Abstract
Rational design of modifications to the interior and exterior surfaces of virus-like particles (VLPs) for future therapeutic and materials applications is based on structural information about the capsid. Existing cryo-electron microscopy based models suggest that the C-terminus of the bacteriophage P22 coat protein (CP) extends towards the capsid exterior. Our biochemical analysis through genetic manipulations of the C-terminus supports the model where the CP C-terminus is exposed on the exterior of the P22 capsid. Capsids displaying a 6xHis tag appended to the CP C-terminus bind to a Ni affinity column, and the addition of positively or negatively charged coiled coil peptides to the capsid results in association of these capsids upon mixing. Additionally, a single cysteine appended to the CP C-terminus results in the formation of intercapsid disulfide bonds and can serve as a site for chemical modifications. Thus, the C-terminus is a powerful location for multivalent display of peptides that facilitate nanoscale assembly and capsid modification.
Introduction
Viruses are increasingly recognized as useful templates for both hard and soft materials applications1–3. The multifunctional, and highly symmetric organization of subunits in the viral capsids provide for a unique environment for functional group display and modification4–5. Knowledge about the structure and assembly of virus capsids aids their use as powerful platforms for functional nanomaterials design and synthesis6–12. The assembly of repeating subunits into capsids generates highly monodisperse multivalent nanoparticles with distinct interior and exterior surfaces that can be used for encapsulation and/or display13–14. Encapsulated cargo is sequestered from the surrounding environment, while modifications on the capsid exterior display functionalities that interact directly with the bulk environment.
The capsid derived from the bacteriophage P22 is a robust nanocontainer that has been extensively used for directed encapsulation15 and surface display16, and its use as a biomaterial is completely decoupled from the infectious virus. The non-infectious P22 procapsid (PC) assembles in vivo (in E. coli) from 420 copies of coat protein with the aid of the P22 scaffold protein (SP)17, which can be truncated to include only residues 141–303 and still template assembly of a T=7 virus like particle18. The truncated SP (SP141) is packaged on the interior of the 60 nm diameter procapsid and by fusing cargo proteins to the N-terminus can act as a means to direct the packaging inside the P22 capsid19. Previously we have genetically engineered cysteine residues into the P22 coat protein on the interior (S39C, K118C)20–21 and exterior (T183C)16 of the assembled P22 capsid to utilize as sites for chemical conjugation.
For the surface display of peptides, a genetic fusion approach is advantageous over chemical conjugation because the stoichiometry of the peptide display is defined by the quaternary structure, and the capsid self-assembles in vivo requiring less processing and a more homogeneous end product. One flexible loop on the capsid exterior has been identified as a potential site for the display of short peptides but does not tolerate large inserts or highly charged peptides16. The availability of another, more robust, site for genetic display on the capsid exterior would expand the versatility and application of the P22 nanoplatform. The available models of the P22 capsid (from cryo-TEM)22–23 suggest that the C-terminal residues of the CP extend toward the exterior of the capsid. However, the models do not include the location of the last few residues of the P22 CP.
Here we have investigated the location of the CP C-terminus in the P22 procapsid by genetically engineering fusion peptides to the C-terminal of the CP and investigating the biochemical presentation of peptides to the exterior environment of the capsid. Natural and synthetic systems utilize coiled coil motifs24–30 to promote hierarchical assembly, so we have explored the presentation of coiled coil peptides on the C-terminus for directed inter-particle interactions. Through these studies, we show that fusions to the C-terminus of the P22 CP do indeed sample the exterior environment, and we demonstrate the utility of this approach for display of peptides that allow manipulation of the biophysical properties of the capsid and suggest further development of the P22 platform as a functional nanomaterial.
Materials and Methods
Generation of P22 Structural Images
The cryo EM reconstruction coordinates were obtained from the protein data bank (PDB) entries for the procapsid coat protein (PDB: 2XYY), the expanded shell (virion coat protein, PDB: 2XYZ and P22 expanded head coat protein, PDB: 3IYI), and the wiffleball morphology (PDB: 3IYH). Images were created using UCSF Chimera (version 1.6.2) from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco31.
Expression Vectors
For expression of wtP22/wtSP particles, the previously described assembler plasmid32 that contains genes corresponding to wt scaffold, wt coat protein, and ampicillin resistance was used. The genes for the novel mCherry capsids presented were generated using a previously described pET 11a+ template containing mCherrySP141, K118C CP, and ampicillin resistance19. From 5′ to 3′, the mCherrySP141 DNA sequence in the template vector coded for mCherry fluorescent protein, a thrombin proteolytic site, a cysteine (SP-C140), and SP141–303 all in a single reading frame. Established polymerase chain reaction protocols33 using the C118K, SP-C140A, and CP-431C primers (S1) were followed to change or add codons in the template. The C118K primer changes a previously described CP variant (K118C) back to wtCP, the SP-C140A substitutes an alanine for the cysteine to create mCherrySP141(C140A), and the CP-431C adds a cysteine to the CP C-terminus (CP-431C).
Genes for the CP C-terminal additions were ligated into the Novagen pRSFDuet™-1 (KanR) vector to allow for expression of P22 virus-like particles. The coat protein (CP) from the Bacteriophage P22 and a truncated form of the P22 scaffold protein SP(141–303), hereafter refered to as SP141, were incorporated into each of the two multiple cloning sites. Initially, the gene corresponding to the SP141 was amplified from the assembler plasmid19 and ligated into the first multiple cloning site BamHI and SacI. Ligation into this location yields a 6xHis tag present in the expression vector reading frame directly upstream of the SP141 gene.
