Abstract
The HIV/AIDS field is gaining momentum in the goal of finding a functional cure for HIV infection by utilizing strategies that specifically reactivate the latent viral reservoir in combination with the HAART regimen to prevent further viral spread. Small-molecule inhibitors such as histone deacetylase (HDAC) and bromodomain and extraterminal (BET) inhibitors can successfully activate HIV transcription and reverse viral latency in clonal cell lines. However, in resting CD4+ T cells, thought to be the principal physiological reservoir of latent HIV, their effect in reactivating the viral reservoir is more variable. It is possible that the discrepant responsiveness of quiescent primary CD4+ T cells to HDAC and BET inhibitors could be attributed to the limiting levels of P-TEFb, a key viral transcription host cofactor, in these cells. In this review, we discuss the role of P-TEFb and the necessity for its mobilization in stimulating viral reactivation from latency upon treatment with HDAC and BET inhibitors.
Keywords: BET inhibitors, HDAC inhibitors, JQ1, latency, NF-κB, P-TEFb, resting CD4+ T cells, SAHA, viral reactivation
In the global fight against the AIDS pandemic, the success of HAART in suppressing viral replication below conventional limits of detection and prolonging the lifespan of infected individuals has been a significant scientific and medical feat. HAART, a combination of drug inhibitors that target various viral enzymes in the HIV life cycle [1] and the activity of a single viral host cofactor CCR5 [2], has significantly decreased the morbidity and mortality associated with the AIDS epidemic. However, HAART is not completely without challenges or drawbacks. Notwithstanding the numerous published reports on the side effects and toxicities associated with HAART (reviewed in [3]), the great disappointment of HAART has been its inability to eradicate a persistent population of integrated and transcriptionally silent provirus found in resting CD4+ T memory cells [4]. These cells are the best-characterized viral latent reservoirs [5,6], thought to be formed when an infected CD4+ T cell escapes virus-mediated cytopathic effects and returns to a resting memory state. Thus, the resultant memory CD4+ T cell now carries an integrated but transcriptionally silent virus, which is rendered replication competent upon cellular reactivation [7]. The inherent property of memory CD4+ T cells to reactivate upon encountering their cognate antigen explains the necessity for HIV-infected individuals to adhere to a strict regimen of HAART, because any interruption in treatment could potentially lead to viral rebound from latent viral reservoirs [8] and perpetuate the infection of bystander CD4+ T cells.
Latently infected cells are virtually indistinguishable from uninfected cells because they do not produce any viral proteins, facilitating immune evasion. Moreover, the extremely low frequency of latently infected cells in vivo further complicates their isolation, detection and quantification. Additionally, understanding the molecular mechanisms underlying the establishment of latency has also been impeded due to the lack of a representative in vitro latency model [9]. Recent estimates of the latent viral pool obtained through analysis of residual viral sequences in patient samples following reactivation ex vivo indicate that the latent viral population is approximately 50-times larger than previous estimates of 1 in a million cells [6,10,11]. Mathematical modeling by the same group further indicates that eliminating 99.9% of the infected viral reservoir will be essential to achieve a lasting functional cure [10,11].
Given the toxicity, various side effects and emergence of resistance associated with chronic HAART, the development of therapeutics to eradicate the latent reservoir is warranted [12]. Previous attempts at purging the viral reservoir with T-cell activators such as anti-CD3 and IL-2 have resulted in nonspecific activation of T cells with unacceptable physiological toxicities [13]. Therefore, a treatment strategy termed ‘shock and kill’ has been proposed, which involves selective reactivation of the viral reservoirs in combination with HAART to prevent any de novo infections [12].
Because the establishment of HIV latency in resting memory CD4+ T cells involves multiple replication blocks to productive viral replication at both the transcriptional and post-transcriptional levels [14], a combinatorial approach aimed at alleviating several such blocks will likely be critical for successful viral reactivation in vivo. Current small-molecule and cell-permeable compounds under investigation, particularly histone deacetylase (HDAC) and bromo-domain and extraterminal (BET) inhibitors, have sought to reactivate latent virus by alleviating the transcriptional block to productive viral replication [15].
HIV latency
To integrate or not to integrate: pre- & post-integration latency
Viral latency can be segregated into pre- and post-integration latency depending on whether or not viral DNA has integrated into the host genome. HIV-1 entry into the host cell is mediated by interactions with cell surface receptor CD4 and the chemokine receptors CCR5 or CXCR4 [16]. Following entry into the host cell cytoplasm, the viral RNA is reverse transcribed into linear dscDNA, which assembles with host and viral proteins to form a preintegration complex (PIC) [17]. The PIC mediates the trafficking of the viral dscDNA through the nuclear pore complex into the nucleus, where the cDNA either circularizes or integrates into the host genome [17,18]. Activation of the CD4+ T cell is necessary for the recruitment of cellular transcription factors to initiate transcription of the integrated viral genome. In resting nondividing CD4+ T cells, several blocks to productive viral integration and subsequent gene expression exist [19]. Viral reverse transcription is inefficient in resting CD4+ T cells due to limiting levels of dNTPs [20,21], and the restrictive actions of host proteins such as APOBEC3G, a cytidine deaminase that catalyzes G to A mutations in the viral genome [22]. Additionally, the intracellular trafficking of viral PICs into the host cell nucleus in resting cells is also impeded because of insufficient ATP [18]. The cumulative result of these factors is the accumulation of unintegrated, linear forms of viral cDNA inside the host cell cytoplasm. Since such unintegrated forms of viral cDNA are highly unstable and have very short half-lives, preintegration latency is probably clinically irrelevant as it does not contribute to the formation of long-term stable viral reservoirs [23,24]. Post-integration latency occurs in some activated CD4+ T cells, with fully integrated proviruses that survive and transition to a quiescent memory state [7]. In such cells the integrated provirus remains transcriptionally silent, subject to activation of the host cell [7]. This reversible postintegration formation of a latent viral reservoir is a complex multifactorial process and is determined chiefly by the activation status of the cell.
