to prevent further bacterial growth and disease development (Iriti and Faoro 2007; Nicaise et al. 2009; Zipfel 2009).
The efficacy and significance of PTI appear to have been a driving force for successful pathogens to evolve virulence factors to overcome PTI, leading to effector-triggered susceptibility (ETS) (Jones and Dangl 2006). In Pseudomonas syringae pv. tomato DC3000, type III effectors suppress innate immunity by using specific enzyme activities (Abramovitch et al. 2006; Bretz et al. 2003; Espinosa et al. 2003; Fu et al. 2007; Lopez-Solanilla et al. 2004; Wang et al. 2010) or by blocking early defense gene transcription and signaling pathways (Boudsocq et al. 2010; He et al. 2006; Wang et al. 2010). In addition to their roles as suppressors of immunity-associated programmed cell death, some virulence factors may enhance plant susceptibility via modulating hormone signaling pathways (Chen et al. 2007; Cohn and Martin 2005; de Torres-Zabala et al. 2007; Zhao et al. 2003) or by suppressing microRNA pathways involved in plant immunity (Navarro et al. 2008). This phytopathogen can also facilitate apoplast invasion by inducing the reopening of stomata via the action of the phytotoxin coronatine (Melotto et al. 2006; Zhang et al. 2008).
To counter the effects of ETS caused by pathogens, some plants have evolved resistance (R) proteins to recognize type III effectors, by which a robust defense system, termed effector-triggered immunity (ETI), is initiated (Jones and Dangl 2006). ETI is also known as gene-for-gene resistance or cultivar-specific resistance, in which plant R proteins interact with one or more host proteins, which act as a guardee or a decoy to detect the presence of an effector or a perturbation in the host cell caused by an effector (van der Biezen and Jones 1998; van der Hoorn and Kamoun 2008).
One of the best studied gene-for-gene interactions, tomato resistance against P. syringae pv. tomato, is determined by the interaction between the Pto kinase in tomato and AvrPto or AvrPtoB from P. syringae pv. tomato (Kim et al. 2002; Lin et al. 2006; Martin et al. 1993; Ronald et al. 1992; Salmeron and Staskawicz 1993). P. syringae, a γ-proteobacterium, is notorious for its ability to cause destructive diseases on a wide variety of plants. The impact of P. syringae in agriculture and its wide host range have made it one of the most important model pathosystems for understanding the molecular mechanisms underlying disease development and plant immunity (Cunnac et al. 2009; Lindeberg et al. 2008, 2009; Mansfield 2009; Staskawicz 2009; van der Biezen and Jones 1998). Like many other phytopathogenic bacteria, for successful infection of host tissues, P. syringae relies on a type III secretion system (T3SS) which translocates certain proteins, termed type III effectors, into the host cell (Cunnac et al. 2009; Greenberg and Vinatzer 2003; Hueck 1998; Tang et al. 2006).
The availability of genome sequences of several representative strains of P. syringae, as well as the development of new experimental methods, is accelerating the progress in the search for functions of type III effectors (Almeida et al. 2009; Feil et al. 2005; Joardar et al. 2005; Lindeberg et al. 2008; Studholme et al. 2009; Vinatzer and Greenberg 2007; Zwiesler-Vollick et al. 2002). It is now known that certain P. syringae effector proteins act to enhance pathogen growth and promote disease development by co-opting host processes to suppress the plant immune response (Dodds and Rathjen 2010).
Upon translocation into the host cell by the T3SS, AvrPto and AvrPtoB physically interact with Pto and trigger Prf-mediated resistance in tomato. Mutational studies revealed that interaction between Pto and AvrPto/AvrPtoB is required for this tomato resistance because disruption of the specific interaction between Pto and AvrPto or AvrPtoB abolishes their ability to elicit Pto-dependent tomato resistance against P. syringae pv. tomato expressing AvrPto or AvrPtoB (Kim et al. 2002; Scofield et al. 1996; Tang et al. 1996). In addition to their ability to elicit Pto-dependent resistance in tomato, AvrPto and AvrPtoB also have virulence activity that enhances plant susceptibility to P. syringae pv. tomato (Gimenez-Ibanez et al. 2009; He et al. 2006; Lin et al. 2006; Rosebrock et al. 2007; Xiang et al. 2008; Xiao et al. 2007; Yeam et al. 2010; Zong et al. 2008).
Pto was originally identified in a wild relative of tomato, Solanum pimpinellifolium, and is a member of a multigene family that is clustered within a 60-kb region containing SpimPtoA, Prf, Fen, SpimPtoC, SpimPtoD, Pto, and the pseudogene SpimPtoF (Chang et al. 2002; Pedley and Martin 2003; Riely and Martin 2001). The Fen protein not only confers sensitivity to an organophosphorous insecticide, fenthion, but also accounts for the tomato resistance suppressed by AvrPtoB C-terminus (Rsb) phenotype. It is now known that the Fen protein is ubiquitinated by the E3 ligase domain present at the C terminus of AvrPtoB, leading to degradation of Fen and suppression of ETI triggered by the interactions between the N-terminal domain of AvrPtoB and Fen (Abramovitch et al. 2003, 2006; Rosebrock et al. 2007). Moreover, avoidance of Fen degradation via Pto phosphorylation of AvrPtoB within the E3 ligase domain has been reported to be responsible for the reinitiation of specific tomato resistance (Ntoukakis et al. 2009). These data suggest that an “arms race” between tomato expressing Fen and P. syringae resulted in the acquisition of the E3 ligase domain at the C terminus of AvrPtoB to overcome the Fen-mediated resistance and, subsequently, in the duplication of a Fen-like sequence with the ability to counteract the E3 ligase activity during the co-evolutionary process (Rosebrock et al. 2007).
Both Pto- and Fen-mediated resistances require Prf, a nucleotide-binding leucine-rich-repeat protein (Mucyn et al. 2006; Salmeron et al. 1996). Heterocomplexes containing Prf and Pto or other Pto-like kinases were identified using stable or transient expression of these genes in Nicotiana benthamiana (Gutierrez et al. 2010), suggesting that, in tomato and N. benthamiana, Prf may form a regulatory switch with Pto-type kinases in the presence of AvrPto or AvrPtoB, controlling a specific immune signaling in an incompatible interaction (Gutierrez et al. 2010; Mucyn et al. 2006; Ntoukakis et al. 2009; Salmeron et al. 1994).
The fact that Pto- and Prf-dependent recognition of AvrPto and AvrPtoB homologs can restrict the ability of diverse P. syringae pathovars to infect tomato suggests that these “nonhost resistance” phenotypes are actually due to gene-for-gene recognition events (Lin and Martin 2007). It also implies that Pto- and Prf-mediated resistance might be a common defense mechanism in protecting tomato plants from attack by a wide array of bacterial pathogens (Lin and Martin 2007). Although functional Pto and Prf genes are widespread among wild tomato and other Solanaceous species, the Pto allele has not been found in any accessions of cultivated tomato (S. lycopersicum) (Chang et al. 2002; Martin et al. 1993; Riely and Martin 2001; Rose et al. 2005, 2007).
Here, we report that tomato cultivars VFNT Cherry and Moneymaker exhibit nonhost resistance to the bean pathogen P. syringae pv. syringae B728a but not to P. syringae pv. syringae 61. To investigate the basis of this resistance in tomato, putative effector genes of B728a (Feil et al. 2005; Greenberg and Vinatzer 2003; Guttman et al. 2002; Vinatzer et al. 2006) were individually expressed in strain 61 and screened for their ability to elicit defense on ‘VFNT Cherry’ and Moneymaker. Surprisingly, only avrPtoB from B728a was found to convert strain 61 to an avirulent strain on ‘VFNT Cherry’ and ‘Moneymaker’ tomato, neither of which expresses Pto. This resistance required Prf, and a Pto family member in tomato. Further investigation revealed that the C-terminal E3 ligase domain of AvrPtoBB728a has very weak E3 ligase activity and is unable to ubiquitinate Fen in an in vitro assay. This diminished E3 ligase activity likely hinders the ability of AvrPtoBB728a to cause degradation of a Pto family member in ‘VFNT Cherry’ and Moneymaker, thereby explaining the nonhost resistance to B728a in these tomato lines.