Subsequently, the gene for the CP-6xHis was amplified out of the assembler plasmid32, and was inserted into the second multiple cloning site of this vector using BglII and XhoI. An SpeI site incorporated upstream of the 6xHis sequence to allow for the insertion of K-coil and E-coil sequences at this site. The primer used for the amplification is shown in Table S1. K-coil and E-coil flanked by SpeI and XhoI were purchased in a pUC57-Kan vector from Genscript, isolated and ligated downstream of the coat protein gene. Novagen pRSFDuet™-1 vectors containing a gene for kanamycin resistance, a truncated P22 scaffold protein (6xHis-SP141), and a P22 coat protein (CP-6xHis, CP-K-coil, or CP-E-coil) were transformed into E.coli strain XL1 electrocompetent cells (Agilent Technologies). Colonies resulting from each ligation were screened for CP amplification by colony PCR. Isolated plasmids from positive colonies were sequenced (Seqwright, Tx) for verification.
Capsid Expression and Purification
Expression vectors for wtP22/wtSP, wtP22/mCherrySP141, wtP22/mCherrySP141(C140A), CP-431C/mCherrySP141(C140A), wtP22/6xHis-SP141, CP-6xHis/6xHis-SP141, CP-E-Coil/6xHis-SP141, and CP-K-Coil/6xHis-SP141 P22 capsids were transformed into electrocompentant E.coli strain BL21 (DE3, Novagen). One milliliter from an overnight culture was used to inoculate 1 L of LB supplemented with the appropriate antibiotic (ampicillicin or kanamycin). Cells were grown at 37°C, and induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at an OD600 of 0.6. Approximately 4h later, cells were collected by centrifugation and lysed with DNase, lysosyme, and RNase (Sigma Aldrich). Sonication was used to disrupt the cells (3× for 2.5 min at 50% amplitude, pulse 0.5 sec on/off, Branson Digital Sonifier 250, 200W, 20kHz). Large cellular debris were removed through centrifugation (45 min at 12,000 × g). Subsequently, P22 capsids were pelleted through a 35% sucrose cushion for 50 min at 45,000 × g using ultracentrifugation (Sorval WX Ultra 80, Thermo Scientific). Pellets were resuspended in phosphate buffered saline (10–30 mg/mL, pH 7.6), and dialyzed to remove excess sucrose. Size exclusion chromatography using a Sephacryl-500 column was completed as a final purification step using phosphate buffered saline (pH 7.6). The P22 capsid concentration was determined with the absorbance at 280 nm using an extinction coefficient A280 = 1.4(mg/mL)−1.
Liquid Chromatography-Mass Spectroscopy (LC-MS)
Masses of the P22 coat proteins prior to and after modification were determined by using a micro-TOF (Bruker Daltonics) mass spectrometer. Aproximately 1.5 micrograms (2–4 μL volume) of each P22 sample in PBS buffer (25 mM phosphate, 50 mM NaCl, pH 7.6, purified as described previously) was injected via an Agilent 1100 series high performance liquid chromatography (HPLC) system equipped with a PolyHydroxyethyl A sizing column (100 × 4.6 mm, 5 μm, 500 A from PolyLC, Inc.). Samples were continuously flowed to the ESI source from the HPLC using an isocratic elution (25% acetonitrile, 75% water, and 0.1% formic acid) and a flow rate of 0.3 mL-min−1. Source parameters were as follows: drying gas 6.0 L-min−1, nebulizer 3.5 Bar, capillary voltage 3500 V, capillary exit 100 V. Spectra were collected in positive mode from 200 to 3000 m/z at a rate of 2 Hz. The resulting multiple charge state distributions for protein were deconvoluted using a maximum entropy deconvolution algorithm in the Bruker Compass Analysis software.
Ni-NTA affinity chromatography
To test for binding of the capsids with 6xHis tags, approximately 0.5 mg (1 mL vol) of each sample was applied to a 1 mL Ni HisTrap® column (Pharmacia) equilibrated in PBS. Bound capsids were eluted from the column at a flow rate of 0.5 mLmin−1 with a buffer gradient containing 0 M to 1 M imidazole in 25 mM HEPES, 100 mM NaCl, pH 7.1. Protein elution was monitored via the absorbance at 280 nm, and fractions containing protein were analyzed by SDS-PAGE.
SDS-PAGE and Western blot analysis
Samples were mixed with 4× loading buffer containing DTT and heated in a boiling water bath for 10 minutes prior to loading on tris-glycine gels with a 4% polyacrylamide stacking gel and a 15% polyacrylamide running gel (stock solutions made from: Biorad 30% Acrylamide/Bis Solution 29:1, Ammonium persulfate, TEMED, and TRIS base from Fisher). Gels were run at a constant current of 35 mA for approximately 1 hour. Upon completion, gels were either stained with coomassie and imaged with a UVP MultDoc-IT Digital Imaging System, or proteins were transferred (2 h at 200 mA) to a Hybond C nitrocellulose membrane (Amersham Biosciences) for Western blot analysis. Nitrocellulose membranes were incubated in blocking solution (5% milk powder in tris-buffered saline with 0.01% Tween-20) overnight at 4°C, followed by incubation with a 1:2,000 dilution of anti-6xHis epitope tagged antibody in blocking solution for 3 hrs at room temperature. Subsequently, a 1:10,000 dilution of anti-mouse HRPO conjugate in blocking solution was added for 30 min at room temperature. The membrane was washed with TBS, 0.01% Tween 20 prior to each incubation step and an Opti-4CN kit (Biorad) was used for detection.
