Summary
Obligate anaerobes are periodically exposed to oxygen, and it has been conjectured that on such occasions their low-potential biochemistry will predispose them to rapid ROS formation. We sought to identify scavenging enzymes that might protect the anaerobe Bacteroides thetaiotaomicron from the H2O2 that would be formed. Genetic analysis of eight candidate enzymes revealed that four of these scavenge H2O2 in vivo: rubrerythrins 1 and 2, AhpCF, and catalase E. The rubrerythrins served as key peroxidases under anoxic conditions. However, they quickly lost activity upon aeration, and AhpCF and catalase were induced to compensate. The AhpCF is an NADH peroxidase that effectively degraded low micromolar levels of H2O2, while the catalytic cycle of catalase enabled it to quickly degrade higher concentrations that might arise from exogenous sources. Using a non-scavenging mutant we verified that endogenous H2O2 formation was much higher in aerated B. thetaiotaomicron than in Escherichia coli. Indeed, the OxyR stress response to H2O2 was induced when B. thetaiotaomicron was aerated, and in that circumstance this response was necessary to forestall cell death. Thus aeration is a serious threat for this obligate anaerobe, and to cope it employs a set of defenses that includes a repertoire of complementary scavenging enzymes.
Keywords: Hydrogen peroxide, Bacteroides thetaiotaomicron, catalase, alkyl hydroperoxide reductase, rubrerythrin, OxyR
Introduction
Microbes exhibit stark differences in their abilities to tolerate oxygen, and this fact profoundly shapes microbial communities throughout the biosphere. Open seas and surface soils are typically saturated with air, and the microbes that dwell in these environments can not only tolerate the ambient 200–300 μM oxygen, but they exploit its oxidizing potential in driving their metabolism. In contrast, oxygen has limited access into many other environments, including subsurface sediments and eukaryotic host tissues; in these habitats the resident oxygen-respiring microbes drive the local oxygen concentration to low micromolar levels. Found here are microaerophilic bacteria, which rely upon oxygen for aerobic-style metabolism but which cannot tolerate the higher levels of oxygen that pertain in fully open environments. At submicromolar oxygen levels obligate anaerobes predominate. These bacteria and archaea do not employ oxygen as a substrate at all, and their growth quickly ceases when even small amounts are introduced.
These community structures are unstable in nature, however, because episodic events expose anaerobes to more oxygen than their metabolism can tolerate. Rainstorms cause air-saturated waters to penetrate hypoxic sediments. Hosts periodically excrete microbial flora, which must then survive oxygen exposure long enough to find a new host to colonize. The evolutionary success of obligate anaerobes attests that they have ways to cope with these stresses.
To unravel how anaerobes withstand these events, it will be necessary to consider why oxygen is toxic to them. In some cases molecular oxygen can directly inactivate a few specialized enzymes that employ glycyl radicals or exposed low-potential metal centers for activity. Pyruvate:formate lyase and anaerobic ribonucleotide reductase are examples of the first group, and pyruvate:ferredoxin oxidoreductase and nitrogenase are examples of the second (Sawers & Watson, 1998, Pieulle et al., 1997).
Perhaps a more-common mechanism of damage involves the generation and reactivity of reactive oxygen species (ROS) such as superoxide (O2−) and hydrogen peroxide (H2O2). These species are formed in the extracellular bulk solution when molecular oxygen chemically oxidizes reduced metals and thiols, a likely event upon the mixing of erstwhile oxic and anoxic water columns. Although O2− is charged and cannot enter cells (Lynch & Fridovich, 1978, Korshunov & Imlay, 2002, H2O2 can readily do so {Seaver, 2001 #2572), and so it may comprise a threat to the flora in abruptly oxygenated habitats.
ROS are also formed intracellularly when oxygen adventitiously steals electrons from the reduced flavins and metal centers of redox enzymes (Ballou et al., 1969, Messner & Imlay, 1999, Massey et al., 1969). Both O2− and H2O2 are potent univalent oxidants, and they readily attack iron cofactored enzymes that are widespread in metabolism (Kuo et al., 1987, Flint et al., 1993, Jang & Imlay, 2007, Sobota & Imlay, 2011, Anjem & Imlay, 2012, Gu & Imlay, 2013). The inactivation of these enzymes imposes bottlenecks in the pathways to which they belong and is responsible for catabolic and biosynthetic problems in oxidatively stressed aerobes. Strikingly, obligate anaerobes may have high titers of low-potential flavoproteins, such as fumarate reductase, that are easily oxidized by molecular oxygen (Imlay, 1995, Messner & Imlay, 2002). This circumstance suggests that obligate anaerobes might suffer especially high rates of intracellular ROS formation when they are aerated.
Thus obligate anaerobes might be prone to acute ROS stress when they are exposed to oxygen. If enzyme damage were the sole problem, bacteria could enter a period of stasis and ride out oxygen exposure until they enter a more permissive hypoxic environment. However, a more existential threat is the formation of hydroxyl radicals (HO·), which are generated through the Fenton reaction between intracellular ferrous iron and H2O2:
Hydroxyl radicals react at nearly diffusion-limited rates with virtually all organic molecules, and their reactions with DNA enable oxidative stress to be lethal. Therefore, even quiescent bacteria must avoid intracellular H2O2 accumulation.
Bacteroides thetaiotaomicron is a gram-negative obligate anaerobe that is among the most prominent microbes in the human gastrointestinal tract. It also is an opportunistic pathogen that can invade and colonize erstwhile oxic tissues when trauma releases it from the intestine, a phenomenon that confirms that this anaerobe can tolerate some period of oxygen exposure. Early surveys (McCord et al., 1971) led to the notion, still propounded in some textbooks, that obligate anaerobes are deficient in scavenging enzymes. However, more-recent work has identified a number of potential peroxidases that might keep H2O2 in check. Examination of the B. thetaiotaomicron genome reveals a large number of plausible candidates: monofunctional catalase and Ahp (NADH peroxidase), which have been demonstrated to be the primary scavengers in Escherichia coli (Seaver & Imlay, 2001a); rubrerythrins, which are iron-dependent enzymes typically found in obligate anaerobes (LeGall et al., 1988, Kurtz, 2006); homologs of bacterioferritin comigratory protein (Bcp) (Jeong et al., 2000), thiol peroxidase (Cha et al., 1995, Hall et al., 2009), and bacterial glutathione peroxidase (Arenas et al., 2010), which are found among aerobes as well; and cytochrome c peroxidase (Schutz et al., 2011), which may redirect respiratory electron flow towards H2O2 elimination. However, the evidence supporting these assignments is mostly limited to biochemical assays (Mishra & Imlay, 2012). Several of these enzymes are found in E. coli but evidently do not confer detectable scavenging activity in vivo, at least under the conditions that have been tested (Seaver & Imlay, 2001a).
In this study we sought to understand how B. thetaiotaomicron copes with intracellular H2O2. We establish that this anaerobe uses a surprising number of enzymes to scavenge this oxidant, although their expression patterns and specific roles apparently differ. Evidence shows that H2O2 formation is indeed rapid upon aeration, and so the ensuing induction of H2O2 defenses is critical for the continued viability of the cell. These results support the emerging view that, contrary to initial conjecture, obligate anaerobes are highly evolved to deal with oxidative stress.
Results
Scavenging of H2O2 in anaerobic cells
Bacteroides thetaiotaomicron is regarded as an obligately anaerobic bacterium because it does not form colonies on either rich or defined medium plates when incubated in the presence of air. Genomic inspection reveals that its sole ribonucleotide reductase is of the oxygen-sensitive NrdD type; the lack of any member of the oxygen-tolerant classes of ribonucleotide reductase should preclude replication when oxygen is present and consign B. thetaiotaomicron to anaerobic niches. When log-phase anoxic cells were aerated, growth immediately slowed, and it ceased shortly thereafter (Fig. 1A). Nevertheless, the cells remained viable for at least 20 hours (Fig. 1B). When cells were returned to anoxic conditions, growth resumed after a one-hour lag (Fig. 1C).
Fig. 1. Wild-type cells of B. thetaiotaomicron fail to grow yet survive prolonged exposure to air.
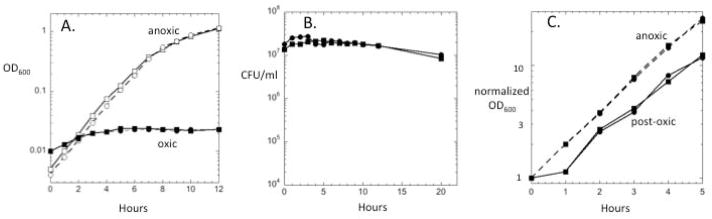
Exponentially growing cells in anoxic medium were subcultured at time zero into anoxic or oxic medium. Wild-type cells (BT5482, squares) and its Δtdk derivative (circles) are shown. (A) Biomass. (B) Viable cells. (C) After 4 hours of aeration, cells were collected and resuspended in anoxic medium (“post-oxic”). Anoxic cells are presented for comparison; all data were normalized to the initial OD600 to facilitate comparison. Data shown are representative of three independent experiments.
Survival during prolonged hours of aeration is presumably necessary for B. thetaiotaomicron to leave one mammalian host and then successfully colonize the gut of another. An important consequence of air exposure is the intracellular formation of oxygen-derived species such as O2− and H2O2. Of these, H2O2 is the greater threat to survival, since the sole known mechanism by which oxidants kill bacteria is through H2O2-mediated DNA damage. B. thetaiotaomicron is an iron-rich bacterium, and it has been proposed that H2O2 formation might be rapid when oxygen is introduced into the medium. In this scenario H2O2 scavenging might be an important element of cell survival.
Measurements confirmed that B. thetaiotaomicron quickly degrades H2O2 that is added to the growth medium (Fig. 2). BLAST analysis of the B. thetaiotaomicron genome sequence, using the protein sequences of both heme- and mangano-catalase homologs as queries, revealed a single catalase homolog, of the monofunctional class (Table S1). A homolog of the primary scavenging enzyme in E. coli, AhpCF, was also apparent. However, a mutant that lacked these enzymes (ΔahpC ΔkatE) still efficiently degraded H2O2 (Fig. 2). Alpha-ketoacids such as pyruvate chemically degrade H2O2; therefore, in order to rule out the possibility that any excreted metabolite conferred the scavenging activity, we checked whether H2O2 was degraded over time after it was added to the spent medium of BT5482 wild-type and ΔkatE ΔahpC strains. The H2O2 concentration did not change over 30 minutes (data not shown).