RESULTS
Tomato cultivars that do not express Pto are resistant to P. syringae pv. syringae B728a but not to P. syringae pv. syringae 61
We discovered that ‘VFNT Cherry’ and ‘Moneymaker’ tomato, which do not have Pto, are resistant to the bean pathogen P. syringae pv. syringae B728a but not to strain 61 (Supplementary Fig. S1). To investigate the basis of this resistance in tomato, we examined putative effector genes of B728a that had been identified by bioinformatic analysis or functional screens (Feil et al. 2005; Greenberg and Vinatzer 2003; Guttman et al. 2002; Lin and Martin 2007; Vinatzer et al. 2006). This analysis revealed that B728a expresses homologs of avrPto and avrPtoB whereas strain 61 does not have either of these effector genes.
Ectopic expression of avrPtoBB728a but not avrPtoB728a allows strain 61 to be recognized by ‘VFNT Cherry’ and ‘Moneymaker’ tomato
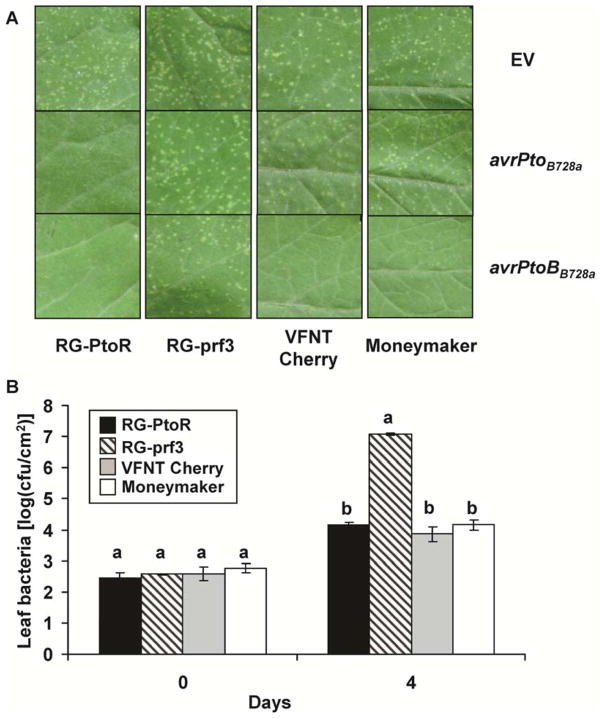
AvrPto and AvrPtoB are the only two type III effectors known to elicit resistance to P. syringae pv. tomato in tomato cultivars, such as ‘Rio-Grande PtoR’ (RG-PtoR), which express Pto and Prf (Kim et al. 2002; Ronald et al. 1992). Homologs of these two effector proteins, which occur in many other P. syringae pathovars, also elicit resistance in Pto- and Prf-expressing tomato cultivars (Lin and Martin 2007; Lin et al. 2006; Nguyen et al. 2010). To examine whether these effectors elicit resistance in VFNT Cherry, a broad-host-range vector expressing each effector was introduced into strain 61, and the transformants were used in an inoculation assay. As expected, the avrPto and avrPtoB (hopAB) homologs from strain B728a (called avrPtoB728a and avrPtoBB728a hereafter) allowed strain 61 to be recognized in leaves of RG-PtoR expressing the Pto and Fen genes, and this resistance was Prf dependent (Fig. 1A). However, unexpectedly, strain 61 expressing avrPtoBB728a was also recognized by ‘VFNT Cherry’ and ‘Moneymaker’ (Fig. 1A). The bacterial numbers in leaves of RG-PtoR, VFNT Cherry, and ‘Moneymaker’ 4 days after infiltration with designated Psy61(avrPtoBB728a) were approximately 104 CFU/cm2 but reached 107 to 108 CFU/cm2 in the susceptible RG-prf3 line (Fig. 1B). Thus, although both avrPtoB728a and avrPtoBB728a elicit Pto- and Prf-dependent resistance in RG-PtoR, only avrPtoBB728a elicits resistance in ‘VFNT Cherry’ and ‘Moneymaker’.
Fig. 1.
AvrPtoBB728a elicits resistance on tomato cultivars that do not express the Pto gene. A, Disease symptoms on leaves of tomato ‘Rio-Grande’ (RG)-PtoR (Pto/Pto and Prf/Prf), RG-prf3 (Pto/Pto and prf3/prf3), VFNT Cherry, and Moneymaker (both have Prf but not Pto) after vacuum infiltration with strain 61 carrying pCPP45 (empty vector [EV]), pCPP45::avrPtoB728, or pCPP45::avrPtoBB728a at 2 × 104 CFU/ml. All photographs were taken 7 days after inoculation. B, Bacterial populations were measured at day 0 and 4 after inoculation with strain 61 expressing avrPtoBB728a. Each value represents the mean of the log of CFU/cm2 obtained from three plants, and error bars represent the standard deviation. Data analysis was performed using Duncan’s multiple range test. Means with the same letter above the bars indicate no difference at a significance level of 5%. Each experiment was performed at least three times with similar results.
Deletion of avrPtoB but not avrPto from strain B728a abolishes its ability to elicit resistance on VFNT Cherry
To confirm that avrPtoBB728a is the only effector in P. syringae pv. syringae B728a that elicits resistance in VFNT Cherry, marker-exchange mutagenesis was performed to replace avrPtoB728a and avrPtoBB728a with the neo gene and ΩSp/Sm, respectively (Supplementary Fig. S2). The single mutants were designated PsyB728aΔavrPto and PsyB728aΔavrPtoB while the double mutant is referred to as PsyB728aΔavrPtoΔavrPtoB. The deletions of avrPto or avrPtoB in B728a in these mutants were confirmed by DNA gel blotting. The growth rates of PsyB728aΔavrPto and PsyB728aΔavrPtoB in liquid King’s B (KB) medium containing rifampicin were the same as the wild type (data not shown).
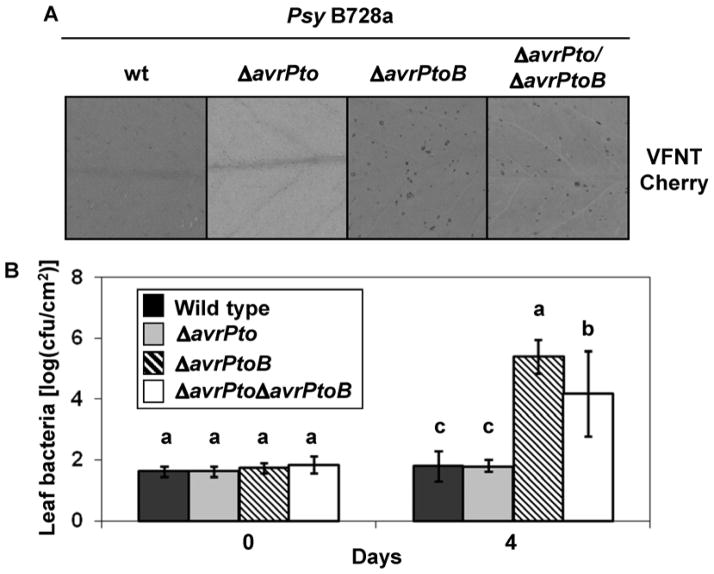
Next, the mutants PsyB728aΔavrPto, PsyB728aΔavrPtoB, or PsyB728aΔavrPtoΔavrPtoB at 2 × 104 CFU/ml were individually inoculated into leaves of ‘VFNT Cherry’ by vacuum infiltration. Four days after inoculation, no symptoms were observed on leaves of ‘VFNT Cherry’ infiltrated with PsyB728a ΔavrPto or wild-type B728a. In contrast, PsyB728aΔavrPto ΔavrPtoB and PsyB728aΔavrPtoB caused bacterial speck-like symptoms on leaves of ‘VFNT Cherry’ (Fig. 2A) (the same result was observed with Moneymaker; data not shown). Bacterial populations in the tomato leaves were measured at 4 days postinoculation to confirm the loss of recognition of PsyB728aΔavrPtoB and PsyB728aΔavrPtoΔavrPtoB. The bacterial population in the leaves of ‘VFNT Cherry’ infiltrated with the wild type or PsyB728aΔavrPto were close to 100 CFU/cm2 but increased to 105 to 106 CFU/cm2 in tomato leaves infiltrated with PsyB728aΔavrPtoB or PsyB728aΔavrPtoΔavrPtoB (Fig. 2B). We also observed that AvrPtoB728a contributes to bacterial virulence in tomato because PsyB728aΔavrPto ΔavrPtoB grew less than PsyB728aΔavrPtoB (Fig. 2B). The reduced bacterial populations of PsyB728aΔavrPtoΔavrPtoB in susceptible tomato leaves could be restored when a broad-host-range vector carrying avrPtoB728a under its native promoter was transformed into this double mutant (Supplementary Fig. S3). Moreover, disruption of the avirulence activity of B728a due to deletion in avrPtoBB728a was complemented when avrPtoBB728a was expressed from a broad-host-range vector under its native promoter and this strain was inoculated on tomato. These results indicate that avrPtoBB728a is responsible for eliciting resistance to B728a in ‘VFNT Cherry’ as well as Moneymaker.