Enzyme Linked Immunosorbent Assay (ELISA)
A 1:10 dilution of a commercially available mouse anti-6xHis tag antibody was made into a 50 mM Carbonate buffer, pH 9.6. Fifty microliters of this antibody solution was loaded into each well on a coated 96 well ELISA plate (Nunc) and incubated for approximately 2.5 hrs at 37°C. A PBS control containing no protein and each P22 capsid sample at 0.5 mg/mL were diluted 10 fold into buffer (containing 25 mM phosphate, 50 mM NaCl, 0.05% Tween 20, 2% w/v polyvinylpyrrolidone (MW 40,000), 0.2% w/v bovine serum albumin, and 0.02% sodium azide), 150 μL was added to each well, and the plate was incubated overnight at 4°C. The plate was subsequently incubated for 3 hrs at room temperature with a rabbit antibody recognizing the P22 coat protein in the same buffer used for the capsid incubation. One hundred microliters of anti-rabbit HRP conjugate (Biorad, 1:3,000 fold dilution in 25 mM phosphate, 50 mM NaCl, 0.05% Tween 20, 2% w/v polyvinylpyrrolidone (Mw 40,000), 0.2% w/v bovine serum albumin) was added to each well and incubated at room temperature for 1 hour. The plate was washed 5× with PBS, 0.05% Tween 20 after each incubation step. The presence of P22 capsids was detected with a OptEIA ELISA TMB Substrate Reagent Set (BD Biosciences), and the relative absorbance at 350 nm was recorded using a Spectra Max Plus 384 plate reader (Molecular Devices) with a SoftMax Pro 5.4.4 software package. A background absorbance value for the PBS control was subtracted from the each average recorded absorbance to yield the data presented in Figure 2C.
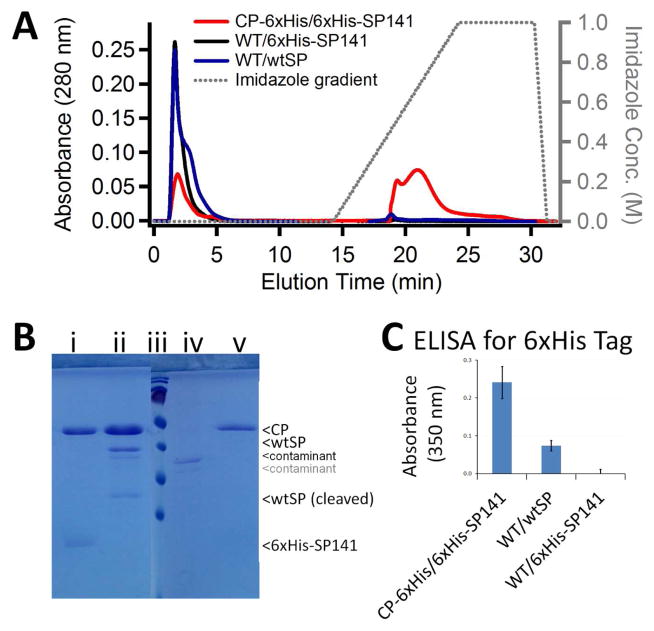
Figure 2.
Nickel chromatography and ELISA assays with assembled P22 procapsid to determine the exposure of 6xHis tags to the capsid exterior. A) Elution chromatograms of CP-6xHis/6xHis-SP141, wtCP/6xHis-SP141, and wtCP/wtSP from a Ni HisTrap column with increasing imidazole concentration. B) SDS-PAGE analysis of peaks in affinity chromatogram A: i) 2 min elution peak: wtCP/6xHis-SP141, ii) 2 min elution peak: wtCP/wtSP, iii) Page Ruler Molecualar Weight standard, and iv) 2min elution peak: CP-6xHis/6xHis-SP141 and v) 21 min elution peak: CP-6xHis/6xHis-SP141. C) Enzyme Linked Immunabsorbent Assay displaying the relative amount of each assembled P22 capsid that interacts with an anti-His antibody.
Size Exclusion Chromatography Coupled to Multi-Angle Light Scattering (SEC-MALS)
Separation on a size exclusion column (Wyatt Technologies, WTC-0200S) preceded detection using a Wyatt HELEOS Multi Angle Laser Light Scattering (MALLS) detector, equipped with a quasi-elastic light scattering detector (QELS) and an Optilab rEX differential refractometer (Wyatt Technology Corporation). Twenty five microliters of each capsid at approximately 1.5 mg/mL was injected through the SEC column on an Agilent 1200 HPLC with buffer (50mM phosphate, 100mM NaCl buffer, 3mM sodium azide) at a flow rate of 0.7 mL/min. Astra 5.3.14 software from Wyatt Technology Corporation was used to calculate the number average molecular weight (Mn) from the molecular weight distribution.