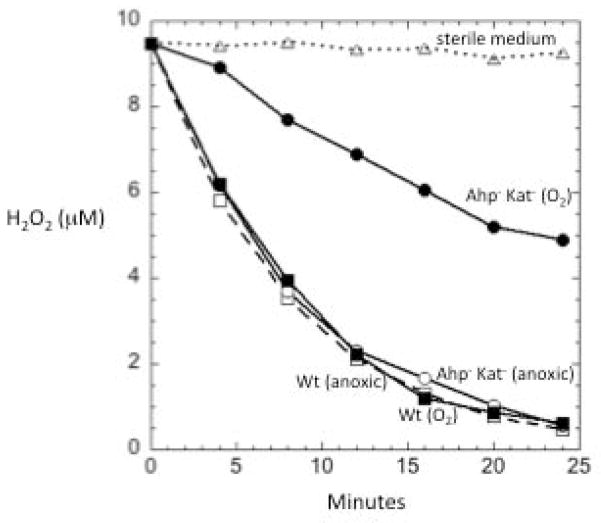
Fig. 2. Anaerobically growing cells of B. thetaiotaomicron possess substantial scavenging activity.
Anaerobically growing cells (OD = 0.1) in BHIS medium were washed and suspended in anoxic PBS/glucose, and 10 μM of H2O2 was added. At time-points the residual H2O2 was measured. Oxic samples (O2) represent cultures that had been aerated for 1 hour in BHIS prior to suspension in PBS/glucose and the addition of H2O2. Squares, wild-type cells; circles, Ahp− Kat− mutants. Solid symbols: oxic; open symbols, anoxic. The data are representative of at least three replicates.
Searches were then conducted for homologs of a wide range of enzymes that have been proposed to serve as peroxidases, including bacterioferritin comigratory protein (BCP), cytochrome c peroxidase (CcP), thioredoxin peroxidase (Tpx), glutathione peroxidase (Gpx), rubrerythrin (Rbr), and reverse rubrerythrin (rev-Rbr). The target sequences for BCP, CcP, Tpx and Gpx were taken from E. coli, whereas Rbr and rev-Rbr targets were chosen from Clostridium spp. and Desulfovibrio spp. protein sequences, as these enzymes are exclusively present in anaerobic bacteria and archaea. We found ORFs homologous to these six peroxidases (in addition to Ahp) (Table S1). Sund and co-workers reported the presence of thioredoxin peroxidase and Dyp-peroxidase in a related bacterium, Bacteroides fragilis (Sund et al., 2008), but these genes were absent from B. thetaiotaomicron. B. fragilis is a notably more oxygen-tolerant bacterium, and its genome includes a ribonucleotide reductase of the oxygen-dependent NrdAB type; therefore, its strategies for dealing with oxygen may differ substantially from those of B. thetaiotaomicron, a committed obligate anaerobe.
The authenticity of these various enzymes as peroxidases has not yet been verified in vivo. While E. coli encodes BCP, CcP, Tpx, and Gpx, the elimination of the catalases and of Ahp is sufficient to eliminate all significant H2O2 scavenging under the conditions that have been tested to date, suggesting either that the other enzymes were not expressed or that their true physiological role is not to degrade H2O2 (Seaver & Imlay, 2001a). In contrast, katE ahpCF mutants of B. thetaiotaomicron continued to degrade H2O2. As a preliminary approach to identify genes that might be involved in H2O2 scavenging, we measured the transcript level of candidate genes coding for catalase and the seven peroxidases in response to H2O2 treatment. The katE, ahpC, tpx, gpx, rbr1 and rbr2 genes were each induced 5- to 40-fold (Fig. 3). Interestingly, transcription of all these genes except rbr2 was significantly increased upon air exposure as well. In contrast, the transcript levels of bcp and ccp remain unchanged in the stressed cultures, which suggested that they are less likely to encode dedicated H2O2 scavengers.
Fig. 3. Responses of potential scavenging enzymes to H2O2 stress and aeration.
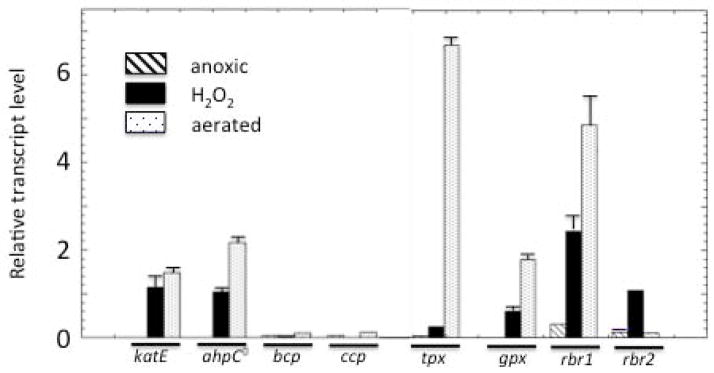
Exponentially growing wild-type cells in anoxic BHIS medium remained anoxic, were exposed to two doses of 50 μM H2O2 at 15 minute intervals, or were aerated with vigorous shaking for 30 minutes. mRNA was extracted and quantified. Error bars represent the SD from the mean of three independent experiments.
The catalytic activity of catalase can be assayed easily in the cell extracts. Activity was not detected in lysates of anaerobically cultured cells, but 2–3 U/mg activity was present in lysates of cultures that had been aerated for 1.5 hours prior to harvesting (Fig. S1); this pattern matches that of katE expression. This activity level is similar to that of aerobic E. coli (Linn & Imlay, 1987). Mutants with deletions in katE had no detectable activity, confirming that this gene encodes the sole catalase. In general, the strong induction of certain scavenger genes in response to aeration suggested that B. thetaiotaomicron might use different scavenging strategies under anoxic and oxic conditions.
In order to identify the key scavenging enzymes in anaerobic cells, we started by making single deletions of the candidate genes. Log-phase cells were washed and suspended in anaerobic PBS containing 0.05% glucose. The complete growth medium was not used because some components interfere with the assay system; however, glucose was included so that bacterial metabolism would be capable of providing electrons to the putative cellular peroxidases. Under these conditions, Δrbr2 cells degraded H2O2 more slowly than did the parent strain, although significant scavenging persisted (Fig. 4A). No other single knockout mutant showed any negative effect on H2O2 scavenging activity. It is important to note that the loss of a minor scavenger may have no effect on the rate at which H2O2 is cleared from the suspension medium: when intracellular scavenging is fast, the rate-limiting step in H2O2 clearance is the influx into the cell(Seaver & Imlay, 2001b). Thus these data tentatively implicated Rbr2, but they also indicated the participation of at least one other scavenger that could not be identified from single-mutant data.
Fig. 4. Roles of scavenging enzymes in H2O2 degradation under anoxic conditions.
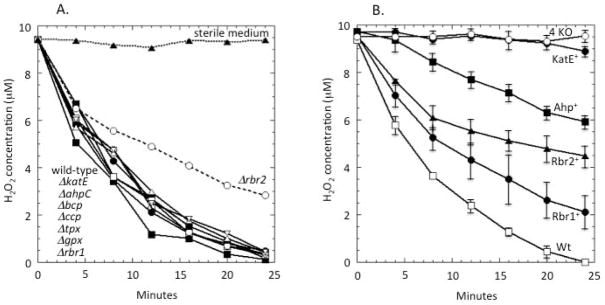
(A) H2O2 was added to anoxic cells, and degradation was monitored. Deletion of rbr2 lowers the H2O2 scavenging rate; the other single mutants and the wild-type strain still scavenge H2O2 at the rate at which it enters the cell. (B) A strain lacking all four scavenging enzymes (ΔkatE ΔahpC Δrbr1 Δrbr2, labelled 4 KO) does not scavenge H2O2. Other curves represent wild-type (WT) or triple-mutant strains that retain only the indicated scavenging enzyme. Data presented are representative of three independent experiments.
The various candidate mutations were assembled in numerous combinations. The final positive results are shown in Fig. 4B. Quadruple mutants lacking katE, ahpC, rbr1, and rbr2 were devoid of H2O2 scavenging activity under anaerobic conditions. Catalase provided very little activity, but strains containing any one of ahp, rbr1, or rbr2 did degrade H2O2 at a significant rate. Assuming that the absence of one gene had no effect upon the expression of the others, the data imply that in anaerobic cells the rubrerythrins are the primary scavengers, while Ahp plays a minor role and catalase is virtually absent.
The quadruple mutant strain (katE ahpC rbr1 rbr2) grew at normal rates under anaerobic conditions, supporting the idea that the primary or exclusive role of these proteins is to protect the cell from H2O2. The mutant exhibited sensitivity to H2O2 on zone-of-inhibition plates (Fig. S2).
Many bacteria induce the synthesis of scavenging enzymes as they enter stationary phase (Schellhorn & Hassan, 1988). The accepted rationale is that starved microbes need pre-synthesized enzymes to deal with any H2O2 that comes along, since they would be ineffective at enzyme induction. Catalase is the most prominent of the stationary-phase induced scavengers. However, stationary-phase B. thetaiotaomicron still depended almost exclusively upon rubrerythrins and Ahp for scavenging (Fig. S3).
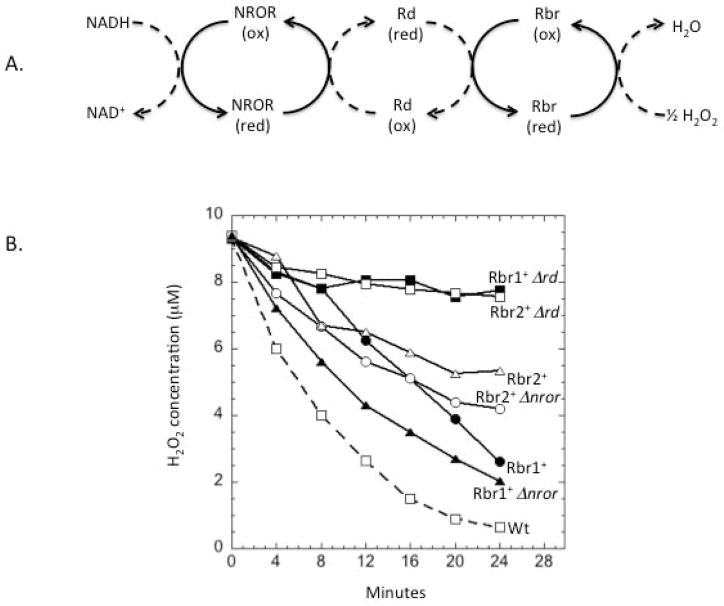
Like all peroxidases, rubrerythrins rely upon an electron donor for turnover. Workers have proposed that electrons can flow from the NADH pool through NADH:rubredoxin oxidoreductase (NROR) to rubredoxin, and from this metalloprotein to rubrerythrins. This pathway (Fig. 5A) has been successfully reconstituted in vitro with purified proteins from C. acetobutylicum (Kawasaki et al., 2009, Riebe et al., 2009).