Fig. 2.
Deletion of avrPtoB but not avrPto from Pseudomonas syringae pv. syringae strain B728a abolishes its ability to elicit resistance on VFNT Cherry. A, Elicitation of Pto-independent resistance by infiltration of tomato leaves of VFNT Cherry with B728a and its derivative mutants at 2 × 104 CFU/ml. Photographs were taken 7 days after inoculation, and specks were observed 3 days after vacuum infiltration with PsyB728aΔavrPtoB or PsyB728aΔavrPtoΔavrPtoB. B, Bacterial populations in tomato leaves infiltrated with B728a and its derivative mutants. Error bars indicate standard deviation for three replicates. Data analysis was performed using Duncan’s multiple range test. Means with the same letter above the bars indicate no difference at a significance level of 5%. Each experiment was performed three times with similar results.
It has been reported that strain B728a can also elicit Pto- and Prf-dependent resistance (Lin and Martin 2007). Hence, this tomato resistance was also evaluated, and only PsyB728a ΔavrPtoΔavrPtoB caused disease symptoms and showed increased bacterial numbers in leaves of RG-PtoR when bacteria were infiltrated at a level of 2 × 104 CFU/ml, implying that avrPtoB728a and avrPtoBB728a function redundantly in B728a to elicit Pto-dependent resistance (Supplementary Fig. S4).
Resistance to B728a requires Prf in ‘VFNT Cherry’ and Moneymaker
The Pto and Fen kinases, originally identified in S. pimpinellifolium, are two well-known R proteins required for AvrPto/AvrPtoB-triggered immunity (Kim et al. 2002; Ronald et al. 1992) and the Rsb phenotype (Abramovitch et al. 2003; Rosebrock et al. 2007), respectively. Resistance conferred by each of these proteins requires Prf. Although there is no Pto allele in ‘VFNT Cherry’ or Moneymaker, these cultivars do have other members of the Pto family, referred to as SlPtoA, SlFen (SlPtoB), SlPtoC, SlPtoD, and SlPtoF, as well as Prf (Chang et al. 2002). Therefore, we used virus-induced gene silencing (VIGS) to investigate the possibility that the resistance of ‘VFNT Cherry’ to B728a depends on a member of the Pto family or Prf.
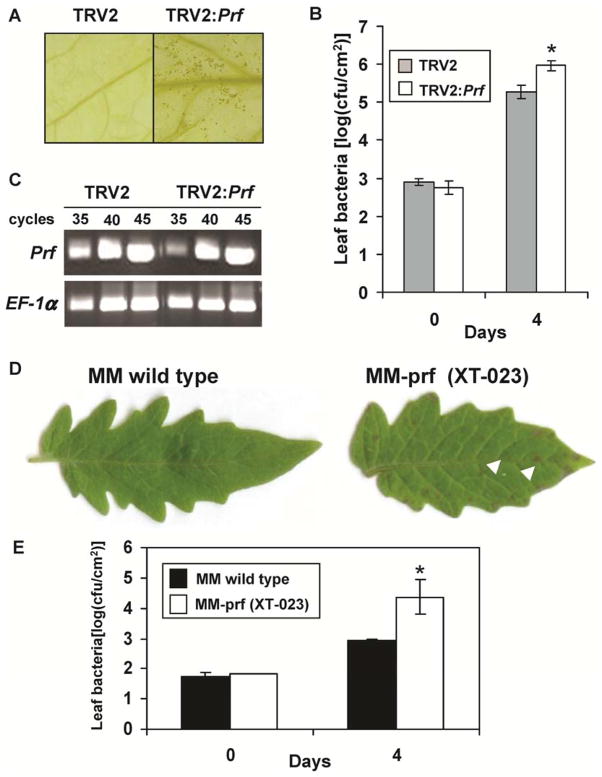
Due to high DNA sequence similarity of Prf in tomato lines and the similarity among Pto family members, Tobacco rattle virus (TRV) constructs generated in previous studies were effective in silencing these genes in ‘VFNT Cherry’ (Ekengren et al. 2003). Four days after inoculation with B728a, speck-like symptoms started to develop on leaves of ‘VFNT Cherry’ silenced for either Prf (Fig. 3A) or members of the Pto family (Supplementary Fig. S5A), and bacterial populations increased 5- to 10-fold in these plants (Fig. 3B). The abundance of Prf transcripts was reduced in leaves showing symptoms upon infection with B728a (Fig. 3C). In addition, transcript abundance of SlFen (SlPtoB) and SlPtoF was decreased in leaves silenced for the Pto family members but transcript abundance of SlPtoC and SlPtoD was unaffected (note that SlPtoA is not detectably expressed in leaves). Inoculation of a ‘Moneymaker’ line that has a mutation in Prf also indicated a requirement of this gene for recognition of B728a (Fig. 3D and E). These data support the hypothesis that Prf and one or more Pto family members (SlFen, SlPtoF, or both) are involved in ‘VFNT Cherry’ resistance against B728a.
Fig. 3.
Resistance elicited by avrPtoBB728a in VFNT Cherry is Prf-dependent. A, Tobacco rattle virus (TRV2) was used to silence Prf in VFNT Cherry. The silenced plants were compromised in their resistance to Pseudomonas syringae pv. syringae strain B728a and specks were observed 4 days after inoculation. At 7 days postinoculation (dpi), chlorophyll was removed from the leaves with ethanol before photographs were taken. B, Bacterial populations in VFNT Cherry leaves silenced for Prf and infiltrated with B728a. C, Silencing of Prf in VFNT Cherry leaves was evaluated using semi-quantitative reverse-transcription polymerase chain reaction. D, Symptoms on leaves after vacuum infiltration of B728a into wild-type Moneymaker (MM) and a prf mutant of Moneymaker (XT-023). Disease symptoms started to develop 3 dpi and photographs were taken 7 days after inoculation. E, Bacterial populations in MM and XT-023 leaves at 0 and 4 dpi. Error bars indicate the standard deviations for three replicates. Data analysis was performed using Student’s t test, and the asterisk (*) indicates significant difference at a level of 5%. Each experiment was performed three times with similar results.
AvrPtoBB728a interacts with SlFen, SlPtoC, and SlPtoD of ‘VFNT Cherry’ in a yeast two-hybrid assay
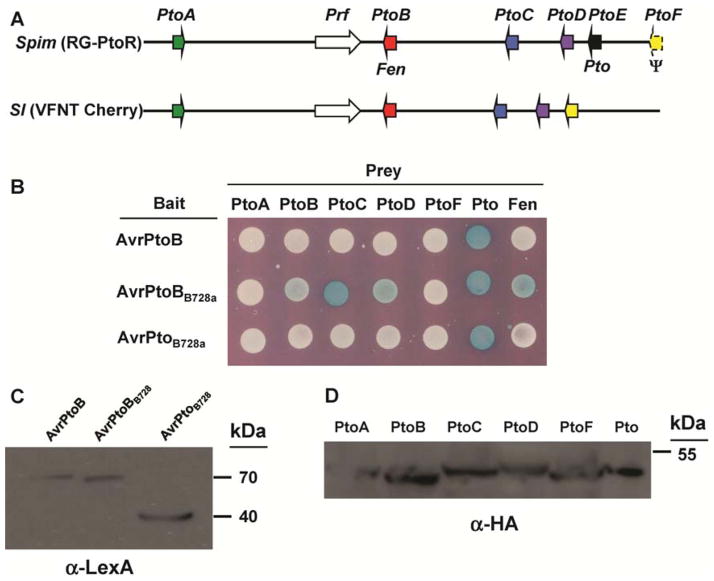
‘VFNT Cherry’ tomato has five members of the Pto family (SlPtoA, SlFen/SlPtoB, SlPtoC, SlPtoD, and SlPtoF; it lacks PtoE [Pto]), each with 80 to 90% sequence homology to its ortholog in the Pto family present in S. pimpinellifolium (Fig. 4A). Moreover, each of the Pto family members except SlPtoA is expressed in leaves of ‘VFNT Cherry’ (Chang et al. 2002). It is known that AvrPto and AvrPtoB both physically interact with Pto in a yeast two-hybrid system, and this interaction is correlated with their ability to trigger resistance in tomato. Therefore, we tested possible interactions between the AvrPto or AvrPtoB homologs from B728a and each Pto family member from ‘VFNT Cherry’ to find clues as to which Pto family member is involved in AvrPtoBB728a-triggered immunity.