Reaction of P22 Cysteines with Fluorescein-5-Maleimide (F5M)
Fluorescein-5-Maleimide (Pierce) was reacted with 2 mg (3.1 mg−mL−1) of each: wtP22/mCherrySP141, wtP22/mCherrySP141(C140A), and CP+431C/mCherrySP141(C140A) procapsid. A 20 fold molar excess of F5M per P22 subunit was added dropwise from a stock solution in DMSO, and the reaction was stirred vigerously for 2 hours at room temperature. Pelleting of the procapsids through a sucrose cushion, followed by resuspension and an additional pelleting by ultracentrifugation in PBS buffer removed excess F5M from the samples. Unmodified and labeled samples were diluted 20× into denaturing buffer (6 M Guanidine HCl, 50mM Phosphate, 100mM NaCl at pH 7.7), mixed, and UV-visible spectra of the denatured protein samples were recorded approximately 10 minutes later. The UV-visible absorbance of a series of F5M concentrations in the denaturing buffer was measured to calculate the extinction coefficient of the F5M in these conditions.
Reaction of P22 Cysteines with N-ethyl Maleimide
N-ethylmaleimide (NEM, Pierce) was reacted with P22 capsids to block inter-capsid disulfide formation. From a stock solution of 50mg/mL NEM dissolved in dimethylformamide(DMF), a fifty fold molar excess of NEM per P22 subunit was added dropwise to 2 mg (0.5 mg mL−1) of each wtP22/mCherrySP141(C140A) and CP+431C/mCherrySP141(C140A) capsids in PBS pH 7.0. Controls for the labeling experiments were carried out by the addition of a corresponding volume of neat DMF. After stirring rapidly for 3 hours at room temperature, P22 capsids were diluted into pH 7.0 PBS buffer, mixed for 1 hr at 4°C, and pelleted through a 35% sucrose cushion in the ultracentrifuge (50 min at 45,000 × g) to remove unreacted NEM. Samples were subsequently resuspended and pelleted (50 min at 45,000 × g) a second time in pH 7.3 PBS buffer, and left rocking in PBS pH 7.3 overnight at 4°C. Samples of resuspended protein were analyzed from the supernatant solution after a 5 minute centrifugation step at 17,000 × g. Subsequently, 20 uL of DTT stock solution was added to reduce disulfides, resulting in a final concentration of 5mM DTT, and samples were left to resuspend overnight at 4°C. Samples of resuspended protein were again analyzed from the supernatant solution after a 5 minute centrifugation step at 17,000 × g.
Quartz Crystal Microbalance with Dissipation (QCM-D)
CP-K-coil and CP-E-Coil capsids were injected onto gold coated quartz crystal (Q-Sense, QSX301) positioned in a Q-Sense D300 (Q-Sense AB) quartz crystal microbalance with dissipation. Additions of 600 μL of each sample (CP-K-coil or CP-E-coil) at a concentration of 3.3 μg/mL in 50 mM phosphate, 100 mM NaCl, pH 7.0 was loaded onto the crystal and let equilibrate for 20 min at 25°C. A 20 minute buffer equilibration step at 25°C followed each capsid addition. The frequency and dissipation values were recorded using QSoft 301 software program (version 1.6.16.69) interfaced to the instrument.
Results
To probe the location of the structurally unresolved C-terminus of the coat protein (CP) in the bacteriophage P22 capsid, a series of genetic mutations were made and biochemically characterized. In the structural model of the P22 procapsid coat protein, the last 5 amino acids are not resolved and their location is ambiguous (Fig 1). However, the published models of the P22 capsid22–23 suggest that the C-terminus of the CP is directed towards the exterior of the capsid (S2).
Figure 1.

A representation of the assembled P22 procapsid (A) from the published cyroEM structure, which includes residues 1–425 of the 430 amino acid P22 coat protein. The residues 419–425 are highlighted in the zoomed versions of the pentamer(B) and hexamer(C) units. In both, it appears that the CP C-terminus extends toward the capsid exterior.
Genetic fusion of short peptides to the C-terminus of the CP was used to test the accessibility of this region to the exterior environment of the P22 procapsid. A 6x-histidine tag, a single cysteine residue, and two coiled coil peptides, were individually fused to the CP C-terminus to create the following new P22 coat protein constructs: CP-431C, CP-6xHis, CP-E-coil, and CP-K-coil. The genetic manipulation was confirmed by DNA sequencing. Each of the four new P22 coat proteins (CP-6xHis, CP-431C, CP-Ecoil, and CP-K-coil) were individually co-expressed with a P22 scaffolding protein (SP) to facilitate the self assembly of the procapsids in the E.coli expression system. The novel procapsids were purified using the same methodology as used for purification of the wtP22 procapsids19, which included ultracentrifugation through a sucrose cushion, followed by size exclusion chromatography (Sephacryl S-500, S3). Analysis of these materials by SDS page gel electrophoresis confirms the presence of both P22 coat protein and scaffold protein (S4).