Fig. 5. Rubredoxin is an essential component of Rbr1 and Rbr2 scavenging pathways.
(A) Schematic of electron transfer pathway of rubrerythrins anticipated from the literature (Riebe et al., 2007, Kawasaki et al., 2009). NROR, NADH:rubredoxin oxidoreductase; Rd, rubredoxin. (B) Measured scavenging of H2O2 by anaerobic cells containing only Rbr1 or Rbr2 as scavenging enzymes. The data are representative of four independent experiments.
A BLAST search revealed that BT_2539 is 88% homologous to C. acetobutylicum rubredoxin and BT_2434 is 41% homologous to C. acetobutylicum NROR. The addition of a Δrd deletion to a ΔkatE ΔahpC Δrbr1 strain eliminated Rbr2-dependent scavenging, and the addition of the same deletion to a ΔkatE ΔahpC Δrbr2 strain eliminated Rbr1-dependent scavenging (Fig. 5B). These data confirm that these enzymes rely upon rubredoxin as their electron source. However, ΔkatE ΔahpC Δrbr1 Δnror and ΔkatE ΔahpC Δrbr2 Δnror mutants scavenged H2O2 as efficiently as did their nror+ counterparts. This indicates that NROR is not an obligatory component of the rubrerythrin reduction pathways. Based on these results, a modified version of the rubrerythrin redox pathway is shown in Fig. S4.
Scavenging in aerated cells
The strong induction of genes upon aeration led to the expectation that scavenging mechanisms might differ in aerobic cells. This idea was especially attractive because peroxidases require active metabolism in order to acquire the electrons they need for turnover, and we thought that electron delivery might be compromised when oxygen poisons parts of cell metabolism.
H2O2 scavenging in cells that had been aerated for 1 hour relied more strongly upon Ahp than upon rubrerythrins (Fig. 6A). The capabilities of the rubrerythrins progressively declined after aeration, with Rbr1 activity disappearing within 10 min and Rbr2 activity failing within 20 min in cells that had no other scavengers (Fig. 6B). In contrast, wild-type cells—containing Ahp and catalase—continued to degrade H2O2. Surprisingly, Ahp continued to scavenge H2O2 even 12 hours after aeration (Fig. 6C), refuting our hypothesis that oxidative stress would eradicate all peroxidase activities. Clearly the cell is capable of generating enough NADH to drive this enzyme even when normal metabolism is poisoned by oxygen. In that light the strong induction of ahpCF upon aeration makes sense.
Fig. 6. Roles of scavenging enzymes in H2O2 degradation under fully oxic conditions.
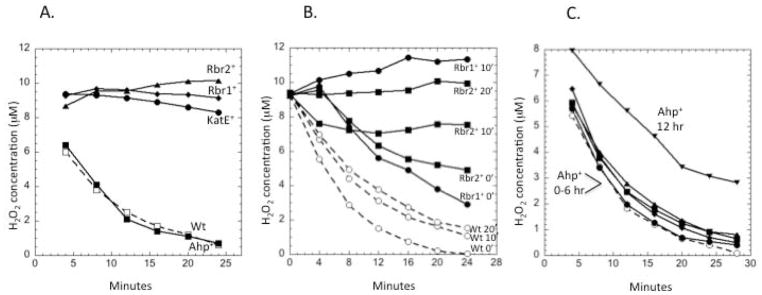
(A) Strains were aerated for 1 hour prior to measurements of H2O2 scavenging. The data represent wild-type (wt) cells plus triple mutants expressing only the indicated scavenging enzyme. (B) Rubrerythrins become non-functional upon aeration. Cells were aerated for the indicated times before H2O2 scavenging was tested. (C) Triple mutants (ΔkatE Δrbr1 Δrbr2) expressing only Ahp as a scavenging enzyme were aerated for 0, 2, 4, 6, and 12 hours prior to the testing of scavenging activity. The time courses of cells aerated for 0–6 hours did not differ significantly. The data presented are representative of multiple replicates.
If Ahp retains function in aerobic cells, why don’t the rubrerythrins? One possibility was that the induction of rubredoxin:oxygen oxidoreductase (ROO) might divert electron flow away from the rubrerythrins by competing for reduced rubredoxin. ROO scavenges oxygen, presumably to help restore an anoxic environment that would allow cell growth to resume (Wildschut et al., 2006, Hillmann et al., 2009, Kawasaki et al., 2009). We knocked out roo in ahpC katE mutants and tested whether the rubrerythrin activities would persist after aeration. However, while some improvement was observed, their scavenging action still diminished (Fig. S5). We speculate instead that rubrerythrin activities may primarily fail either because metabolic blocks hinder electron delivery to rubredoxin or because H2O2 itself reacts with off-cycle intermediates of the enzyme (see Discussion).
Why does B. thetaiotaomicron express both catalase and peroxidase systems to scavenge H2O2?
The presence of multiple enzymes to degrade H2O2 seems unnecessary. None of the B. thetaiotaomicron Ahp, catalase, or rubrerythrin enzymes have secretory leader sequences, and so they all appear to be devoted to scavenging H2O2 in the cytoplasmic compartment. However, a key difference between catalase and peroxidase enzymes lies in their catalytic cycles. Peroxidases require only a single H2O2 substrate molecule to complete their catalytic cycle, whereas catalases require two:
The problem with the latter arrangement is that when H2O2 levels fall as a result of scavenging, the intermediate ferryl/radical form of the enzyme will accumulate. These species are strong oxidants, and in the absence of H2O2 they can abstract electrons from the protein polypeptide, irreversibly inactivating it. Monofunctional catalases of the KatE type typically bind NAD(P)H as a reductant that helps quench this intermediate (Hillar & Nicholls, 1992). Nevertheless, catalases are therefore non-optimal scavengers when H2O2 levels decline. Peroxidases are typically used in their place. The complementary problem with peroxidases is that when H2O2 levels are high, the rate of H2O2 entry into the cell can outstrip the rate at which cells can provide electrons to the peroxidase. Thus H2O2 scavenging reaches a limit, and continuing turnover of peroxidases is not only fruitless but expends reductants that might be more productively used to generate energy or drive biosynthetic pathways. Perhaps for this reason, when H2O2 levels are high Ahp has been shown to be inactivated by overoxidation of its catalytic thiol to a sulfinic acid. This tendency towards overoxidation has been argued to be an evolutionarily selected trait (Sayed & Williams, 2004, Saccoccia et al., 2012).
Saturation of turnover of Ahp from E. coli has been documented (Seaver & Imlay, 2001a). To evaluate whether catalase is the favored scavenger at high concentrations of H2O2, we evaluated the rate at which aerobic B. thetaiotaomicron degraded 1 mM H2O2. Wild-type cells were able to do so, whereas katE mutants could not, indicating that Ahp was inactive (Fig. 7A). When H2O2-treated cells were returned to anaerobic conditions in the presence of chloramphenicol, they did not regain the ability to use Ahp or Rbr2, indicating that they had been damaged (Fig. 7B). We conclude that B. thetaiotaomicron employs catalase as the preferred scavenger of high concentrations of H2O2. Rubrerythrins are used to scavenge low levels of H2O2 that enter anaerobic cells, whereas Ahp is the primary scavenger of trace H2O2 in oxic habitats.
Fig. 7. Catalase E is the sole scavenger that acts on high concentrations of H2O2.
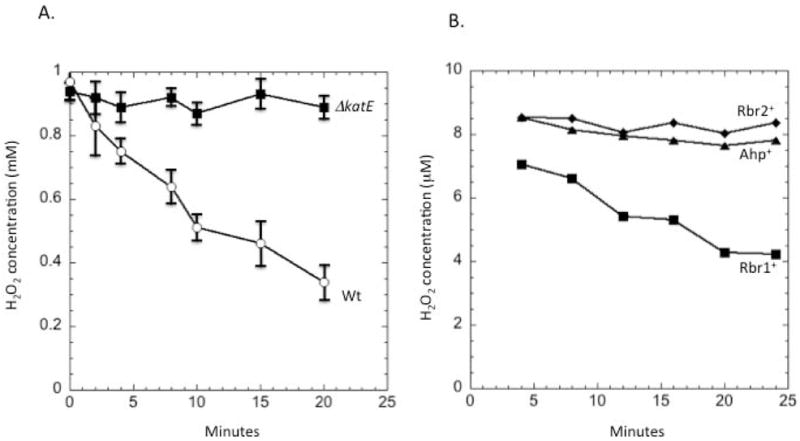
(A) Wild-type and ΔkatE mutant strains were aerated for 1.5 hours to induce aerobic expression of scavenging enzymes. H2O2 (1 mM) was then added, and degradation was monitored. (B) Triple mutants retaining Ahp, Rbr1, or Rbr2 as sole scavenger enzyme were grown under anoxic conditions, and 1 mM H2O2 added. After 5 min H2O2 was removed by washing, and H2O2 scavenging ability was tested using 10 μM H2O2. To avoid any effect of oxygen toxicity, cells were not aerated.
This analysis leaves unexplained the roles of putative CcP, Tpx, Gpx, and BCP peroxidases, which exhibited no H2O2 scavenging activity inside B. thetaiotaomicron under any condition we tested. We considered that these enyzmes might actually operate upon organic hydroperoxides. However, mutants were not hypersensitive to either cumene hydroperoxide or t-butylhydroproxide (data not shown). It seemed possible that poor expression prevented their scavenging activities from being apparent under our typical growth conditions, and so the genes were placed under control of the SusA promoter in a katE ahpC rbr1 rbr2 mutant strain. Even when the promoter was induced, no scavenging occurred (Fig. S6). We infer that these proteins probably have some other physiological role.
Regulation of scavenging enzymes
The OxyR protein regulates the H2O2 stress response in most Gram-negative bacteria, including the related organism Bacteroides fragilis (Rocha et al., 2000). A BLAST search of the B. thetaiotaomicron genome sequence revealed that the BT_4716 gene product shares 57% and 89% homology with the OxyR proteins of E. coli and B. fragilis, respectively. The cysteine residues at positions 199 and 208 of the putative B. thetaiotaomicron OxyR correlate with the sensory and resolving cysteines at the identical positions of E. coli OxyR. BT_4716 lies immediately downstream from a homolog of Dps, and this genomic context matches that of B. fragilis. Thus we anticipated that BT_4716 would direct the response of scavenging enzymes to H2O2. The upstream sequences of katE, ahpC, rbr1, oxyR, and dps each indicated the presence of OxyR recognition sequences (Fig. S7). This was not true of the upstream sequence of rbr2.