Fig. 4.
Interactions in a yeast two-hybrid system between AvrPtoBB728a and Pto family members. A, Genome organization of Pto region in ‘Rio-Grande’ (RG)-PtoR (from Solanum pimpinellifolium or Spim) (Pedley and Martin 2003) and ‘VFNT Cherry’ (S. lycopersicum, Sl). Arrows indicate the location and the orientation of genes in these regions, and open reading frames with the same color are orthologs. B, Yeast two-hybrid analysis. AvrPtoB, AvrPtoB728a, and AvrPtoBB728a all interact with Pto but differentially interact with SlFen, SlPtoC, and SlPtoD (labeled as PtoB, PtoC, and PtoD, respectively). Expression of the bait and prey proteins was confirmed by Western blotting using an C, anti-LexA or D, anti-HA antibodies, respectively.
AvrPtoBB728a interacted weakly with SlFen (SlPtoB) and SlPtoD and more strongly with SlPtoC (Fig. 4B). It also interacted with Pto and Fen from S. pimpinellifolium. It is noteworthy that, in contrast to AvrPtoB, AvrPtoBB728a was unable to mask its interaction with Fen, suggesting that it is unable to ubiquitinate this protein efficiently (Rosebrock et al. 2007). As expected, AvrPtoB and AvrPtoB728a only interacted with Pto in a yeast two-hybrid assay. Western blot analysis indicated that all bait or prey proteins were expressed in the yeast cells (Fig. 4C and D). Therefore, these data suggested that AvrPtoBB728a might be recognized by SlFen, SlPtoC, or SlPtoD in ‘VFNT Cherry’ to elicit R-gene-mediated resistance.
AvrPtoBB728a has weaker E3 ligase activity than AvrPtoB
AvrPtoBB728a shares 52% amino acid sequence identity overall with AvrPtoB and, notably, has reasonably high sequence similarity to the E3 ligase domain of AvrPtoB (the amino acid sequences share 69% identity with and 73% similarity to that of AvrPtoB). It also has residues known in AvrPtoB to be required for ubiquitination activity (Supplementary Fig. S6A). Based on this similarity, it might be expected that the C-terminal domain of AvrPtoBB728a is an active E3 ligase, allowing AvrPtoBB728a to facilitate degradation of the tomato resistance protein Fen (Rosebrock et al. 2007). At the same time, the fact that ‘VFNT Cherry’ and ‘Moneymaker’ both recognize AvrPtoBB728a in a Prf-dependent manner might suggest that its E3 ligase activity is ineffective, thereby allowing a Pto family member in these cultivars to recognize B728a.
To assess AvrPtoBB728a E3 ligase activity, we purified AvrPtoBB728a and AvrPtoB-GST fusion proteins and used them in an in vitro ubiquitination assay in the presence or absence of E2 conjugating enzyme. AvrPtoB rapidly synthesized large amounts of high molecular weight polyubiquitin species (Fig. 5A). In contrast, AvrPtoBB728a showed greatly reduced E3 ligase activity. Although polyubiquitin accumulation reached a maximum after 5 min in the presence of AvrPtoB, AvrPtoBB728a synthesized much less even after 15 min of incubation.
Fig. 5.
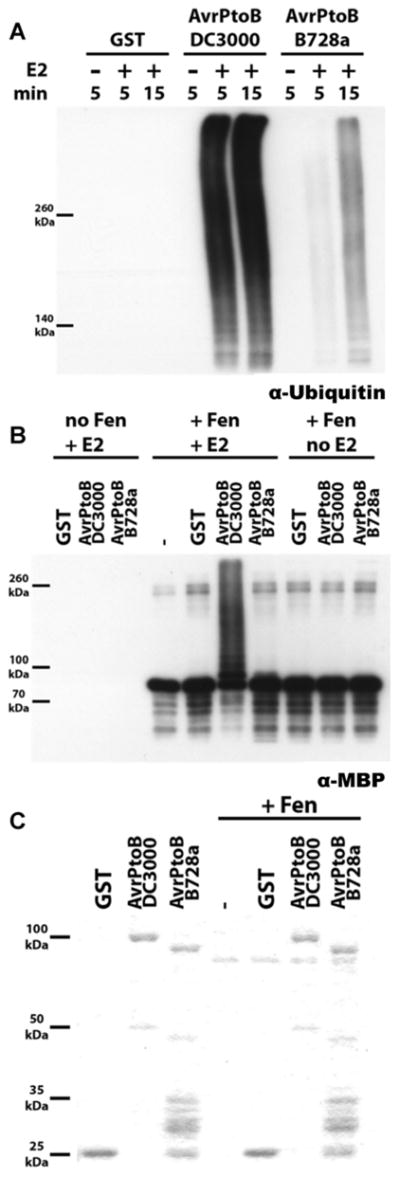
AvrPtoBB728a has reduced E3 ligase activity compared with AvrPtoB. A, N-terminal glutathione-S-transferase (GST) fusions of AvrPtoB and AvrPtoBB728a were purified from Escherichia coli and subjected to an in vitro ubiquitination assay with or without an E2 conjugating enzyme for 5 min and with E2 for 15 min. B, Purified maltose-binding-protein (MBP):Fen protein was used as a substrate for AvrPtoB and AvrPtoBB728a E3 ligases. Only AvrPtoB efficiently ubiquitinated SlFen. C, The remaining protein master-mixes used in the experiments presented in A and 5B were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis and stained with Coomassie Brilliant Blue to show equal abundance of the different proteins. Both experiments were conducted in parallel using the same master mixes.
To test whether our AvrPtoBB728a protein preparation contained possible inhibitors that might interfere specifically with the E3 ligase activity in this assay, we conducted a titration experiment. Mixing of AvrPtoBB728a with AvrPtoB protein in our assay conditions showed no decrease in AvrPtoB E3 ligase activity, ruling out the presence of an inhibitor in the AvrPtoBB728a preparation. Thus, the E3 ligase activity of the AvrPtoBB728a C-terminus was reduced compared with that of AvrPtoB. In conjunction with the yeast-two-hybrid analysis, this suggests that AvrPtoBB728a might be unable to degrade a Pto family member, possibly SlFen, efficiently enough, resulting in robust resistance in VFNT Cherry.
To test this hypothesis, we next subjected a purified maltose-binding-protein–Fen fusion protein (MBP-Fen) to a cross-ubiquitination assay using either AvrPtoB or AvrPtoBB728a. Only AvrPtoB from P. syringae pv. tomato DC3000 was able to efficiently polyubiquitinate Fen in the presence of E2. In contrast, no ubiquitinated forms of MBP-Fen were detectable in the presence of AvrPtoBB728a (Fig. 5B). These observations support the hypothesis that the greatly reduced general E3 ligase activity of AvrPtoBB728a results in an inability to degrade cellular targets and, consequently, to suppress resistance mediated by a Pto family member in conjunction with Prf. The abundance of AvrPtoB proteins and Fen used in the above experiments is shown in Figure 5C.
DISCUSSION
The bean pathogen P. syringae pv. syringae can cause “syringae disease” on tomato (Jones 1991), and our observation that S. lycopersicum cultivars Moneymaker and VFNT Cherry recognize only one of two P. syringae pv. syringae strains suggested a race specificity for tomato resistance against this pathogen. By means of heterologous expression of strain B728a effectors in strain 61 and mutation analysis in B728a, we identified AvrPtoBB728a as the sole effector protein that is responsible for eliciting nonhost disease resistance to B728a in tomato. In addition, results from our yeast two-hybrid and VIGS experiments indicated that one or more members of the Pto gene family in ‘VFNT Cherry’ and ‘Moneymaker’ are responsible for this resistance to B728a. Importantly, we found that AvrPtoBB728a has weaker E3 ligase activity compared with AvrPtoB. This lower E3 ligase activity was found to be incapable of ubiquitinating the Fen kinase and, therefore, likely hinders the ability of AvrPtoBB728a to cause degradation of the responsible Pto family member, resulting in disease resistance.