No significant changes in particle assembly were observed upon the addition of amino acids to the C-terminus. The addition of the residues to the CP C-terminus, on the purified P22 constructs, was confirmed by liquid chromatography -mass spectroscopy (LC-MS) analysis (S5 and S6). By size exclusion chromatography (SEC), all constructs eluted at approximately 65 mL, which is consistent with the elution volume of the assembled wtP22 procapsid (S3). The fractions from this peak were pooled and subjected to HPLC-size exclusion chromatography coupled to multi-angle light scattering (MALS) and refractive index detection. Analysis of the light scattering revealed packaged procapsids with particle diameters reflecting that of assembled capsids for each sample (S7) and all of the capsids had particle RMS radii in the 23–30 nm range and hydrodynamic radii between 25 and 33nm. The mass of each procapsid calculated from multi-angle light scattering was observed to be between 22 and 30 MDa. (S7) The radii and mass ranges reflect slight differences in the construct size and packaging of cargo; each individual construct is highly monodisperse (S7).
To determine if the coat protein 6xHis tag is exposed on the capsid exterior, three different variants of the P22 procapsid were studied. These P22 variants included CP with C-terminal 6xHis tag combined with a scaffolding protein having a N-terminal 6xHis tag, a wild type CP combined with a scaffolding protein having a N-terminal 6xHis tag, and a wild type CP combined with a wild type scaffold protein (CP-6xHis/6xHis-SP141, wtCP/6xHis-SP141, and wtCP/wtSP, respectively). In each case, the scaffold protein is packaged on the interior of the P22 procapsid. The exposure of the CP-6xHis tag to the exterior of the capsids was probed by binding to a Ni chelate affinity column. The only construct that demonstrated binding to the Ni column was the CP-6xHis/6xHis-SP141, and these capsids could be eluted by an increasing imidazole gradient (Fig 2A). In contrast, the wtCP/6xHis-SP141 and wtCP/wtSP samples did not bind to the column (Fig 2A). The presence of P22 VLPs in all fractions was confirmed by SDS-PAGE gel (Fig 2B). The CP-6xHis/6xHis-SP141 sample exhibited a peak at 2mL corresponding to protein that didn’t bind to the column. Analysis of this peak by SDS-PAGE indicates both the absence of P22 CP-6xHis/6xHis-SP141 and the presence of a protein contaminant (Fig 2Biv) that appears on an SDS-PAGE gel of the sample prior to affinity chromatography (S8). The binding of CP-6xHis/6xHis-SP141, but not wtCP/6xHis-SP141 to the Ni column is a clear indication that the 6xHis tag on the CP is presented to the exterior of the P22 capsid. The lack of binding by the wtCP/6xHis-SP141 construct also strongly suggests that the scaffold protein is sequestered on the interior and does not sample the outside to any significant extent.
Additional data supporting the exposure of the CP-6xHis on the exterior of the procapsid was obtained via an ELISA sandwich assay utilizing a 6xHis tag antibody and a CP specific antibody. A 96 well plate was coated with anti-His tag antibody and each well was subsequently incubated with CP-6xHis/6xHis-SP141, wtCP/6xHis-SP141, or wtCP/wtSP capsids. The binding of each P22 construct to the anti-His antibody was probed by the CP antibody (rabbit) and subsequently quantified by a secondary goat-anti-rabbit antibody via a horseradish peroxidase/TMB colorimetric assay34. The highest absorbance was observed for the CP-6xHis/6xHis-SP141 construct, while the wtCP/6xHis-SP141 was similar to the background measurement of the assay (Fig 2C).
A western blot confirmed that the 6xHis tags were present on 6xHis-SP141, and that they could be recognized by the same anti-His antibody used in the ELISA (S8) but only after capsid denaturation. The coat and scaffold proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane, which was incubated with the mouse anti-His antibody. Bands containing 6xHis tags were subsequently detected using a rabbit-anti-mouse HRP conjugated secondary antibody. Bands corresponding to 6xHis tags on the coat and scaffold were both detectable, while neither the wtCP nor wtSP were highlighted (S8). Together, these data are consistent with the suggestion from the structural models and with binding of this construct to the Ni column, indicating that peptides fused to the CP C-terminus are accessible to the capsid exterior while the scaffolding protein is sequestered on the capsid interior. Demonstrating the display of the CP-6xHis on the capsid exterior provides the foundation for future utilization of the CP C-terminal location for the display of targeting peptides or antigenic epitopes.
To further investigate the accessibility of the C-terminus to the capsid exterior, a single cysteine was appended to the C-terminus of the CP (CP-431C/mCherrySP141(C140A)). The addition of a single cysteine residue is less likely to cause alterations in the procapsid structure than longer charged peptides. Successive quick change mutagenesis steps were carried out on a previously described vector19, resulting in the creation of two unique P22 constructs (wtCP/mCherrySP141 and wtCP/mCherrySP141(C140A), in route to obtaining CP-431C/mCherrySP141(C140A).
To demonstrate the utility of both the C-terminal CP-431C and scaffold protein cysteine (SP-C140) as a sites for chemical conjugation, wtCP/mCherrySP141, wtCP/mCherrySP141(C140A), and CP-431C/mCherrySP141(C140A) were each reacted with fluorescein-5-maleimide (F5M). The F5M labeling of each capsid was quantified by monitoring the UV-visible absorbance of unmodified and F5M reacted capsids under denaturing conditions (S9), after separation of free F5M from the capsids by ultracentrifugation through a sucrose cushion. Calculations indicated that F5M labeling resulted in 176 F5M per wtCP/mCherrySP141 capsid and 272 F5M per CP-431C/mCherrySP141(C140A), while only 40 F5M per capsid were observed for the wtCP/mCherrySP141(C140A) capsid (S9). Liquid chromatography-mass spectroscopy confirmed the labeling of CP-431C with F5M, while no F5M labeling of the CP was observed for the wtCP/mCherrySP141 or wtCP/mCherrySP141(C140A)(S10). These data demonstrate the cysteine packaged inside the wtCP/mCherrySP141 is a useful site for chemical conjugation, and the utility of the procapsid CP C-terminal cysteine chemical conjugation with imaging agents.