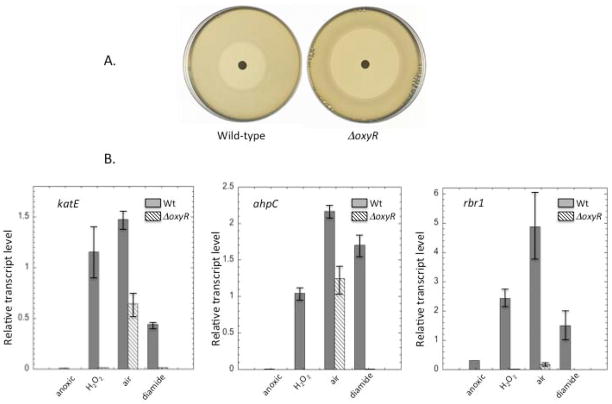
A ΔBT_4716 strain exhibited sensitivity to H2O2 on zone-of-inhibition plates (Fig. 8A). The transcript levels of katE, ahpC, and rbr1 all increased when H2O2 was added to wild-type cultures, but this response did not occur in the ΔBT_4716 mutant (Fig. 8B). Diamide, which activates OxyR proteins by chemically modifying its sulfhydryl residues (Privalle & Fridovich, 1990), also activated expression of these genes in wild-type but not the mutant strain. We conclude that BT_4716 does encode an OxyR transcription factor and that it is responsible for driving the responses of these genes to H2O2.
Fig. 8. Role of OxyR in H2O2 stress.
(A) Wild-type and ΔoxyR strains were spread on anoxic BHIS plates and exposed to a filter disk containing H2O2. The zone of inhibition after 24 hours of incubation was 26 +/− 2 mm for wild-type cells and 50 +/− 3 mm for the ΔoxyR strain. (B) The katE, ahpC, and rbr1 genes were induced in an OxyR-dependent manner upon anoxic treatment with H2O2 or diamide. Induction upon aeration had OxyR-dependent and –independent components. Transcripts were measured by qRT-PCR under anoxia, after anoxic treatment with 100 μM H2O2 for 30 min, after aeration for 45 min, or after anoxic treatment with 1 mM diamide for 15 min. Error bars represent SD from the mean of three independent experiments.
The OxyR regulon members were also activated by simple aeration of the culture. The response of rbr1 was almost completely lost in the oxyR mutant, which implies that OxyR is activated when B. thetaiotaomicron is aerated. This behavior is not seen in E. coli. In contrast, the responses of katE and ahpC were diminished in the ΔoxyR (ΔBT_4716) strain but were still substantial. A similar effect was reported for at least one OxyR-controlled gene, dps, in B. fragilis (Sund et al., 2008). These genes may be additionally controlled by a transcription factor that responds either to oxygen directly or, perhaps, to the stasis that it imposes.
Endogenous H2O2 is a hazard when B. thetaiotaomicron is exposed to oxygen
One plausible rationale for the oxygen sensitivity of obligate anaerobes is that endogenous H2O2 might toxify them. Obligate anaerobes are more likely than aerobes to transfer electrons to low-potential acceptors, and the enzymes that do so may be prone to the adventitious transfer of electrons to molecular oxygen, generating superoxide and hydrogen peroxide. One enzyme that is predisposed to this error is fumarate reductase (Imlay, 1995, Korshunov & Imlay, 2010). This enzyme lies in the center of metabolism in B. thetaiotaomicron and accordingly handles a very high electron flux, and so we posited that aeration might trigger substantial formation of H2O2. This idea could be tested using the katE ahpC rbr1 rbr2 mutant, since H2O2 that is formed in non-scavenging bacteria will flow out of the cell into the medium, where it can be quantified.
This experiment was performed. Indeed, H2O2 rapidly accumulated in the medium when the katE ahpC rbr1 rbr2 mutant was aerated (Fig. 9). The rate of H2O2 generation was much higher than for an analogous katG katE ahp mutant of E. coli. This difference probably explains why the OxyR regulon is activated upon aeration of B. thetaiotaomicron but not E. coli.
Fig. 9. B. thetaiotaomicron cells generate endogenous H2O2 at a faster rate than does E. coli.
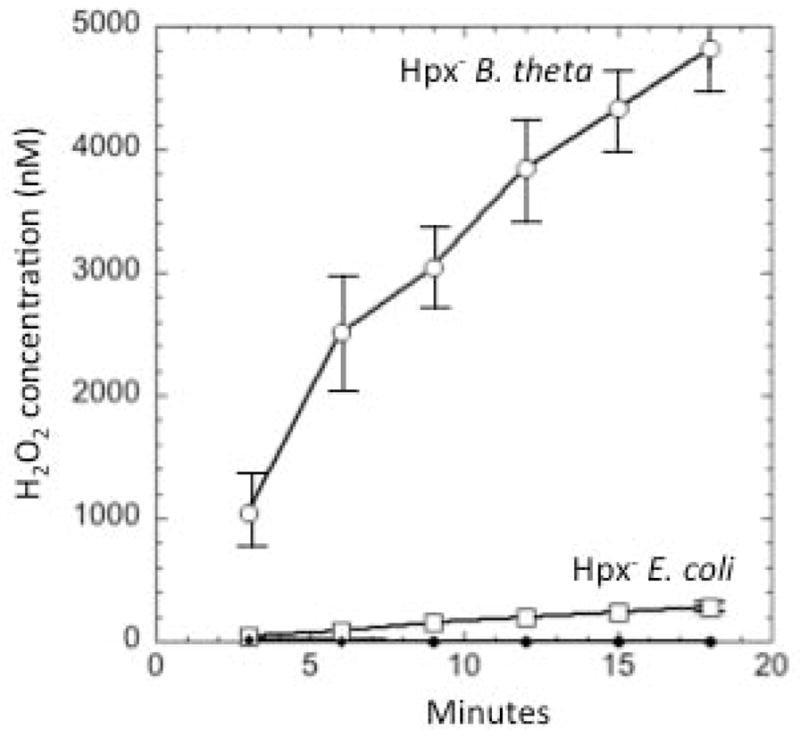
Exponentially growing non-scavenging (“Hpx−”) mutants of B. thetaiotaomicron (ΔkatE ΔahpC Δrbr1 Δrbr2) and E. coli (ΔkatG ΔkatE ΔahpCF) were grown in anoxic BHIS or LB, respectively, to exponential phase. They were then washed and suspended in aerated PBS/glucose, and H2O2 accumulation in the growth medium was monitored. No detectable H2O2 accumulated either in sterile PBS/glucose or in cultures containing either wild-type organism.
The induction of the OxyR regulon might therefore comprise a key defense against this endogenous H2O2. Wild-type cells survived aeration, but the oxyR mutants did not, with > 99% mortality in 10 hours (Fig. 10A). By comparison, aeration does not kill oxyR mutants of E. coli. Interestingly, the non-scavenging katE ahpC rbr1 rbr2 mutant did not lose viability, indicating that OxyR-controlled activities in addition to scavenging enzymes were sufficient to keep the cell alive (Fig. 10B). The most likely cause of cell death is DNA damage from the Fenton reaction, in which H2O2 is reduced by unincorporated intracellular iron. In all organisms examined to date this iron pool is diminished by one or more components of the OxyR regulon, including Dps, an iron-sequestering storage protein (Grant et al., 1998, Ilari et al., 2002, Altuvia et al., 1994). To test whether Fenton chemistry underlay the rapid death of the B. thetaiotaomicron oxyR mutants, we added dipyridyl, an iron chelator that can penetrate cells and trap ferrous iron in non-reactive sandwich complexes. Dipyridyl prevented aerobic oxyR death (Fig. 10A). We conclude that aeration promotes rapid H2O2 formation inside B. thetaiotaomicron and that the robust induction of the OxyR regulon is necessary to keep this H2O2 from killing the cell.
Fig. 10. OxyR activation is necessary to allow B. thetaiotaomicron to survive aeration.
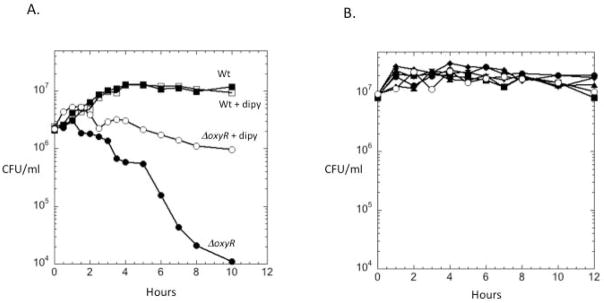
(A) Exponentially growing wild-type and ΔoxyR strains were aerated in BHIS medium at time zero, and cell viability was tracked by plating. Where indicated, 2.5 mM of the cell-penetrating iron chelator dipyridyl (dipy) was added prior to aeration (open symbols). (B) Survival of mutant lacking all H2O2 scavengers (katE, ahpC, rbr1 and rbr2, open symbols) in aerated BHIS did not differ detectably from that of wild-type strains or triple mutants. The data presented are representative of three independent experiments.
Discussion
B. thetaiotaomicron employs multiple enzymes to degrade endogenous H2O2
It has taken awhile to recognize the extent of oxidative defenses in anaerobic bacteria. For many decades catalase was the sole known microbial scavenger of H2O2, and Clostridial-oriented surveys supported a notion that obligate anaerobes lacked any scavenging capacity at all (McCord et al., 1971). This perspective began to change when catalase activity was found in extracts of Desulfovibrio gigas, Desulfovibrio vulganis, and Bacillus fragilis (Santos et al., 2000, Fareleira et al., 2003, Rocha & Smith, 1997), and it was completely reconsidered when Ahp was recognized as the predominant scavenger of H2O2 in model enteric bacteria (Seaver & Imlay, 2001a). Genomic studies find that Ahp is widespread, including among anaerobes.
Still, some oxygen-tolerant anaerobes lack both catalase and Ahp, which raised the prospect that they are protected by other scavenging systems. In vitro searches for peroxidases ultimately presented a number of candidates: thiol peroxidase, bacterioferritin comigratory protein, glutathione peroxidase, rubrerythrins, and cytochrome c peroxidase (for review, see (Mishra & Imlay, 2012). Each of these has either reactive sulfhydryl residues (Tpx, Bcp, Gpx) or iron atoms (Rbr, Ccp) that are arranged so that reduction of H2O2 occurs efficiently. The turnover number of each of these enzymes is sufficiently high that H2O2 reduction is plausibly its biological role. The purpose of the present study was to determine which of these enzymes are responsible for H2O2 degradation in B. thetaiotaomicron. What we learned is that rubrerythrins appear to be the primary scavengers under anaerobic conditions. Previous work by Bahl and coworkers with Clostridium acetobutylicum had demonstrated a correlation between its Rbr content and oxygen tolerance, strongly suggesting that Rbr protects cells by degrading H2O2 (Riebe et al., 2007, Hillmann et al., 2009). In that example workers were unable to confirm this idea by generating Rbr-deficient mutants; the present data fill that gap.