Resting CD4+ T cells: the realm of HIV latency
In resting CD4+ T cells, transcription of the integrated provirus and post-transcriptional viral mRNA processing and export are restricted due to a complex interplay between various host cofactors. For a detailed overview of the transcriptional and post-transcriptional mechanisms of viral latency, please refer to two recent comprehensive reviews [7,14]. RNAPII transcription initiation and elongation of viral transcripts are both actively regulated processes. Transcription initiation is inhibited in quiescent CD4+ T cells due to sequestration in the cytoplasm of key host transcriptional activators such as NF-κB and NFAT [25]. Additionally, the chromatin organization around the HIV long terminal repeat (LTR) also inhibits viral transcription due to epigenetic repression. The degree of chromatin condensation, regulated by post-translational reversible modifications such as acetylation and deacetylation, is an important determinant of cellular gene expression. Chromatin acetylation, mediated by histone acetyl transferases (HATs), is associated with euchromatin or regions of actively transcribed chromatin. HDACs deacetylate chromatin, and this is associated with hetero-chromatin or repressed transcription [25]. There are four different classes of HDACs, of which HDACs belonging to class I have been shown to be particularly important in regulating viral latency [26,27]. The combined actions of HDACs and HATs creates a histone code, which serves as a recognition site for the recruitment of various regulatory proteins [28]. The 5′ HIV LTR, irrespective of the host integration site, has two nucleosomes, Nuc0 and Nuc1, located upstream and downstream of the transcription start site, which reduces the accessibility of transcription factors to the viral promoter [29]. Since HDAC inhibitors make important contributions to the reactivation of latent viruses, it is very likely that the nucleosomes around the viral LTR are actively deacetylated by HDACs. Transcription factors such as AP-4 and CBF-1 have previously been shown to recruit HDACs to the viral LTR in latently infected cells [25]. As a result of these repressive epigenetic processes, the levels of the HIV master regulator protein Tat are too low to promote productive viral transcription [30].
Multiple extracellular stimuli, such as T-cell receptor engagement, cytokines or PKC agonists, activate resting CD4+ T cells [25]. Cellular reactivation results in recruitment of NF-κB to the nucleus, where it recruits HATs such as p300/CBP to the viral LTR, resulting in chromatin decompaction and the stimulation of transcription initiation by RNAPII, thus elevating the production of the viral protein Tat above a threshold [25]. However, the transition from transcription initiation to elongation is ineffective due to the association of two negative elongation factors, NELF and DSIF, with RNAPII [31]. The HIV protein Tat bypasses this block to processive elongation of viral transcripts by recruiting a host transcriptional elongation factor, called P-TEFb, to the TAR RNA element formed at the 5′ end of viral transcripts [31]. P-TEFb is a kinase, the core of which is composed of the catalytic subunit CDK9 and a regulatory subunit Cyclin T1. Enzymatically active CDK9, which requires phosphorylation of Thr-186 in its T-loop for catalytic function, phosphorylates the C-terminal domain (CTD) of the stalled RNAPII and the negative elongation factors, thereby stimulating processivity of RNAPII [31]. In addition to recruiting P-TEFb, Tat also recruits several other proteins – AFF4, ENL, AF9 and an elongation factor ELL2 – to the viral LTR, leading to the formation of a super elongation complex (SEC) [32,33], thereby potently activating RNAPII elongation.
Regulation of P-TEFb in resting CD4+ T cells: complexity of 7SK RNA & BRD4
Extensive studies conducted in resting CD4+ T cells, which are a mixture of naive and memory CD4+ T cells, have revealed that the expression and enzymatic activity of P-TEFb subunits Cyclin T1 and CDK9 are stringently modulated by post-transcriptional regulation and post-translational modifications, respectively [34,35]. In resting CD4+ T cells, Cyclin T1 protein levels are low due to miRNA-mediated translational repression of Cyclin T1 mRNA [36]. Upon cellular activation, the levels of Cyclin T1-targeting miRNAs are reduced, leading to increased expression of Cyclin T1 protein [36]. The kinase activity of CDK9 is regulated by the differential phosphorylation of Thr-186 in the T-loop [37]. We have previously shown that in resting CD4+ T cells, CDK9 T-loop phosphorylation is inhibited by the cellular Mg2+-dependent phosphatase PPM1A [38]. Activation of T cells results in increased CDK9 T-loop phosphorylation by undetermined mechanisms, although CDK7 has recently been identified as a kinase that phosphorylates the T-loop of CDK9 [39]. We have also recently shown a similar pattern of regulation for both Cyclin T1 and the CDK9 T-loop in latently infected and reactivated memory CD4+ T cells in an in vitro central memory CD4+ T-cell model of HIV latency [40].