In an effort to further confirm the labeling of the wtCP/mCherrySP141, the mass spectroscopy data was analyzed. Under the chromatography and ionization conditions utilized, the P22 CP ionizes much more efficiently than mCherrySP141 or mCherrySP141(C140A). This results in very poor signal intensity and resolution for the peaks at corresponding to the mCherrySP141 (with a predicted mass of 45962 Da) and mCherrySP141(C140A) (with a predicted mass of 45994 Da) (S10). Therefore, any labeling of the scaffold protein could easily be in the baseline noise of the spectrum.
We probed the formation of disulfide bonds in this construct to investigate the ability of the single CP C-terminal cysteine to protrude to the exterior of the capsid, and result in the formation of intercapsid interactions. SEC purified CP-431C/mCherrySP141(C140A) was pelleted by ultracentrifugation and it was observed that, in contrast to the wtCP/mCherrySP141(C140A), this construct did not readily resuspend when incubated at 4°C overnight. Since the fluorescent protein mCherry is packaged on the capsid interior the difference in the sample behavior was easily observed qualitatively by eye (S11), and a comparison of the yield of resupended protein was quantified by UV-visible spectroscopy (S11). Relative to the wtCP/mCherrySP141(C140A), only 3% of CP-431C/mCherrySP141(C140A) was recovered (Fig 3A).
Figure 3.
Evidence for disulfide bond formation between CP-431C/mCherrySP141(C140A) capsids. A) Bar graph showing the relative yield of soluble CP-431C/mCherrySP141(C140A) protein compared to wtCP/mCherrySP141(C140A) after resuspension in PBS buffer, PBS buffer after blocking CP-431C/mCherrySP141(C140A) with N-ethylmaleimide (NEM) and in PBS buffer and with 5mM DTT. B) Mass spectrum of CP-431C/mCherrySP141(C140A) before and after labeling with NEM. C) Mass spectrum of wtCP/mCherrySP141(C140A) before and after labeling with NEM. The original m/z data is shown in S5 and S6.
To confirm that disulfide bond formation played a role in the inefficient resuspension of the CP-431C/mCherrySP141(C140A) construct, the capsid was treated with N-ethyl maleimide to block cysteine thiols. Labeling of the CP subunits was confirmed via liquid chromatography mass spectroscopy analysis, in which all CP subunits were observed to be modified for the CP-431C/mCherrySP141(C140A) construct (Fig 3B) while the wtCP/mCherrySP141(C140A) showed no labeling under the same reaction conditions (Fig 3C). Excess N-ethyl maleimide (NEM) was purified from the capsids via ultracentrifugation through a sucrose cushion, and SEC-MALS analysis revealed that the morphology of the capsids was retained after labeling (S7). Upon pelleting of the NEM labeled CP-431C/mCherrySP141(C140A), the capsid resuspended in PBS buffer with a recovery identical to wtCP/mCherrySP141(C140A) under these conditions (Fig 3A). This is supporting evidence for the creation of an inter-capsid network from exposed cysteines in CP-431C/mCherrySP141(C140A).
Having evidence for disulfide formation between capsids, we hypothesized that recovery of the CP-431C/mCherrySP141(C140A) construct might be accomplished through the addition of a reducing agent. Upon addition of dithiotheatol (DTT, 5 mM), the CP-431C/mCherrySP141(C140A) construct resuspended with an apparent 114% recovery compared to wt capsid under the same conditions (Fig 3A and S11). Dynamic light scattering measurements indicated the presence of intact procapsids (average radius of hydration, Rh= 58.6 ± 0.3 nm for wtCP/mCherrySP141(C140A); Rh= 59.3 ± 1.6 nm for CP-431C/mCherrySP141(C140A)) under these reducing conditions for both wtCP/mCherrySP141(C140A) and CP-431C/mCherrySP141(C140A) capsids (S12). This provides additional evidence for the existence of disulfide association between CP-431C/mCherrySP141(C140A) capsids.
Building on our demonstrations that peptide extensions to the CP C-terminus are displayed on the capsid exterior and can mediate interparticle interaction, we explored coiled coil motifs to direct interparticle association and assembly. Coiled coil peptides can be described as heptad repeats of amino acids that form superhelical structural motifs when mixed. Strong associations between peptides arise from packing of hydrophobic residues at the interface between the helices and stabilization of the supercoiled structure through electrostatic interactions.
A pair of anti-parallel heterodimeric coiled coil petides was used to transform individual P22 procapsids into building blocks for an extended network structure. Amino acid sequences corresponding to either a positive (CP-K-coil, +TR(VAALKEK)3) or negatively charged (CP-E-coil, +TS(VAALEKE)3) alpha helix of a three heptad heterodimeric coiled coil motif were appended to the C-terminus of the coat protein. Peptide sequences, VAALKEK3 and VAALEKE3, have been previously described to form a anti-parallel coiled coil heterodimer upon association25. The genetic amenability of the C-terminal location is reflected in the addition of these 23 residues without noticable alterations to the assembly of the P22 procapsid.