Upon aeration, the scavenging activity of rubrerythrins subsides, and both Ahp and catalase are strongly induced. In this situation Ahp becomes the primary scavenger of endogenous H2O2, while catalase excels at degrading higher concentrations that might enter the cell from external sources. Thus the key difference between the scavenging plans of B. thetaiotaomicron and E. coli is that the anaerobe relies upon rubrerythrins under anoxic conditions. It is pertinent to ponder why this is so, and why Rbr activity fades upon aeration.
The active site of rubrerythrins positions two ferrous iron atoms to concertedly bind H2O2 (for review see Kurtz, 2006). Electron transfer follows, with consequent divalent reduction of H2O2 to water and oxidation of the iron atoms to their ferric forms. A second rubrerythin domain holds a rubredoxin-like Fe(Cys)4 center within electron-transfer distance of the binuclear iron site, and soluble rubredoxin itself can bind near that center to deliver electrons. Two such univalent deliveries restore the protein to its di-ferrous ready state, completing the catalytic cycle.
This system was first discovered in a microaerophile (Alban et al., 1998), and it is commonly found in obligate anaerobes. Notably, it is absent from microbes that grow in fully oxic environments—a distribution that dovetails with our observation that rubrerythrin function stopped after B. thetaiotaomicron was aerated. We have considered some explanations. First, prior study suggested that rubredoxin:oxygen oxidoreductase (ROO) might compete with Rbrs for the electrons carried by rubredoxin. Work in Desulfovibrio vulgaris and B. fragilis (Wildschut et al., 2006, Meehan et al., 2012) determined that these anaerobes remained viable in microaerobic media for longer periods of time if ROO was inactivated, which might be consistent with enhanced scavenging by Rbrs. Our experiments gave some limited support to this idea: genetic elimination of ROO consistently improved the degradation of H2O2 by Rbr in aerobic medium, but it did so only to a modest extent, and ultimately the enzymes failed regardless.
A second possibility is that Rbrs are oxidatively damaged. There is a potential problem inherent to the Rbr catalytic cycle: reduction of the oxidized (Fe3+/Fe3+) form requires two consecutive encounters with reduced rubredoxin. What happens if H2O2 enters the active site of the mixed-valence (Fe2+/Fe3+) intermediate? A univalent Fenton reaction seems likely, generating a ferryl radical that could either decompose into a hydroxyl radical or else directly abstract an electron from surrounding residues. In fact, workers have observed the formation of nearby tyrosyl radicals during turnover with excess H2O2 (Kurtz, 2006). Such polypeptide oxidation could inactivate the enzyme. The risk would run higher when H2O2 becomes competitive with reduced rubredoxin for the mixed-valence enzyme—that is, when H2O2 concentration is high or the metabolic provision of reducing equivalents is low. Both situations apply when B. thetaiotaomicron enters oxic environments, due to rapid endogenous H2O2 formation and oxidative poisoning of metabolism. In our experiments Rbr scavenging activity invariably slowed over the course of H2O2 exposure, and it slowed more quickly in strains lacking any other scavenger (Fig. 6B), as if H2O2 itself were the problem. Scavenging did not resume when cells were restored to anoxic conditions, which would be consistent with irreversible enzyme damage.
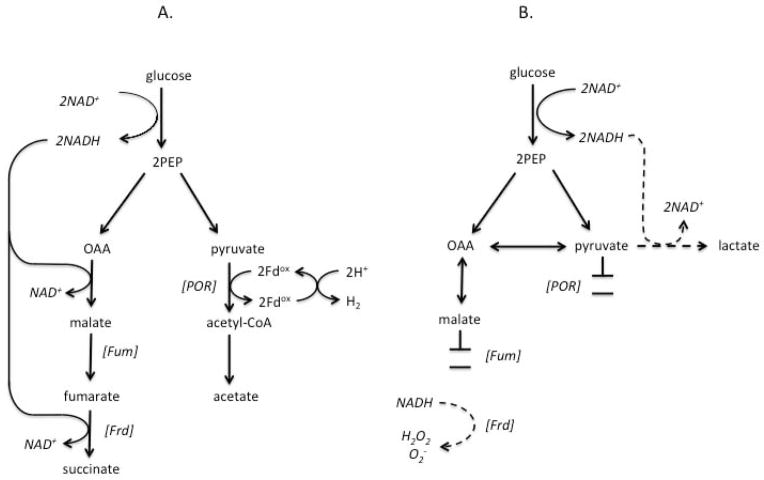
A non-exclusive alternative is that electron flow to rubredoxin, and thus to Rbr, slows and stops when B. thetaiotaomicron metabolism is poisoned by oxygen. In particular, molecular oxygen inactivates its pyruvate:acceptor oxidoreductase (POR), the central metabolic enzyme that transfers electrons to univalent acceptors (Pan & Imlay, 2001). Univalent electron flow is necessary for hydrogen production, a keystone fermentative action of B. thetaiotaomicron (Figure 11A). POR activity is substantially diminished within 40 min of aeration and becomes undetectable within two hours, a time frame that is roughly congruent with the diminution of Rbr turnover. If POR is an important conduit for univalent electrons that find their way, directly or indirectly, to rubredoxin, then this inactivation might compromise Rbr function.
Fig. 11. Schemes of metabolic flow in anoxic and aerated B. thetaiotaomicron.
(A) Anaerobic metabolism. NADH generated during glycolysis is reoxidized through reduction of PEP to succinate. Other PEP is oxidatively decarboxylated by pyruvate:acceptor oxidoreductase (POR), with transfer of electrons via a carrier (here, ferredoxin, Fd) to hydrogenase. (B) Metabolism upon aeration (Pan & Imlay, 2001). The inactivation of fumarase blocks NADH reoxidation, and in the absence of fumarate the residual electron flow to fumarate reductase is likely to allow its autoxidation with ROS formation. POR loses activity, eliminating its reduction of univalent acceptors. NADH reoxidation through pyruvate reduction is not adequate to sustain glucose catabolism. Abbreviations: PEP, phosphoenolpyruvate. OAA, oxaloacetate. Fum, fumarase. Frd, fumarate reductase. POR, pyruvate:acceptor oxidoreductase. Fd, ferredoxin.
Further work is necessary to test these hypotheses. In any case, the inability of rubrerythrin to function in oxic habitats is presumably the raison d’etre of the induction of Ahp. In contrast to Rbrs, Ahp is directly driven by NADH, a divalent electron donor. The significance is that NADH is produced by the sections of B. thetaiotaomicron central metabolism that remain functional even in oxic environments: glycolysis during the catabolism of carbohydrates (Fig. 11B), and lactate dehydrogenase during the catabolism of lactate (not shown). Indeed, when glucose-fed B. thetaiotaomicron are exposed to oxygen, the succinate/acetate production shifts to lactate production, as the cell reduces pyruvate in order to reoxidize its accumulating NADH (Pan & Imlay, 2001). Thus, despite disruption of downstream metabolism, NADH pools remain available to drive H2O2 reduction by Ahp. Our data showed that Ahp remained active for at least 12 hours, long after POR was inactive and cell growth had ceased.
Finally, we note that Bcp, Gpx, Tpx, and Ccp turned out to be ineffective at H2O2 degradation in vivo, even when measures were taken to ensure that their structural genes were expressed. This outcome mirrors the situation in E. coli, which also encodes each of these. Of the four, only Tpx was induced upon H2O2 exposure or aeration, and none of the mutants exhibited sensitivity to H2O2 or organic hydroperoxides. These proteins may have roles in signalling, analogous to Gpx3 of yeast (Delaunay et al., 2002), or in the degradation of a particular organic peroxide species. This outcome, however, emphasizes that the demonstration of peroxidase activity in vitro is not sufficient evidence of function in vivo.
How does oxygen damage obligate anaerobes?
The isolation of a non-scavenging mutant will facilitate efforts to uncover how obligate anaerobes struggle with oxygen. Previous studies indicated that aeration disrupts B. thetaiotaomicron metabolism through the inactivation of POR and of fumarase (Pan & Imlay, 2001). The former might be damaged by oxygen per se, but the latter is more likely to be inactivated when superoxide and/or hydrogen peroxide attack its solvent-exposed iron-sulfur cluster. Since homologous fumarase enzymes remain functional in oxygen-tolerant bacteria such as E. coli, it was suggested that the rate of endogenous ROS formation must be unusually high inside aerated B. thetaiotaomicron. A compelling explanation is the autoxidizability of fumarate reductase, an anaerobic respiratory enzyme which, in its reduced form, accumulates a high electron density on its flavin moiety (Messner & Imlay, 2002, Yandovskaya et al., 2003). Flavins are prone to oxidation by oxygen, with commensurate formation of O2− and H2O2. Indeed, studies with scavenger-deficient E. coli demonstrated that fumarate reductase is the preponderant source of ROS when E. coli transits from anoxic to oxic medium (Korshunov & Imlay, 2010). The situation may be exacerbated in B. thetaiotaomicron, both because electron flux through fumarate reductase is far higher in B. thetaiotaomicron than in E. coli, and because the progressive inactivation of fumarase in the anaerobe blocks fumarate formation—leaving fumarate reductase without the substrate that is designed to accept its electrons (Fig. 11B). Because the TCA cycle runs in the opposite direction in aerated E. coli, in the facultative bacterium fumarate accumulates upon fumarase inactivation, thereby competing with oxygen for the reduced enzyme and suppressing fumarate reductase autoxidation.
Meehan and Malamy used B. fragilis to test this idea (Meehan & Malamy, 2012). A katE ahp tpx mutant strain was generated in an effort to use a scavenger-deficient strain to quantify H2O2 production. The mutant retained scavenging activity, but that activity diminished upon extended aeration. At that point, the rate of endogenous H2O2 formation was measured, and it exceeded what is customarily seen for E. coli by about 2.5-fold, and fumarate reductase appeared to be a significant contributor. The data of the present study explain and extend that outcome. It now seems probable that the residual scavenging activity in the B. fragilis mutant was due to Rbr, which that organism contains, and that its dissipation was due to the oxidative inactivation of Rbrs discussed above. Our experiments with katE ahp rbr1 rbr2 mutants differed from those in B. fragilis in that we were able to measure H2O2 release immediately upon aeration and without any residual scavenging activity. Most importantly, those experiments confirmed that the pace of H2O2 formation in B. thetaiotaomicron is far higher than in E. coli.