Why do these two AvrPtoB homologs have such different ubiquitination activities despite the relatively high amino acid sequence similarities in their E3 ligase domains? One possibility is that they differ for certain amino acids that play a specific role in E3 ligase activity. In this regard, it is interesting that one of the three E2-binding sites (located at position 476) is a histidine in AvrPtoBB728a whereas, in AvrPtoB, it is phenylalanine. We used site-directed mutagenesis to modify this site in AvrPtoBB728a and observed that the AvrPtoBB728a (H476F) variant protein still elicited resistance in ‘VFNT Cherry’ against B728a (C.-F. Chien and N.-C. Lin, unpublished data). Thus, this single polymorphism in AvrPtoBB728a is not solely responsible for the reduced E3 ligase activity we observed. The second, more likely possibility is that the 36 residues that are different in the E3 ligase domains of the two proteins alter the overall conformation of the enzyme, interfering with protein folding or leading to gross changes hindering interaction with the E2 conjugating enzyme.
The presence of other Pto family kinases linked to Prf in ‘VFNT Cherry’ and ‘Moneymaker’ suggests that, through recruitment of different kinases, the functions of Prf can be expanded to detect several type III effectors to trigger specific defense responses (Gutierrez et al. 2010). In addition to SlPto, AvrPtoBB728a also interacts with Fen, suggesting that AvrPtoBB728a probably elicits both Fen- and Pto-dependent resistance in the resistant tomato line RG-PtoR. This is supported by the observation that more severe symptoms develop on tomato line RG-prf3 (a Prf-deficient line) than on RG-pto11 (a Pto-deficient line) when they are inoculated with B728a (Lin and Martin 2007). The fact that AvrPtoBB728a triggers Pto-independent resistance provides additional evidence that the existence of multiple Pto-like genes is likely the result of evolution for recognition of a continuously evolving effector (AvrPtoB homolog in this case) to activate robust tomato immunity.
It is possible that AvrPtoBB728a is a transitional version of AvrPtoB during its evolution when it had acquired the C-terminal domain but had not yet developed E3 ligase activity. However, it is perhaps more likely that AvrPtoBB728a is a version of AvrPtoB that has lost its E3 ligase activity because its primary host (bean) lacks a Pto family member (e.g., Fen) that recognizes it and maintenance of this activity carries a metabolic cost (Martin 2012). This possibility is further supported by the existence of HopPmaL, a C-terminal truncated version of AvrPtoB that occurs in P. syringae pv. maculicola ES4326. This variant has no E3 ligase domain and yet retains its full virulence activity (Guttman et al. 2002; Lin et al. 2006).
Our data provide further support for the hypothesis that R-gene-mediated ETI is the mechanism that underlies nonhost resistance in many plant interactions with bacterial pathogens and possibly those involving other pathogens. We reported earlier that a broad array of P. syringae pathovars that are nonhost pathogens of tomato are, in fact, able to infect tomato if the Prf gene is mutated (Lin and Martin 2007). The basis for these nonhost interactions was found to be recognition of either an AvrPto or AvrPtoB protein in the diverse P. syringae pathovars. Another compelling example is the finding that mutation of a single effector gene, hopQ1-1, in the P. syringae pv. tomato DC3000 allows it to become a pathogen on the nonhost N. benthamiana (Wei et al. 2007). More recently, it has been reported that a single effector, HopAS1, in the P. syringae pv. tomato T1 is recognized by Arabidopsis, giving rise to the apparent nonhost resistance to this tomato pathogen (Sohn et al. 2012). As additional effector repertoires of bacterial pathogens are investigated, we predict that many more examples of such gene-for-gene interactions underlying nonhost resistance will be identified.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions
The bacterial strains and plasmids used in this study are listed in Table 1. Strains of P. syringae were grown in KB medium at 28°C with appropriate antibiotics. Escherichia coli DH5α used for DNA maintenance and triparental mating was grown in Luria-Bertani (LB) medium at 37°C. Agrobacterium tumefaciens GV3101 was grown in LB medium at 28°C, unless indicated otherwise. The concentrations of antibiotics used in each selective medium were as follows: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; rifampicin, 100 μg/ml; spectinomycin (Sp) and streptomycin (Sm), 25 μg/ml; and tetracycline, 25 μg/ml.
Table 1.
Bacterial strains and plasmids used in this study
| Strains and plasmids | Relevant characteristicsa | Source or reference |
|---|---|---|
| Bacterial strains | ||
| Escherichia coli DH5α | … | Invitrogen |
| E. coli BL21(DE3) | … | Invitrogen |
| Pseudomonas syringae pv. syringae | Rifampicin derivative of field-isolated strain | |
| B728a | Rifampicin derivative of field-isolated strain | Loper and Lindow 1987 |
| B728aΔavrPto | ΔavrPto::nptII, Rif r Kanr | This study |
| B728aΔavrPtoB | ΔavrPtoB::Ω Spr/Smr, Rif r Spr/Smr | This study |
| B728aΔavrPtoΔavrPtoB | ΔavrPto::nptII, ΔavrPtoB::Ω Spr/Smr, Rif r, Kanr, Spr/Smr | This study |
| Agrobacterium tumefaciens GV3101 | Rif r | Holsters et al. 1980 |
| Plasmids | ||
| pGilda | HIS3, Ampr, under GAL1 promoter for Y2H with LexA | Clontech Laboratories |
| pJG4-5 | TRP1, Ampr, under GAL1 promoter for Y2H with HA epitope tag | Invitrogen |
| pCPP45 | Broad-host-range vector with RP4 par region, Tcr | Charkowski et al. 1997 |
Rif r, Kanr, Spr, Smr, Ampr, and Tcr indicate resistant to rifampicin, kanamycin, spectinomycin, streptomycin, ampicillin, and tetracycline, respectively.
DNA manipulation
Plasmid DNA was isolated using a High-Speed Plasmid DNA Purification Mini Kit (Geneaid Biotech Ltd., Taiwan). The Gel Extraction/PCR Clean-up Kit (Geneaid Biotech Ltd.) was utilized to recover DNA fragments from the agarose gel or the reaction mixture after polymerase chain reaction (PCR). Oligonucleotide primers (Supplementary Table S1) were synthesized at Genomics Biosci & Tech (Taipei, Taiwan) and sequencing was performed at Genomics Biosci & Tech with an ABI 3730XL DNA Analyzer (Applied Biosystems, Foster City, CA, U.S.A.). DNA sequences were analyzed using the Vector NTI (Invitrogen Co, Carlsbad, CA, U.S.A.).
A broad-host-range vector expressing avrPtoB728a, pavrPtoB728a, was obtained by cloning a 0.7-kb KpnI/XbaI fragment containing the hrp-box, open reading frame (ORF), and 0.2-kb downstream sequences of avrPtoB728a into pCPP45. pavrPtoB728a or pAvrPtoBB728a constructed previously (Lin et al. 2006) was then introduced into P. syringae pv. syringae 61 by triparental mating to generate Psy61(avrPtoB728a) or Psy61(avrPtoBB728a) for further analysis.
Construction of pΔavrPtoB728a and pΔAvrPtoBB728a for marker exchange mutagenesis
To delete avrPto in PsyB728, a 2.5-kb fragment containing the upstream region and a 2-kb fragment containing the downstream region of avrPto were PCR amplified using primer pairs B728amut1-F/B728mut1-R and B728mut2-F/B728amut2-R, respectively. A DNA fragment of the nptII gene without terminator was released from pCPP2988 (Alfano et al. 1996) digested with HindIII and XhoI. These three fragments were sequentially cloned into pBluescript II SK(−), and the KpnI-XbaI fragment was subcloned into pRK415 to generate pΔavrPtoB728a, which was then transformed into B728a to screen for a PsyB728aΔavrPtoB728a mutant whose avrPto was exchanged with the nptII selectable marker. pΔavrPtoBB728a was constructed as pΔavrPtoB728a, because the KpnI-BamHI fragment containing an ΩSp fragment from pHP45ΩSp and the flanking regions of avrPtoBB728a amplified with primer pairs B728a-mt1/B728a-mt2 and B728a-mt3/B728a-mt4 was subcloned into pRK415. pΔavrPtoBB728a was then introduced into B728a and PsyB728aΔavrPtoB728a to generate PsyB728aΔavrPtoB728a and PsyB728aΔavrPtoB728aΔavrPtoBB728a, whose avrPtoB loci were replaced by a ΩSp fragment which confers spectinomycin and streptomycin resistance to the marker-exchanged mutants. Marker-exchanged mutants PsyB728aΔavrPtoB728a, PsyB728aΔavrPtoBB728a, and PsyB728aΔavrPtoB728aΔavrPtoBB728a were all confirmed using Southern blot analysis.