The inherent multivalent interactions between the shorter three heptad-coiled coils on each capsid promote significant capsid interactions, despite having a less thermally stable heterodimer than longer heptad coiled coil repeats35. We used the three-heptad system in an effort to avoid kinetic traps in material assembly, which are less probable in the particle-particle assembly based upon the cumulative effect of many weak interactions. Upon mixing of solutions containing 0.5 mg/mL of the CP-E-coil and CP-K-coil capsids, a significant increase in light scattering was observed within seconds (Fig 4A), indicating interaction and association of capsids within this mixture to form larger particulates. No increase in scattering was observed with control samples containing each of CP-E-coil or CP-K-coil individually, or the mixture of each coil construct with wtP22 capsid (Fig 4A). Single spectra of the CP-E-coil and CP-K-coil capsids at 1mg/mL individually, and after mixing are shown in S13.
Figure 4.

Interaction of P22 procapsids displaying Kcoil (+TR(VAALKEK)3) or E-coil (+TR(VAALEKE)3) peptides appended to the CP c-terminus. A) UVvisible spectroscopy showing an increase in scattering upon mixing of the E-coil and K-coil capsids relative to each capsid individually or mixtures with wtP22. B) Assembly of K-coil and Ecoil capsids on a surface was observed via quartz crystal microbalance with dissipation.
The capacity of the coiled coil interaction between the C-termini of adjacent procapsids to facilitate assembly on a surface, using a layer-by-layer approach, was monitored via quartz crystal microbalance with dissipation (QCM-D). The CP-K-coil (20 μg) was deposited on a gold-coated quartz sensor followed by a buffer wash and equilibration and subsequent addition of CP-E-coil (20 μg) and another buffer equilibration. This process was repeated two more times in the layer-by-layer deposition process (Fig 4B). Each addition of P22 resulted in a decrease in frequency (Δf) indicating deposition of the protein on the crystal surface and was also accompanied by an increase in the dissipation (D) at each step.
Conclusions
Here we have shown that the C-terminal region of the coat protein from the bacteriophage P22 is indeed exposed to the exterior environment of the capsid. From the available cryo-electron microscopy models, the exact location of the C-terminus of the CP is slightly ambiguous. Using genetic manipulation of the C-terminus, including the addition of a single cysteine residue to the C-terminus, incorporation of a 6xHis motif, and genetic fusion of complementary coiled-coil peptides we have established that these modifications to the C-terminus are exposed to the exterior of the capsid. This greatly enhances the ability to use the P22 capsid system as a functional nanomaterial and we have demonstrated this unique site on the capsid can be used for small molecule chemical conjugation, as well as for fabrication of capsid-based extended materials through the directed interparticle interaction mediated by disulfide bond formation and directed coiled-coil interactions.
Supplementary Material
Acknowledgments
The authors wish to thank Rui Li for the anti-CP antibody used in the ELISA and Western Blots, Dustin Patterson for helpful discussions about the assembly of materials via coiled coil interactions, and Ravi Kant for assistance with the QCM-D instrumentation. This work was supported in part by grants from the National Institutes of Health (NIBIB R01-EB012027) and the U.S. Department of Energy, Office of Basic Energy Sciences, Division of Materials Science and Engineering (DE-FG02-07ER46477).
Footnotes
Supporting Information Available: Additional results are available in the Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Chen C, Daniel MC, Quinkert ZT, De M, Stein B, Bowman VD, Chipman PR, Rotello VM, Kao CC, Dragnea B. Nano Lett. 2006;6:611–615. doi: 10.1021/nl0600878. [DOI] [PubMed] [Google Scholar]
- 2.Ma YJ, Nolte RJM, Cornelissen J. Adv Drug Delivery Rev. 2012;64:811–825. doi: 10.1016/j.addr.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 3.de la Rica R, Matsui H. Chem Soc Rev. 2010;39:3499–3509. doi: 10.1039/b917574c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrico IS, Kirshenbaurn K. Nat Nanotechnol. 2009;4:14–15. doi: 10.1038/nnano.2008.389. [DOI] [PubMed] [Google Scholar]
- 5.Kirshenbaum K, Zuckermann RN, Dill KA. Curr Opin Struct Biol. 1999;9:530–535. doi: 10.1016/S0959-440X(99)80075-X. [DOI] [PubMed] [Google Scholar]