The abundant H2O2 that ensues can damage enzymes, but more significantly it can participate in Fenton chemistry that lethally damages DNA. The induction of the OxyR system confirmed that H2O2 accumulates even in scavenger-proficient cells, and the death of oxyR mutants confirmed that this stress is potentially lethal. We have not identified the crucial OxyR-controlled proteins whose induction is necessary to maintain function. However, Dps, an iron-sequestration protein that circumvents Fenton chemistry (Grant et al., 1998, Ilari et al., 2002), is the strongest candidate. The oxyR mutants were fully protected by exogenous chelators, so it makes sense that the bacterium would use the same tactic. We confirmed that dps was strongly induced upon aeration, and in fact it is an invariant member of the OxyR regulon in the bacterial world (Altuvia et al., 1994, Chiancone & Ceci, 2010). E. coli scavenging mutants that lack dps similarly lose viability upon aeration (Park et al., 2005).
These studies only scratch the surface of how this obligate anaerobe reacts to oxygen. This bacterium, like some others, has in addition to OxyR a PerR homolog. In B. subtilis PerR controls a similar set of genes as does OxyR in E. coli (Herbig & Helmann, 2001, Lee & Helmann, 2006); however, in B. thetaiotaomicron these regulators evidently control distinct regulons. The rationale is not at all obvious. Further, the OxyR-controlled regulon is substantially, although not fully, induced upon aeration even in an oxyR-deficient strain, indicating the existence of an OxyR- and PerR-independent mechanism of activation. Whether this is due to an oxygen-sensing regulator, or a regulator that senses the metabolic disturbance that oxygen creates, remains to be determined.
Experimental procedures
Reagents
All antibiotics (ampicillin, erythromycin, gentamicin, and chloramphenicol), 5′-fluorodeoxyuridine, potassium dihydrogen phosphate, sodium chloride, magnesium(II) chloride heptahydrate, calcium(II) chloride dihydrate, cobalt(II) chloride heptahydrate, manganese(II) chloride tetrahydrate, ammonium chloride, sodium sulfate, hemin chloride, iron(II) sulfate heptahydrate, L-cystine hydrochloride, L-methionine, D-glucose, 2,2′-bipyridyl, horseradish peroxidase, and 30% hydrogen peroxide were purchased from Sigma. AmplexUltra red reagent was purchased from Molecular Probes. DEPC water, 20% SDS, 0.5 M sodium acetate, 0.5 M EDTA, TE buffer, water-saturated phenol, acid phenol:chloroform, and nuclease free water were from Ambion. Random hexamer primer, superscript(III) reverse transcriptase, and DNAse were from Invitrogen.
Bacterial strains, plasmids and growth conditions
Bacterial strains and plasmids used in this study are listed in Table S2. The B. thetaiotaomicron strains used in this study were derived from BT5482 Δtdk, which contained a partial deletion of tdk gene (encoding for thymidine kinase, an enzyme in the salvage pathway of pyrimidine biosynthesis). The Δtdk mutation is exploited in the creation of mutations (Koropatkin et al., 2008). BT5482 Δtdk exhibited identical growth, survival, and recovery behavior after aeration as did the wild-type strain, BT5482 (data not shown).
B. thetaiotaomicron cultures were routinely grown at 37°C in an anaerobic chamber (Coy Laboratory Products Inc.) containing 85% N2, 10% H2 and 5% CO2. Whenever cultures were shifted from anoxic to oxic conditions or vice versa, cells were centrifuged and resuspended in fresh anaerobic medium to avoid carry-over of fermentation products. Aerobic growth was carried out at 37°C with vigorous shaking.
Brain heart infusion supplemented (BHIS) medium contained 37 g/L of brain-heart infusion (BHI), 15 μM haemin chloride, 4 mM L-cystine hydrochloride, and 22.6 mM sodium bicarbonate. Cystine was used rather than cysteine to avoid H2O2 formation by the chemical oxidation of the latter during oxygen exposure. Defined medium contained 6.6 mM potassium dihydrogen phosphate, 15.4 mM sodium chloride, 98 μM magnesium(II) chloride heptahydrate, 176.5 μM calcium(II) chloride dihydrate, 4.2 μM cobalt(II) chloride heptahydrate, 50.5 μM manganese(II) chloride tetrahydrate, 9.3 mM ammonium chloride, 1.75 mM sodium sulfate, 15 μM iron(II) sulfate heptahydrate, 3.19 mM L-cystine dihydrochloride, 134 μM L-methionine, 28 mM D-glucose, 15 μM haemin chloride, and 23.8 mM of sodium bicarbonate (Varel & Bryant, 1974). Glucose was replaced with 0.5% maltose, when used for protein expression using a maltose inducible promoter containing plasmid. The pH of BHIS and defined medium was adjusted to 7.0 before sterilization. Antibiotics were supplemented into the medium when required as 20 μg ml−1 erythromycin, 200 μg ml−1 gentamicin, 15 μg ml−1 chloramphenicol, and 200 μg ml−1 5′-fluoroxyuridine (FuDR).
E. coli strains were grown in Luria—Bertani medium consisting of 10 g L−1 bactotryptone, 5 g L−1 yeast extract, and 10 g L−1 NaCl (Miller, 1972). Antibiotics were used as 100 μg ml−1 of ampicillin, and 20 μg ml−1 of chloramphenicol.
Genomic DNA isolation
1.5 ml of culture (O.D.600 ~ 0.7) were harvested by centrifugation (13,000 rpm, 5 minutes, RT). Pellets were resuspended into 50 μl of 30% sucrose solution (in TE buffer, pH 8.0) and cells were lysed by adding 250 μl of 10% SDS solution (in TE buffer, pH 8.0). After lysis, genomic DNA was isolated using DNAeasy blood and tissue kit (Qiagen) following manufacturer’s instruction.
Construction of deletion mutants
BT5482 Δtdk strain served as the parent strain for construction of all the deletion mutants using a published method (Koropatkin et al., 2008). Approximately 700 bp region flanking the region to be deleted from the genome were PCR amplified using primers listed in the Table S3. These flanking halves were joined by sewing PCR and cloned within the XbaI/SmaI sites of a suicide plasmid, pExchange-tdk (Koropatkin et al., 2008). The resulting plasmids were transformed into competent BW19851 E. coli with selection for ampicillin resistance.
Deletion mutants were constructed by a biparental mating method. The parent B. thetaiotaomicron strain and BW19851 cells containing the deletion region as an insert in the pExchange-tdk plasmid were grown to mid log phase (O.D.600 = 0.5). Two ml of both cultures were harvested, and cells were mixed in 1 ml of BHIS. This suspension containing the parent B. thetaiotaomicron strain and BW19851 with plasmid was spread on a BHIS plate and was incubated overnight at 37°C in the anaerobic chamber. The resultant bacterial lawn containing single recombinants was washed with 1 ml of BHIS, and the resultant suspension was selected for erythromycin and gentamicin resistance. Single recombinant colonies were inoculated into 5 ml of BHIS and grown overnight at 37°C in BHIS without any antibiotic to remove the erythromycin and gentamicin selection markers by double recombination. Double recombinants were selected against 5′-fluorodeoxyuridine.
Construction of strains for protein expression
In order to complement the genes encoding catalase and peroxidases, respective genes with 15 bp upstream region were PCR amplified from wild-type genomic DNA using primers listed in the Table S3. PCR products were digested with BamHI and SacI and inserted into the identical sites of pNLY-pSusA plasmid. The 15 bp upstream region of the genes was included in the cloning protocol to provide the ribosome binding site as pNLY-pSusA plasmid lacks it. Plasmids were transformed into the parent B. thetaiotaomicron strains using a biparental mating procedure as in the case of deletion strains, and selected for chloramphenicol resistance. Expression was achieved by growing the recombinant plasmid-containing B. thetaiotaomicron strain in defined medium supplemented with 0.5% maltose in place of glucose.
H2O2 scavenging by whole cells
Overnight grown cultures were diluted to OD600 of 0.005 in BHIS and were grown to O.D.600 = 0.2 in the anaerobic chamber. (When peroxidases were expressed from a maltose-inducible promoter, cultures were instead grown in defined medium containing 0.5% maltose.) Cells were then centrifuged (6,000 rpm, RT, 5 minutes) and washed twice with equal volumes of PBS (50 mM phosphate buffer containing 0.9% NaCl, pH 7.3). Pellets were resuspended into PBS containing 0.05% glucose (or maltose) to OD600 of 0.1 at RT. H2O2 (10 μM) was added to the culture, aliquots were harvested at regular intervals (6,000 rpm, 1 minute, RT), and supernatants were stored on dry-ice/ethanol until sampling was over. Supernatants were thawed, and the remaining H2O2 concentration was immediately determined using the Amplex Red/horseradish peroxidase method (Seaver & Imlay, 2001a) in a Shimadzu RF Mini-150 fluorometer. H2O2 scavenging was measured in PBS and 0.05% carbon source rather than complete medium because medium components interfere with the H2O2 detection system. Technical replicates of H2O2 levels exhibited < 5% variation. Modest variation in scavenging rates occurred in response to different media preparations, but strain-to-strain differences persisted. Thus representative experiments from a single day are presented.
In order to measure H2O2 scavenging by aerated cells, anaerobically grown cells (OD600 = 0.2) were harvested, resuspended in prewarmed, anaerobic BHIS medium to the same density, and shifted to air with vigorous shaking at 37°C for designated periods. After aeration, cells were washed twice with equal volume of PBS and were resuspended in oxic PBS and 0.05% glucose. H2O2 was added, and H2O2 scavenging efficiency was determined as in the case of anaerobic cells.
Recovery of H2O2 scavenging activity of the enzymes on return to anaerobic condition after aeration
Overnight cultures were diluted to OD600 = 0.005 in BHIS and grown till the O.D.600 reached 0.2. Cultures were then centrifuged, resuspended in an equal volume of prewarmed anaerobic BHIS containing 150 μg/ml of chloramphenicol, removed from the chamber, and aerated by shaking at 37°C. At time points cells were centrifuged, the pellets were shifted to the anaerobic chamber, and cells were resuspended into an equal volume of prewarmed BHIS containing 150 μg/ml of chloramphenicol. Chloramphenicol was added into the medium to avoid any new synthesis of enzymes on return to anaerobic condition. In order to allow recovery of the H2O2 scavenging activity of the enzymes on return to anaerobic environment, cells were incubated for either 10 minutes or 1 hour. After recovery period, cells were washed and tested for H2O2 scavenged as outlined above. Chloramphenicol (150 μg ml−1) remained in the washing buffer as well as the scavenging medium to avoid any new synthesis of enzymes.