Pathogenicity assay
Tomato (S. lycopersicum) cultivars Rio-Grande, VFNT Cherry, and Moneymaker were grown in a growth chamber setting of 22°C, a 16-h photoperiod, and 70% relative humidity. Tomato inoculation and determination of bacterial populations in tomato leaves was performed using the procedure described previously (Lin and Martin 2005). Disease development was monitored every day and symptoms were photographed 7 days after inoculation. For clear observation of specks on leaves of ‘VFNT Cherry’ tomato, leaves were treated with Carnoy’s solution (60% ethanol, 30% chloroform, and 10% glacial acetic acid) overnight to remove chlorophylls, and preserved in storage buffer containing 5% acetic acid, 20% ethanol, and 20% glycerol before photographing.
VIGS
VIGS using strains of A. tumefaciens GV3101 containing pTRV2:Pto and pTRV2:Prf obtained previously was done by procedures described before (Ekengren et al. 2003). Plants were kept in the growth chamber for 2 to 3 weeks until the pathogenicity assay was implemented. To monitor bacterial population and silencing efficiency, each leaf disc collected with a cork borer was cut in half because one part was used for bacterial recovery and the other part was kept in liquid nitrogen for semi-quantitative reverse-transcription (RT)-PCR. RNA was extracted using the Trizol reagent (Invitrogen) and cDNA was synthesized with a Superscript First-Strand Synthesis System for RT-PCR (Invitrogen). PCR was performed using primers specific to each Pto ortholog or all Pto-like sequences (designated as ‘Pto’) and Prf (Ekengren et al. 2003), and expression of EF-1α was used as an internal control.
Yeast two-hybrid analysis
A LexA yeast two-hybrid system was used to test interactions between Pto family members and AvrPtoB, AvrPtoB728a, and AvrPtoBB728a. A DNA fragment containing ORF of the Pto family members, including SlPtoA, SlFen, SlPtoC, SlPtoD, or SlPtoF, was PCR amplified from genomic DNA of ‘VFNT Cherry’ tomato with each gene-specific primer pair, digested with BamHI, and cloned into BamHI-polished pJG4-5 vector to generate prey constructs. A DNA fragment containing ORF of avrPtoB, avrPtoB728a, and avrPtoBB728a was cloned into the EcoRI/BamHI site of pGilda (Clontech, Mountain View, CA, U.S.A.) to create a bait construct. The bait and prey constructs subsequently were introduced into Saccharomyces cerevisiae EYG48 containing a reporter vector, and yeast cells containing both bait and prey constructs were streaked on complete medium dropout media containing raffinose/galactose and 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside to investigate interactions between prey and bait proteins. pJG4-5::Pto and pJG4-5::Fen were used as positive controls.
In vitro ubiquitination assay
In vitro ubiquitination assays were performed as described (Andersen et al. 2004), with minor modifications. In brief, 10 μl of protein master mix containing 1 μg of purified glutathione-S-transferase–fusion protein (and, in the case of the SlFen cross-ubiquitination assays, 200 ng of purified MBP:Fen) (Rosebrock et al. 2007) was added to the reaction mixture containing 50 nM E1 (UBE1; BostonBiochem, Cambridge, MA, U.S.A.), 5 μM E2 (UbcH5a; BostonBiochem), and 10 μg of Ubiquitin (BostonBiochem) in reaction buffer (50 mM Tris [pH 7.5], 2 mM dithiothreitol, 5 mM ATP, and 5 mM MgCl2) to a total of 30 μl and incubated at 30°C for the time indicated. The reaction was terminated by the addition of 30 μl of 2× Laemmli buffer and boiling at 98°C for 5 min. Proteins were resolved on sodium dodecyl sulfate (SDS)-polyacrylamide gels (5 or 10%), followed by Western blotting onto polyvinylidene diflouride membranes. Proteins were visualized either by infrared imaging on an Odyssey system (LI-COR, Lincoln, NE, U.S.A.) using primary anti-Ubiquitin (Santa Cruz Biotechnology Inc., Santa Cruz, CA, U.S.A.) and secondary anti-mouse-680 (LI-COR) antibodies or using horseradish peroxidase-coupled secondary anti-mouse and anti-rabbit antibodies (Santa Cruz Biotechnology Inc.,) and Amersham ECL plus (GE Healthcare, Piscataway, NJ, U.S.A.) detection reagent. Primary rabbit anti-MBP antibody was purchased from Sigma-Aldrich (St Louis). Because of high signal intensities, protein samples had to be diluted 1/20 in Laemmli buffer prior to SDS-polyacrylamide gel electrophoresis [spelled “PAGE”] in the case of horseradish peroxidase detection. To show equal loading, the remaining master mix for each sample mix was resolved on a separate gel and stained with Coomassie Brilliant Blue.
Supplementary Material
Acknowledgments
This work was supported by the National Science Council, Taiwan, R.O.C. grant number NSC 97-2311-B-002-006 to N.-C. Lin and by National Institutes of Health grant R01-GM078021 to G. B. Martin. J. Mathieu and P. Boyle were supported by postdoctoral fellowships from the Human Frontier Science Program. We thank F. Xiao and C. Chen for their critical reading of the manuscript and helpful suggestions, and X. Tang for the XT-023 ‘Moneymaker’ line.
Footnotes
The e-Xtra logo stands for “electronic extra” and indicates that six supplementary figures, one supplementary table, and supplementary materials and methods information are published online.
LITERATURE CITED
- Abramovitch RB, Kim YJ, Chen S, Dickman MB, Martin GB. Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO (Eur Mol Biol Organ) J. 2003;22:60–69. doi: 10.1093/emboj/cdg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramovitch RB, Janjusevic R, Stebbins CE, Martin GB. Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc Natl Acad Sci USA. 2006;103:2851–2856. doi: 10.1073/pnas.0507892103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrios GN. Plant Pathology. Elsevier Academic Press; Amsterdam: 2005. [Google Scholar]
- Alfano JR, Bauer DW, Milos TM, Collmer A. Analysis of the role of the Pseudomonas syringae pv. syringae HrpZ harpin in elicitation of the hypersensitive response in tobacco using functionally non-polar hrpZ deletion mutations. Mol Microbiol. 1996;19:715–728. doi: 10.1046/j.1365-2958.1996.415946.x. [DOI] [PubMed] [Google Scholar]
- Almeida NF, Yan S, Lindeberg M, Studholme DJ, Schneider DJ, Condon B, Liu H, Viana CJ, Warren A, Evans C, Kemen E, Maclean D, Angot A, Martin GB, Jones JD, Collmer A, Setubal JC, Vinatzer BA. A draft genome sequence of Pseudomonas syringae pv. tomato T1 reveals a type III effector repertoire significantly divergent from that of Pseudomonas syringae pv tomato DC3000. Mol Plant-Microbe Interact. 2009;22:52–62. doi: 10.1094/MPMI-22-1-0052. [DOI] [PubMed] [Google Scholar]
- Andersen P, Kragelund BB, Olsen AN, Larsen FH, Chua NH, Poulsen FM, Skriver K. Structure and biochemical function of a prototypical Arabidopsis U-box domain. J Biol Chem. 2004;279:40053–40061. doi: 10.1074/jbc.M405057200. [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J. Differential innate immune signalling via Ca2+ sensor protein kinases. Nature. 2010;464:418–422. doi: 10.1038/nature08794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretz JR, Mock NM, Charity JC, Zeyad S, Baker CJ, Hutcheson SW. A translocated protein tyrosine phosphatase of Pseudomonas syringae pvtomato DC3000 modulates plant defence response to infection. Mol Microbiol. 2003;49:389–400. doi: 10.1046/j.1365-2958.2003.03616.x. [DOI] [PubMed] [Google Scholar]
- Chang JH, Tai YS, Bernal AJ, Lavelle DT, Staskawicz BJ, Michelmore RW. Functional analyses of the Pto resistance gene family in tomato and the identification of a minor resistance determinant in a susceptible haplotype. Mol Plant-Microbe Interact. 2002;15:281–291. doi: 10.1094/MPMI.2002.15.3.281. [DOI] [PubMed] [Google Scholar]