- 6.Aniagyei SE, Kennedy CJ, Stein B, Willits DA, Douglas T, Young MJ, De M, Rotello VM, Srisathiyanarayanan D, Kao CC. Nano Lett. 2009;9:393. doi: 10.1021/nl8032476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Y, Chiang CY, Lee SK, Gao Y, Hu EL, Yoreo JD, Belcher AM. Nano Lett. 2005;5:1429–1434. doi: 10.1021/nl050795d. [DOI] [PubMed] [Google Scholar]
- 8.Royston E, Ghosh A, Kofinas P, Harris MT, Culver JN. Langmuir. 2008;24:906–912. doi: 10.1021/la7016424. [DOI] [PubMed] [Google Scholar]
- 9.Stephanopoulos N, Liu M, Tong GJ, Li Z, Liu Y, Yan H, Francis MB. Nano Lett. 2010;10:2714–2720. doi: 10.1021/nl1018468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q, Lin T, Johnson JE, Finn M. Chem Biol. 2002;9:813–819. doi: 10.1016/s1074-5521(02)00166-7. [DOI] [PubMed] [Google Scholar]
- 11.Yoo PJ, Zacharia NS, Doh J, Nam KT, Belcher AM, Hammond PT. ACS Nano. 2008;2:561–571. doi: 10.1021/nn700404y. [DOI] [PubMed] [Google Scholar]
- 12.Kostiainen MA, Hiekkataipale P, Jose Á, Nolte RJ, Cornelissen JJ. J Mater Chem. 2011;21:2112–2117. [Google Scholar]
- 13.Uchida M, Klem MT, Allen M, Suci P, Flenniken M, Gillitzer E, Varpness Z, Liepold LO, Young M, Douglas T. Adv Mater. 2007;19:1025–1042. [Google Scholar]
- 14.Capehart SL, Coyle MP, Glasgow JE, Francis MB. J Am Chem Soc. 2013;135:3011–3016. doi: 10.1021/ja3078472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patterson DP, Prevelige PE, Douglas T. ACS Nano. 2012;6:5000–5009. doi: 10.1021/nn300545z. [DOI] [PubMed] [Google Scholar]
- 16.Kang S, Lander GC, Johnson JE, Prevelige PE. Chembiochem. 2008;9:514–518. doi: 10.1002/cbic.200700555. [DOI] [PubMed] [Google Scholar]
- 17.Earnshaw W, Casjens S, Harrison SC. J Mol Biol. 1976;104:387–410. doi: 10.1016/0022-2836(76)90278-3. [DOI] [PubMed] [Google Scholar]
- 18.Parker MH, Casjens S, Prevelige PE. J Mol Biol. 1998;281:69–79. doi: 10.1006/jmbi.1998.1917. [DOI] [PubMed] [Google Scholar]
- 19.O’Neil A, Reichhardt C, Johnson B, Prevelige PE, Douglas T. Angew Chem, Int Ed. 2011;50:7425–7428. doi: 10.1002/anie.201102036. [DOI] [PubMed] [Google Scholar]
- 20.Lucon J, Qazi S, Uchida M, Bedwell GJ, Lafrance B, Prevelige PE, Jr, Douglas T. Nat Chem. 2012;4:781–8. doi: 10.1038/nchem.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang S, Uchida M, O’Neil A, Li R, Prevelige PE, Douglas T. Biomacromolecules. 2010;11:2804–2809. doi: 10.1021/bm100877q. [DOI] [PubMed] [Google Scholar]
- 22.Chen DH, Baker ML, Hryc CF, DiMaio F, Jakana J, Wu WM, Dougherty M, Haase-Pettingell C, Schmid MF, Jiang W, Baker D, King JA, Chiu W. Proc Natl Acad Sci USA. 2011;108:1355–1360. doi: 10.1073/pnas.1015739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parent KN, Sinkovits RS, Suhanovsky MM, Teschke CM, Egelman EH, Baker TS. Phys Biol. 2010;7 doi: 10.1088/1478-3975/7/4/045004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altunbas A, Pochan DJ. Peptide-Based Materials. Springer; 2012. pp. 135–167. [DOI] [PubMed] [Google Scholar]
- 25.Pechar M, Pola R, Laga R, Ulbrich K, Bednarova L, Malon P, Sieglova I, Kral V, Fabry M, Vanek O. Biomacromolecules. 2011;12:3645–3655. doi: 10.1021/bm200897b. [DOI] [PubMed] [Google Scholar]
- 26.Minten IJ, Nolte RJM, Cornelissen J. Macromol Biosci. 2010;10:539–545. doi: 10.1002/mabi.201000030. [DOI] [PubMed] [Google Scholar]
- 27.Patterson DP, Desai AM, Holl MMB, Marsh ENG. RSC Adv. 2011;1:1004–1012. doi: 10.1039/C1RA00282A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kopecek J, Yang JY. Angew Chem, Int Ed. 2012;51:7396–7417. doi: 10.1002/anie.201201040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fletcher JM, Harniman RL, Barnes FR, Boyle AL, Collins A, Mantell J, Sharp TH, Antognozzi M, Booth PJ, Linden N, Miles MJ, Sessions RB, Verkade P, Woolfson DN. Science. 2013;340:595–599. doi: 10.1126/science.1233936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Apostolovic B, Danial M, Klok H-A. Chem Soc Rev. 2010;39:3541–3575. doi: 10.1039/b914339b. [DOI] [PubMed] [Google Scholar]
- 31.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 32.Prevelige PE, Jr, Thomas D, King J. J Mol Biol. 1988;202:743–757. doi: 10.1016/0022-2836(88)90555-4. [DOI] [PubMed] [Google Scholar]
- 33.Kirsch RD, Joly E. Nucleic Acids Res. 1998;26:1848–1850. doi: 10.1093/nar/26.7.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frey A, Meckelein B, Externest D, Schmidt MA. J Immunol Methods. 2000;233:47–56. doi: 10.1016/s0022-1759(99)00166-0. [DOI] [PubMed] [Google Scholar]
- 35.Pechar M, Pola R. Biotechnol Adv. 2013;31:90–96. doi: 10.1016/j.biotechadv.2012.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.