Recovery of H2O2 scavenging activity after treatment with 1 mM H2O2
Overnight cultures in BHIS were diluted to OD600 = 0.005 in anaerobic BHIS and grown at 37°C. When culture O.D.600 reached 0.2, 150 μg ml−1 of chloramphenicol was added. After 5 min 1 mM H2O2 was added to the cells, which were incubated at 37°C for 5 minutes. Cells were harvested by centrifugation to remove the H2O2, and cells were resuspended into prewarmed BHIS containing chloramphenicol. After 10–60 min recovery period, cells were centrifuged, washed with 50 mM PBS containing chloramphenicol, and assayed for H2O2 scavenging activity in 50 mM PBS + 0.05% glucose + 150 μg ml−1 of chloramphenicol.
Measurement of H2O2 accumulation upon aeration
Cells were diluted to OD600 = 0.005 in BHIS from an overnight grown culture and allowed to grow at 37°C till OD600 reached 0.2. Cells were then centrifuged and washed with 50 mM PBS, pH 7.3 (twice), and the pellets were removed from the anaerobic chamber. The cell pellets were resuspended to OD600 = 0.1 in 50 mM PBS, pH 7.3, supplemented with 0.05% glucose, and grown in the presence of air through vigorous shaking at 37°C. At regular intervals, aliquots were removed and centrifuged, and supernatants were stored on dry ice/ethanol until sample collection was complete. Supernatants were thawed and immediately assayed for H2O2 concentration using Amplex red/horseradish peroxidase (Seaver & Imlay, 2001a).
Quantification of gene expression by real-time PCR
Overnight cultures in BHIS were diluted to OD600 = 0.005 in anoxic BHIS. After growth to OD600 = 0.2, cultures were centrifuged and resuspended in prewarmed anoxic BHIS to OD600 = 0.5. Cultures were split and treated as follows: i) Anoxic incubation for 15 minutes; ii) Two doses of 50 μM H2O2 at 15 minute intervals under anoxic conditions; iii) 1 mM diamide for 15 minutes; and iv) aeration at 37°C for 30 or 45 minutes with vigorous shaking. (Two doses of relatively high amounts of H2O2 were used to compensate for scavenging by the dense cultures.) RNA was extracted by hot phenol method. DNA from the extracted RNA was removed by digestion with DNase (Turbo DNA-free kit, Ambion). cDNA was synthesized from RNA using Superscript III reverse transcriptase (Invitrogen) following the manufacturer’s instructions. Gene expression was analyzed using the primer pairs listed in Table S3. The gapA mRNA was measured as housekeeping control. Real-time PCR consisted of a 25 μl reaction mixture contained 10 ng of cDNA, 12.5 μl of SYBR Green supermix (Bio-Rad), and 1.6 μM of each primer. PCR was carried out in a Mastercycler ep realplex machine (Eppendorf) for 40 cycles in three steps: 95°C for 20 seconds, 56°C for 20 seconds, and 72°C for 20 seconds.
Catalase activity
Log-phase anoxic cultures (OD600 = 0.2) were split into two parts. One part remained anoxic while the other part was centrifuged, resuspended into an equal volume of prewarmed anoxic BHIS, and then aerated by vigorous shaking at 37°C for 1.5 hours. Cells were harvested, washed twice with an equal volume of ice-cold 50 mM potassium phosphate buffer, pH 7.8, and lysed in 1 ml of the same buffer. Lysis of anoxic cells was performed in anoxic buffer by sonication in the anaerobic chamber, whereas aerated cultures were lysed by passage through the French press in aerated buffer. After lysis, all the steps were performed outside the anaerobic chamber. Cell extracts were centrifuged at 13,000 rpm for 20 minutes at 4°C to remove the cell debris. Catalase activity was measured in a 1 ml reaction mixture containing 1 mM H2O2 in 50 mM PBS, pH 7.3, and 30 μl of lysate. Every 5 minutes, appropriate dilutions of reaction mixture in PBS were assayed for H2O2 concentration using the Amplex red/horseradish peroxidase method.
Disc diffusion assay
Overnight cultures in BHIS medium were diluted to OD600 = 0.005 and grown until the OD600 reached 0.2. Cultures were split into two parts. One part was immediately mixed with 2 ml of BHIS top agar and spread on BHIS plates in the anaerobic chamber. The other part of the culture was centrifuged and pellets were resuspended into equal volume of pre-warmed, anoxic BHIS and aerated with vigorous shaking at 37°C for 1 hour. Aliquots of cultures were returned to the anaerobic chamber, mixed in 2 ml of BHIS top agar, and spread on anoxic BHIS plates. A 6 mm sterile filter disc was placed in the center of the plate and spotted with 10 μl of 1 M H2O2. Plates were incubated at 37°C in the anaerobic chamber for ~36 hours, until a lawn appeared as a central zone of inhibition around the filter discs. The diameter of the zone of inhibition measured.
Outgrowth after shift from oxic to anoxic conditions
Overnight cultures in BHIS medium were diluted to OD600 of 0.005 and grown until the culture reached an OD600 of 0.15. Cultures were then centrifuged, and cell pellets were resuspended in an equal volume of pre-warmed, anoxic BHIS. The cultures were shifted out of the anaerobic chamber and incubated in the presence of air with vigorous shaking at 37°C for 4 hours. Cells were then centrifuged, resuspended in pre-warmed, anoxic BHIS to OD600 of 0.005, and grown in the anaerobic chamber at 37°C. Growth was monitored by OD600.
Survival upon aeration
Overnight cultures were diluted to OD600 = 0.005 in anoxic BHIS and allowed to grow at 37°C in the anaerobic chamber. When the OD600 of cultures reached 0.1, cells were centrifuged, resuspended in BHIS to OD600 = 0.01, moved out of the chamber, and grown with vigorous shaking at 37°C. At designated time points, aliquots of cells were collected and shifted to the anaerobic chamber. Appropriate dilutions were made from the collected aliquots in BHIS, mixed with BHIS top agar and spread on BHIS plates. Plates were incubated at 37°C in the anaerobic chamber for 3–4 days until colonies appeared. Survival was measured as number of cfu/ml. In some experiments 2.5 mM of 2,2′-bipyridyl was added to the medium 5 minutes prior to the aeration of cells.
Supplementary Material
Acknowledgments
We thank Jia Liu, Yuan Sui, and Zheng Lu for sharing useful experimental data. This work was supported by GM49640 from the National Institutes of Health.
References
- Alban PS, Popham DL, Rippere KE, Krieg NR. identification of a gene for rubrerythrin/nigerythrin-like protein in Spirillum volutans by using amino acid sequence data from mass spectrometry and NH2-terminal sequencing. J Appl Microbiol. 1998;85:875–882. doi: 10.1046/j.1365-2672.1998.00602.x. [DOI] [PubMed] [Google Scholar]
- Altuvia S, Almiron M, Huisman G, Kolter R, Storz G. The dps promoter is activated by OxyR during growth and by IHF and sigma S in stationary phase. Mol Microbiol. 1994;13:265–272. doi: 10.1111/j.1365-2958.1994.tb00421.x. [DOI] [PubMed] [Google Scholar]
- Anjem A, Imlay JA. Mononuclear iron enzymes are primary targets of hydrogen peroxide stress. J Biol Chem. 2012;287:15544–15556. doi: 10.1074/jbc.M111.330365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas FA, Diaz WA, Leal CA, Perez-Donoso JM, Imlay JA, Vasquez CC. The Escherichia coli btuE gene encodes a glutathione peroxidase that is induced under oxidative stress conditions. Biochem Biophys Res Commun. 2010;398:690–694. doi: 10.1016/j.bbrc.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballou D, Palmer G, Massey V. Direct demonstration of superoxide anion production during the oxidation of reduced flavin and of its catalytic decomposition by erythrocuprein. Biochem Biophys Res Commun. 1969;36:898. doi: 10.1016/0006-291x(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Cha MK, Kim HK, Kim IH. Thioredoxin-linked “thiol peroxidase” from periplasmic space of Escherichia coli. J Biol Chem. 1995;270:28635–28641. doi: 10.1074/jbc.270.48.28635. [DOI] [PubMed] [Google Scholar]
- Chiancone E, Ceci P. The multifaceted capacity of Dps proteins to combat bacterial stress conditions: Detoxification of iron and hydrogen peroxide and DNA binding. Biochim Biophys Acta. 2010;1800:798–805. doi: 10.1016/j.bbagen.2010.01.013. [DOI] [PubMed] [Google Scholar]
- Delaunay A, Pflieger D, Barrault MB, Vinh J, Toledano MB. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell. 2002;111:471–481. doi: 10.1016/s0092-8674(02)01048-6. [DOI] [PubMed] [Google Scholar]
- Fareleira P, Santos BS, Antonio C, Moradas-Ferreira P, LeGall G, Xavier AV, Santos H. Response of a strict anaerobe to oxygen: survival strategies in Desulfovibrio gigas. Microbiology. 2003;149:1513–1522. doi: 10.1099/mic.0.26155-0. [DOI] [PubMed] [Google Scholar]
- Flint DH, Tuminello JF, Emptage MH. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J Biol Chem. 1993;268:22369–22376. [PubMed] [Google Scholar]
- Grant RA, Filman DJ, Finkel SE, Kolter R, Hogle JM. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat Struct Biol. 1998;5:294–303. doi: 10.1038/nsb0498-294. [DOI] [PubMed] [Google Scholar]
- Gu M, Imlay JA. Superoxide poisons mononuclear iron enzymes by causing mismetallation. Mol Microbiol. 2013;89:123–134. doi: 10.1111/mmi.12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A, Sankaran B, Poole LB, Karplus PA. Structural changes common to catalysis in the Tpx peroxiredoxin subfamily. J Mol Biol. 2009;393:867–881. doi: 10.1016/j.jmb.2009.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig AF, Helmann JD. Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol Microbiol. 2001;41:849–859. doi: 10.1046/j.1365-2958.2001.02543.x. [DOI] [PubMed] [Google Scholar]
- Hillar A, Nicholls P. A mechanism for NADPH inhibition of catalase compound II formation. FEBS Lett. 1992;314:179–182. doi: 10.1016/0014-5793(92)80969-n. [DOI] [PubMed] [Google Scholar]
- Hillmann F, Riebe O, Fischer RJ, Mot A, Caranto JD, Kurtz DM, Jr, Bahl H. Reductive dioxygen scavenging by flavo-diiron proteins of Clostridium acetobutylicum. FEBS Lett. 2009;583:241–245. doi: 10.1016/j.febslet.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilari A, Ceci P, Ferrari D, Rossi G, Chiancone E. Iron incorporation into E. coli Dps gives rise to a ferritin-like microcrystalline core. J Biol Chem. 2002;277:37619–37623. doi: 10.1074/jbc.M206186200. [DOI] [PubMed] [Google Scholar]