- Charkowski AO, Huang HC, Collmer A. Altered localization of HrpZ in Pseudomonas syringae pvsyringae hrp mutants suggests that different components of the type III secretion pathway control protein translocation across the inner and outer membranes of gram-negative bacteria. J Bacteriol. 1997;179:3866–3874. doi: 10.1128/jb.179.12.3866-3874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Agnew JL, Cohen JD, He P, Shan L, Sheen J, Kunkel BN. Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc Natl Acad Sci USA. 2007;104:20131–20136. doi: 10.1073/pnas.0704901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn JR, Martin GB. Pseudomonas syringae pv. tomato type III effectors AvrPto and AvrPtoB promote ethylene-dependent cell death in tomato. Plant J. 2005;44:139–154. doi: 10.1111/j.1365-313X.2005.02516.x. [DOI] [PubMed] [Google Scholar]
- Cunnac S, Lindeberg M, Collmer A. Pseudomonas syringae type III secretion system effectors: Repertoires in search of functions. Curr Opin Microbiol. 2009;12:53–60. doi: 10.1016/j.mib.2008.12.003. [DOI] [PubMed] [Google Scholar]
- de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Rodriguez Egea P, Bogre L, Grant M. Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO (Eur Mol Biol Organ) J. 2007;26:1434–1443. doi: 10.1038/sj.emboj.7601575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat Rev Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- Ekengren SK, Liu Y, Schiff M, Dinesh-Kumar SP, Martin GB. Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J. 2003;36:905–917. doi: 10.1046/j.1365-313x.2003.01944.x. [DOI] [PubMed] [Google Scholar]
- Espinosa A, Guo M, Tam VC, Fu ZQ, Alfano JR. The Pseudomonas syringae type III-secreted protein HopPtoD2 possesses protein tyrosine phosphatase activity and suppresses programmed cell death in plants. Mol Microbiol. 2003;49:377–387. doi: 10.1046/j.1365-2958.2003.03588.x. [DOI] [PubMed] [Google Scholar]
- Feil H, Feil WS, Chain P, Larimer F, DiBartolo G, Copeland A, Lykidis A, Trong S, Nolan M, Goltsman E, Thiel J, Malfatti S, Loper JE, Lapidus A, Detter JC, Land M, Richardson PM, Kyrpides NC, Ivanova N, Lindow SE. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv tomato DC3000. Proc Natl Acad Sci USA. 2005;102:11064–11069. doi: 10.1073/pnas.0504930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Guo M, Jeong BR, Tian F, Elthon TE, Cerny RL, Staiger D, Alfano JR. A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature. 2007;447:284–288. doi: 10.1038/nature05737. [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Hann DR, Ntoukakis V, Petutschnig E, Lipka V, Rathjen JP. AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol. 2009;19:423–429. doi: 10.1016/j.cub.2009.01.054. [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Vinatzer BA. Identifying type III effectors of plant pathogens and analyzing their interaction with plant cells. Curr Opin Microbiol. 2003;6:20–28. doi: 10.1016/s1369-5274(02)00004-8. [DOI] [PubMed] [Google Scholar]
- Gutierrez JR, Balmuth AL, Ntoukakis V, Mucyn TS, Gimenez-Ibanez S, Jones AM, Rathjen JP. Prf immune complexes of tomato are oligomeric and contain multiple Pto-like kinases that diversify effector recognition. Plant J. 2010;61:507–518. doi: 10.1111/j.1365-313X.2009.04078.x. [DOI] [PubMed] [Google Scholar]
- Guttman DS, Vinatzer BA, Sarkar SF, Ranall MV, Kettler G, Greenberg JT. A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science. 2002;295:1722–1726. doi: 10.1126/science.295.5560.1722. [DOI] [PubMed] [Google Scholar]
- He P, Shan L, Lin NC, Martin GB, Kemmerling B, Nurnberger T, Sheen J. Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell. 2006;125:563–575. doi: 10.1016/j.cell.2006.02.047. [DOI] [PubMed] [Google Scholar]
- Holsters M, Silva B, Van Vliet F, Genetello C, De Block M, Dhaese P, Depicker A, Inzé D, Engler G, Villarroel R, Van Montagu M, Schell J. The functional organization of the nopaline A. tumefaciens plasmid pTiC58. Plasmid. 1980;3:212–230. doi: 10.1016/0147-619x(80)90110-9. [DOI] [PubMed] [Google Scholar]
- Hueck CJ. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iriti M, Faoro F. Review of innate and specific immunity in plants and animals. Mycopathologia. 2007;164:57–64. doi: 10.1007/s11046-007-9026-7. [DOI] [PubMed] [Google Scholar]
- Joardar V, Lindeberg M, Jackson RW, Selengut J, Dodson R, Brinkac LM, Daugherty SC, Deboy R, Durkin AS, Giglio MG, Madupu R, Nelson WC, Rosovitz MJ, Sullivan S, Crabtree J, Creasy T, Davidsen T, Haft DH, Zafar N, Zhou L, Halpin R, Holley T, Khouri H, Feldblyum T, White O, Fraser CM, Chatterjee AK, Cartinhour S, Schneider DJ, Mansfield J, Collmer A, Buell CR. Whole-genome sequence analysis of Pseudomonas syringae pvphaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J Bacteriol. 2005;187:6488–6498. doi: 10.1128/JB.187.18.6488-6498.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JB. Bacterial speck. In: Jones JB, Jones JP, Stall RE, Zitter TA, editors. Compendium of Tomato Diseases. American Phytopathological Society Press; St. Paul, MN, U.S.A: 1991. pp. 26–27. [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Lin NC, Martin GB. Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell. 2002;109:589–598. doi: 10.1016/s0092-8674(02)00743-2. [DOI] [PubMed] [Google Scholar]
- Lin NC, Martin GB. An avrPto/avrPtoB mutant of Pseudomonas syringae pvtomato DC3000 does not elicit Pto-mediated resistance and is less virulent on tomato. Mol Plant-Microbe Interact. 2005;18:43–51. doi: 10.1094/MPMI-18-0043. [DOI] [PubMed] [Google Scholar]
- Lin NC, Martin GB. Pto- and Prf-mediated recognition of AvrPto and AvrPtoB restricts the ability of diverse Pseudomonas syringae pathovars to infect tomato. Mol Plant-Microbe Interact. 2007;20:806–815. doi: 10.1094/MPMI-20-7-0806. [DOI] [PubMed] [Google Scholar]
- Lin NC, Abramovitch RB, Kim YJ, Martin GB. Diverse AvrPtoB homologs from several Pseudomonas syringae pathovars elicit Pto-dependent resistance and have similar virulence activities. Appl Environ Microbiol. 2006;72:702–712. doi: 10.1128/AEM.72.1.702-712.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeberg M, Myers CR, Collmer A, Schneider DJ. Roadmap to new virulence determinants in Pseudomonas syringae: Insights from comparative genomics and genome organization. Mol Plant-Microbe Interact. 2008;21:685–700. doi: 10.1094/MPMI-21-6-0685. [DOI] [PubMed] [Google Scholar]
- Lindeberg M, Cunnac S, Collmer A. The evolution of Pseudomonas syringae host specificity and type III effector repertoires. Mol Plant Pathol. 2009;10:767–775. doi: 10.1111/j.1364-3703.2009.00587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loper JE, Lindow SE. Lack of evidence for in situ fluorescent pigment production by Pseudomonas syringae pvsyringae on bean leaf surfaces. Phytopathol. 1987;77:1449–1454. [Google Scholar]
- Lopez-Solanilla E, Bronstein PA, Schneider AR, Collmer A. HopPtoN is a Pseudomonas syringae Hrp (type III secretion system) cysteine protease effector that suppresses pathogen-induced necrosis associated with both compatible and incompatible plant interactions. Mol Microbiol. 2004;54:353–365. doi: 10.1111/j.1365-2958.2004.04285.x. [DOI] [PubMed] [Google Scholar]
- Mansfield JW. From bacterial avirulence genes to effector functions via the hrp delivery system: An overview of 25 years of progress in our understanding of plant innate immunity. Mol Plant Pathol. 2009;10:721–734. doi: 10.1111/j.1364-3703.2009.00576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GB. Suppression and activation of the plant immune system by Pseudomonas syringae effectors AvrPto and AvrPtoB. In: Martin F, Kamoun S, editors. Effectors in Plant-Microbe Interactions. Wiley-Blackwell; Oxford: 2012. pp. 123–154. [Google Scholar]
- Martin GB, Brommonschenkel SH, Chunwongse J, Frary A, Ganal MW, Spivey R, Wu T, Earle ED, Tanksley SD. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Janeway CA., Jr Innate immunity: The virtues of a nonclonal system of recognition. Cell. 1997;91:295–298. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006;126:969–980. doi: 10.1016/j.cell.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Mucyn TS, Clemente A, Andriotis VM, Balmuth AL, Oldroyd GE, Staskawicz BJ, Rathjen JP. The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell. 2006;18:2792–2806. doi: 10.1105/tpc.106.044016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Jay F, Nomura K, He SY, Voinnet O. Suppression of the microRNA pathway by bacterial effector proteins. Science. 2008;321:964–967. doi: 10.1126/science.1159505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HP, Yeam I, Angot A, Martin GB. Two virulence determinants of type III effector AvrPto are functionally conserved in diverse Pseudomonas syringae pathovars. New Phytol. 2010;187:969–982. doi: 10.1111/j.1469-8137.2009.03175.x. [DOI] [PubMed] [Google Scholar]
- Nicaise V, Roux M, Zipfel C. Recent advances in PAMP-triggered immunity against bacteria: Pattern recognition receptors watch over and raise the alarm. Plant Physiol. 2009;150:1638–1647. doi: 10.1104/pp.109.139709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntoukakis V, Mucyn TS, Gimenez-Ibanez S, Chapman HC, Gutierrez JR, Balmuth AL, Jones AM, Rathjen JP. Host inhibition of a bacterial virulence effector triggers immunity to infection. Science. 2009;324:784–787. doi: 10.1126/science.1169430. [DOI] [PubMed] [Google Scholar]
- Pedley KF, Martin GB. Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu Rev Phytopathol. 2003;41:215–243. doi: 10.1146/annurev.phyto.41.121602.143032. [DOI] [PubMed] [Google Scholar]
- Riely BK, Martin GB. Ancient origin of pathogen recognition specificity conferred by the tomato disease resistance gene Pto. Proc Natl Acad Sci USA. 2001;98:2059–2064. doi: 10.1073/pnas.98.4.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald PC, Salmeron JM, Carland FM, Staskawicz BJ. The cloned avirulence gene avrPto induces disease resistance in tomato cultivars containing the Pto resistance gene. J Bacteriol. 1992;174:1604–1611. doi: 10.1128/jb.174.5.1604-1611.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose LE, Langley CH, Bernal AJ, Michelmore RW. Natural variation in the Pto pathogen resistance gene within species of wild tomato (Lycopersicon). I Functional analysis of Pto alleles. Genetics. 2005;171:345–357. doi: 10.1534/genetics.104.039339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose LE, Michelmore RW, Langley CH. Natural variation in the Pto disease resistance gene within species of wild tomato (Lycopersicon). II Population genetics of Pto . Genetics. 2007;175:1307–1319. doi: 10.1534/genetics.106.063602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosebrock TR, Zeng L, Brady JJ, Abramovitch RB, Xiao F, Martin GB. A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature. 2007;448:370–374. doi: 10.1038/nature05966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron JM, Staskawicz BJ. Molecular characterization and hrp dependence of the avirulence gene avrPto from Pseudomonas syringae pv. tomato. Mol Gen Genet. 1993;239:6–16. doi: 10.1007/BF00281595. [DOI] [PubMed] [Google Scholar]
- Salmeron JM, Barker SJ, Carland FM, Mehta AY, Staskawicz BJ. Tomato mutants altered in bacterial disease resistance provide evidence for a new locus controlling pathogen recognition. Plant Cell. 1994;6:511–520. doi: 10.1105/tpc.6.4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron JM, Oldroyd GE, Rommens CM, Scofield SR, Kim HS, Lavelle DT, Dahlbeck D, Staskawicz BJ. Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell. 1996;86:123–133. doi: 10.1016/s0092-8674(00)80083-5. [DOI] [PubMed] [Google Scholar]
- Scofield SR, Tobias CM, Rathjen JP, Chang JH, Lavelle DT, Michelmore RW, Staskawicz BJ. Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- Sohn KH, Saucet SB, Clarke CR, Vinatzer BA, O’Brien HE, Guttman DS, Jones JD. HopAS1 recognition significantly contributes to Arabidopsis nonhost resistance to Pseudomonas syringae pathogens. New Phytol. 2012;193:58–66. doi: 10.1111/j.1469-8137.2011.03950.x. [DOI] [PubMed] [Google Scholar]
- Staskawicz B. First insights into the genes that control plant-bacterial interactions. Mol Plant Pathol. 2009;10:719–720. doi: 10.1111/j.1364-3703.2009.00584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studholme DJ, Ibanez SG, MacLean D, Dangl JL, Chang JH, Rathjen JP. A draft genome sequence and functional screen reveals the repertoire of type III secreted proteins of Pseudomonas syringae pathovar tabaci 11528. BMC Genomics. 2009;10:395–410. doi: 10.1186/1471-2164-10-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Frederick RD, Zhou J, Halterman DA, Jia Y, Martin GB. Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- Tang X, Xiao Y, Zhou JM. Regulation of the type III secretion system in phytopathogenic bacteria. Mol Plant-Microbe Interact. 2006;19:1159–1166. doi: 10.1094/MPMI-19-1159. [DOI] [PubMed] [Google Scholar]
- van der Biezen EA, Jones JD. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci. 1998;23:454–456. doi: 10.1016/s0968-0004(98)01311-5. [DOI] [PubMed] [Google Scholar]
- van der Hoorn RA, Kamoun S. From guard to decoy: A new model for perception of plant pathogen effectors. Plant Cell. 2008;20:2009–2017. doi: 10.1105/tpc.108.060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinatzer BA, Greenberg JT. Whole-genome analysis to identify type III-secreted effectors. Methods Mol Biol. 2007;354:19–34. doi: 10.1385/1-59259-966-4:19. [DOI] [PubMed] [Google Scholar]
- Vinatzer BA, Teitzel GM, Lee MW, Jelenska J, Hotton S, Fairfax K, Jenrette J, Greenberg JT. The type III effector repertoire of Pseudomonas syringae pvsyringae B728a and its role in survival and disease on host and non-host plants. Mol Microbiol. 2006;62:26–44. doi: 10.1111/j.1365-2958.2006.05350.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li J, Hou S, Wang X, Li Y, Ren D, Chen S, Tang X, Zhou JM. A Pseudomonas syringae ADP-ribosyltransferase inhibits Arabidopsis mitogen-activated protein kinase kinases. Plant Cell. 2010;22:2033–2044. doi: 10.1105/tpc.110.075697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CF, Kvitko BH, Shimizu R, Crabill E, Alfano JR, Lin NC, Martin GB, Huang HC, Collmer A. A Pseudomonas syringae pvtomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana . Plant J. 2007;51:32–46. doi: 10.1111/j.1365-313X.2007.03126.x. [DOI] [PubMed] [Google Scholar]
- Xiang T, Zong N, Zou Y, Wu Y, Zhang J, Xing W, Li Y, Tang X, Zhu L, Chai J, Zhou JM. Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol. 2008;18:74–80. doi: 10.1016/j.cub.2007.12.020. [DOI] [PubMed] [Google Scholar]
- Xiao F, He P, Abramovitch RB, Dawson JE, Nicholson LK, Sheen J, Martin GB. The N-terminal region of Pseudomonas type III effector AvrPtoB elicits Pto-dependent immunity and has two distinct virulence determinants. Plant J. 2007;52:595–614. doi: 10.1111/j.1365-313X.2007.03259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeam I, Nguyen HP, Martin GB. Phosphorylation of the Pseudomonas syringae effector AvrPto is required for FLS2/BAK1-independent virulence activity and recognition by tobacco. Plant J. 2010;61:16–24. doi: 10.1111/j.1365-313X.2009.04028.x. [DOI] [PubMed] [Google Scholar]
- Zhang W, He SY, Assmann SM. The plant innate immunity response in stomatal guard cells invokes G-protein-dependent ion channel regulation. Plant J. 2008;56:984–996. doi: 10.1111/j.1365-313X.2008.03657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Thilmony R, Bender CL, Schaller A, He SY, Howe GA. Virulence systems of Pseudomonas syringae pvtomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J. 2003;36:485–499. doi: 10.1046/j.1365-313x.2003.01895.x. [DOI] [PubMed] [Google Scholar]
- Zipfel C. Early molecular events in PAMP-triggered immunity. Curr Opin Plant Biol. 2009;12:414–420. doi: 10.1016/j.pbi.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Zong N, Xiang T, Zou Y, Chai J, Zhou JM. Blocking and triggering of plant immunity by Pseudomonas syringae effector AvrPto. Plant Signal Behav. 2008;3:583–585. doi: 10.4161/psb.3.8.5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwiesler-Vollick J, Plovanich-Jones AE, Nomura K, Bandyopadhyay S, Joardar V, Kunkel BN, He SY. Identification of novel hrp-regulated genes through functional genomic analysis of the Pseudomonas syringae pvtomato DC3000 genome. Mol Microbiol. 2002;45:1207–1218. doi: 10.1046/j.1365-2958.2002.02964.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.