- Imlay JA. A metabolic enzyme that rapidly produces superoxide, fumarate reductase of Escherichia coli. J Biol Chem. 1995;270:19767–19777. [PubMed] [Google Scholar]
- Jang S, Imlay JA. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J Biol Chem. 2007;282:929–937. doi: 10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong W, Cha MK, Kim IH. Thioredoxin-dependent hydroperoxide peroxidase activity of bacterioferritin comigratory protein (BCP) as a new member of the thiol-specific antioxidant protein (TSA)/Alkyl hydroperoxide peroxidase C (AhpC) family. J Biol Chem. 2000;275:2924–2930. doi: 10.1074/jbc.275.4.2924. [DOI] [PubMed] [Google Scholar]
- Kawasaki S, Sakai Y, Takahashi T, Suzuki I, Niimura Y. O2 and reactive oxygen species detoxification complex, composed of O2-responsive NADH:rubredoxin oxidoreductase-flavoprotein A2-desulfoferrodoxin operon enzymes, rubperoxin, and rubredoxin, in Clostridium acetobutylicum. Appl Environ Microbiol. 2009;75:1021–1029. doi: 10.1128/AEM.01425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin NM, Martins EC, Gordon JI, Smith TJ. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure. 2008;16:1105–1115. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshunov S, Imlay JA. Two sources of endogenous hydrogen peroxide in Escherichia coli. Mol Microbiol. 2010;75:1389–1401. doi: 10.1111/j.1365-2958.2010.07059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshunov SS, Imlay JA. A potential role for periplasmic superoxide dismutase in blocking the penetration of external superoxide into the cytosol of phagocytosed bacteria. Mol Microbiol. 2002;43:95–106. doi: 10.1046/j.1365-2958.2002.02719.x. [DOI] [PubMed] [Google Scholar]
- Kuo CF, Mashino T, Fridovich I. α,β-dihydroxyisovalerate dehydratase: a superoxide-sensitive enzyme. J Biol Chem. 1987;262:4724–4727. [PubMed] [Google Scholar]
- Kurtz DM., Jr Avoiding high-valent iron intermediates: superoxide reductase and rubrerythrin. J Inorg Biochem. 2006;100:679–693. doi: 10.1016/j.jinorgbio.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Lee JW, Helmann JD. The PerR transcription factor senses H2O2 by metal-catalyzed histidine oxidation. Nature. 2006;440:363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- LeGall J, Prickril BC, Moura I, Xavier AV, Moura JJ, Huynh BH. Isolation and characterization of rubrerythrin, a non-heme iron protein from Desulfovibrio vulgaris that contains rubredoxin centers and a hemerythrin-like binuclear iron cluster. Biochemistry. 1988;27:1636–1642. doi: 10.1021/bi00405a037. [DOI] [PubMed] [Google Scholar]
- Linn S, Imlay JA. Toxicity, mutagenesis and stress responses induced in Escherichia coli by hydrogen peroxide. J Cell Sci Suppl. 1987;6:289–301. doi: 10.1242/jcs.1984.supplement_6.19. [DOI] [PubMed] [Google Scholar]
- Lynch R, Fridovich I. Permeation of the erythrocyte stroma by superoxide radical. J Biol Chem. 1978;253:4697–4699. [PubMed] [Google Scholar]
- Massey V, Strickland S, Mayhew SG, Howell LG, Engel PC, Matthews RG, Schuman M, Sullivan PA. The production of superoxide anion radicals in the reaction of reduced flavins and flavoproteins with molecular oxygen. Biochem Biophys Res Commun. 1969;36:891–897. doi: 10.1016/0006-291x(69)90287-3. [DOI] [PubMed] [Google Scholar]
- McCord JM, Keele BB, Jr, Fridovich I. An enzyme-based theory of obligate anaerobiosis: the physiological function of superoxide dismutase. Proc Natl Acad Sci USA. 1971;68:1024–1027. doi: 10.1073/pnas.68.5.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan BM, Baughn AD, Galegos R, Malamy MH. Inactivation of a single gene enables microaerobic growth of the obligate anaerobe Bacteroides fragilis. Proc Natl Acad Sci USA. 2012;109:12153–12158. doi: 10.1073/pnas.1203796109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan BM, Malamy MH. Fumarate reductase is a major contributor to the generation of reactive oxygen species in the anaerobe Bacteroides fragilis. Microbiology. 2012;158:539–546. doi: 10.1099/mic.0.054403-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner KR, Imlay JA. The identification of primary sites of superoxide and hydrogen peroxide formation in the aerobic respiratory chain and sulfite reductase complex of Escherichia coli. J Biol Chem. 1999;274:10119–10128. doi: 10.1074/jbc.274.15.10119. [DOI] [PubMed] [Google Scholar]
- Messner KR, Imlay JA. Mechanism of superoxide and hydrogen peroxide formation by fumarate reductase, succinate dehydrogenase, and aspartate oxidase. J Biol Chem. 2002;277:42563–42571. doi: 10.1074/jbc.M204958200. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, N.Y: 1972. [Google Scholar]
- Mishra S, Imlay JA. Why do bacteria use so many enzymes to scavenge hydrogen peroxide? Arch Biochem Biophys. 2012;525:145–160. doi: 10.1016/j.abb.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan N, Imlay JA. How does oxygen inhibit central metabolism in the obligate anaerobe Bacteroides thetaiotaomicron? Mol Microbiol. 2001;39:1562–1571. doi: 10.1046/j.1365-2958.2001.02343.x. [DOI] [PubMed] [Google Scholar]
- Park S, You X, Imlay JA. Substantial DNA damage from submicromolar intracellular hydrogen peroxide detected in Hpx− mutants of Escherichia coli. Proc Natl Acad Sci USA. 2005;102:9317–9322. doi: 10.1073/pnas.0502051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieulle L, Magro V, Hatchikian EC. Isolation and analysis of the gene encoding the pyruvate-ferredoxin oxidoreductase of Desulfovibrio africanus, production of the recombinant enzyme in Escherichia coli, and effect of carboxy-terminal deletions on its stability. J Bacteriol. 1997;179:5684–5692. doi: 10.1128/jb.179.18.5684-5692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privalle CT, Fridovich I. Anaerobic biosynthesis of the manganese-containing superoxide dismutase in Escherichia coli. Effects of diazenedicarboxyliic acid bis(N,N′-dimethlamide) (diamide) J Biol Chem. 1990;265:21966–21970. [PubMed] [Google Scholar]
- Riebe O, Fischer RJ, Bahl H. Desulfoferrodoxin from Clostridium acetobutylicum funcitons as a superoxide reductase. FEBS Lett. 2007;581:5605–5610. doi: 10.1016/j.febslet.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Riebe O, Fischer RJ, Wampler DA, Kurtz DM, Jr, Bahl H. Pathway for H2O2 and O2 detoxification in Clostridium acetobutylicum. Microbiology. 2009;155:16–24. doi: 10.1099/mic.0.022756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha ER, Owens G, Jr, Smith CJ. The redox-sensitive transcriptional activator OxyR regulates the peroxide response regulon in the obligate anaerobe Bacteroides fragilis. J Bacteriol. 2000;182:5059–5069. doi: 10.1128/jb.182.18.5059-5069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha ER, Smith CJ. Regulation of Bacteroides fragilis katB mRNA by oxidative stress and carbon limitation. J Bacteriol. 1997;179:7033–7039. doi: 10.1128/jb.179.22.7033-7039.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccoccia F, Micco PD, Boumis G, Brunori M, Koutris I, Miele AE, Morea V, Sriratana P, Williams DL, Bellelli A, Angelucci F. Moonlighting by different stressors: crystal structure of the chaperone species of a 2-Cys peroxiredoxin. Structure. 2012;20:429–439. doi: 10.1016/j.str.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos WGD, Pacheco I, Liu MY, Teixeira M, Xavier AV, LeGall G. Purification and characterization of an iron superoxide dismutase and a catalase from the sulphate-reducing bacterium Desulfovibrio gigas. J Bacteriol. 2000;182:796–804. doi: 10.1128/jb.182.3.796-804.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawers G, Watson G. A glycyl radical solution: oxygen-dependent interconversion of pyruvate formate-lyase. Mol Microbiol. 1998;29:945–954. doi: 10.1046/j.1365-2958.1998.00941.x. [DOI] [PubMed] [Google Scholar]
- Sayed AA, Williams DL. Biochemical characterization of 2-Cys peroxiredoxins from Schistosoma mansoni. J Biol Chem. 2004;297:26159–26166. doi: 10.1074/jbc.M401748200. [DOI] [PubMed] [Google Scholar]
- Schellhorn HE, Hassan HM. Transcriptional regulation of katE in Escherichia coli K-12. J Bacteriol. 1988;170:4286–4292. doi: 10.1128/jb.170.9.4286-4292.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutz B, Seidel J, Sturm G, Einsle O, Gescher J. Investigation of the electron transport chain to and the catalytic activity of the diheme cytochrome c peroxidase CcpA of Shewanella oneidensis. Appl Environ Microbiol. 2011;77:6172–6180. doi: 10.1128/AEM.00606-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaver LC, Imlay JA. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J Bacteriol. 2001a;183:7173–7181. doi: 10.1128/JB.183.24.7173-7181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaver LC, Imlay JA. Hydrogen peroxide fluxes and compartmentalization inside growing Escherichia coli. J Bacteriol. 2001b;183:7182–7189. doi: 10.1128/JB.183.24.7182-7189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobota JM, Imlay JA. Iron enzyme ribulose-5-phosphate 3-epimerase in Escherichia coli is rapidly damaged by hydrogen peroxide but can be protected by manganese. Proc Natl Acad Sci USA. 2011;108:5402–5407. doi: 10.1073/pnas.1100410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sund CJ, Rocha ER, Tzianabos AO, Wells WG, Gee JM, Reott MA, O’Rourke DP, Smith CJ. The Bacteroides fragilis transcriptome response to oxygen and H2O2: the role of OxyR and its effect on survival and virulence. Mol Microbiol. 2008;67:129–142. doi: 10.1111/j.1365-2958.2007.06031.x. [DOI] [PubMed] [Google Scholar]
- Varel VH, Bryant MP. Nutritional features of Bacteroides fragilis subsp. fragilis. Appl Microbiol. 1974;18:251–257. doi: 10.1128/am.28.2.251-257.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildschut JD, Lang RM, Voordouw JK, Voordouw G. Rubredoxin:oxygen oxidoreductase enhances survival of Desulfovibrio bulgaris Hildenborough under microaerophilic conditions. J Bacteriol. 2006;188:6253–6260. doi: 10.1128/JB.00425-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandovskaya V, Horsefield R, Tomroth S, Luna-Chavez C, Miyoshi H, Leger C, Byrne B, Cecchini G, Iwata S. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299:700–704. doi: 10.1126/science.1079605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.