In metabolically active cells, the association of core P-TEFb with the 7SK RNP complex or with the transcriptional activator protein BRD4 is maintained in a state of equilibrium. BRD4 belongs to the BET family of nuclear-localized proteins, which possess two characteristic and highly conserved bromodomain motifs at the N terminus. These proteins also possess an extra-terminal domain at the C-terminus, which possibly serves a regulatory function [41]. BRD4 is a nuclear-localized protein that decodes epigenetic memory by recognizing acetylated lysine residues present on histone H3 and H4 tails through its bromodomain [42]. In interphase nuclei, the majority of BRD4 is associated with euchromatin. However, in response to extracellular stress signals, BRD4 dissociates from the open chromatin and recruits P-TEFb to cellular promoters and facilitates its interaction with the mediator complex [43]. By positively regulating P-TEFb function, BRD4 therefore stimulates transcriptional elongation of a wide-range of cellular genes, particularly those involved in cell cycle progression [44].
In contrast to association with BRD4, association with the 7SK RNP complex inhibits P-TEFb activity [45]. The 7SK RNP complex consists of 7SK snRNA, a noncoding small RNA, which nucleates the assembly of the other components, HEXIM1/2, LARP7 and MePCE [45,46]. In this complex, the enzymatic activity of CDK9 is inhibited due to the obstruction of its ATP-binding catalytic cleft by HEXIM1 [47]. Tat and BRD4 can directly extract P-TEFb from the 7SK RNP complex and recruit it to the integrated provirus to accelerate processive transcriptional elongation [48].
It has been suggested that the sequestration of P-TEFb in the 7SK RNP complex is one of the mechanisms that limits P-TEFb availability in resting CD4+ T cells and thus drives viral latency. However, we have shown that the levels of 7SK RNA and HEXIM1 are very low in resting peripheral blood lymphocytes and resting CD4+ T cells obtained from healthy blood donors [49,50]. Activation of peripheral blood lymphocytes or resting CD4+ T cells upregulates both 7SK RNA and HEXIM1 levels. Using a primary cell in vitro model of HIV latency developed by the Planelles laboratory, we have further shown that HEXIM1 levels are very low in latently infected memory CD4+ T cells, but increase significantly upon cellular reactivation. In these cells, we have also shown that low levels of HEXIM1 preclude the association of P-TEFb with the 7SK RNP complex [40]. While this finding has not been confirmed in resting CD4+ T cells derived from patients on HAART, it is very likely that the association of P-TEFb with the 7SK RNP complex cannot be a significant regulator of viral latency in resting central memory CD4+ T cells.
Viral reactivation from latency
Involvement of P-TEFb in viral reactivation using HDAC inhibitors
Epigenetic modifications of the host chromatin surrounding the proviral transcription start site not only regulate viral transcription, but also the interaction of various host cofactors with the viral promoter in response to extracellular stimuli. Since acetylated chromatin is associated with active transcription, HDAC inhibitors specific for different classes of HDACs are attractive candidates for viral reactivation [51]. Various HDAC inhibitors have been tested for their ability to reactivate the latent viral reservoir with varying degrees of success. The administration of valproic acid (VPA) in combination with intensified HAART therapy to a group of four HIV-infected individuals provided encouraging results in an initial study [52]. Following treatment, a significant reduction in the resting CD4+ T-cell reservoir was observed in three out of the four patients. VPA was subsequently administered to a cohort of 56 HIV-infected patients with suppressed viremia in a randomized clinical trial [53]. However, the results of this trial did not replicate the success of the first study, revealing that VPA was unable to significantly reduce the size of the latently infected CD4+ T-cell pool in the infected individuals. Additional studies with VPA were similarly disappointing and did not reiterate its initial promise [54,55]. In addition to VPA, other HDAC inhibitors such as vorinostat (also called suberoylanilide hydroxamic acid [SAHA]) and ST-80, which target different HDAC classes, can reactivate latent viruses in clonal cell lines independent of NF-κB activation [56–59].
A recently conducted clinical study with SAHA, a HDAC inhibitor approved by the US FDA to treat cutaneous T-cell lymphoma, has renewed interest in HDAC inhibitors to deplete the latent viral reservoir. In this study, SAHA was evaluated for its ability to reactivate latent viruses in a group of eight patients. Following the administration of a single dose of SAHA, all patients showed a statistically significant 4.8-fold induction in viral mRNA from resting CD4+ T cells isolated from circulating blood, indicating successful reactivation of latent viruses [15]. Treatment with SAHA also increased histone acetylation as expected. As a proof-of-concept study, this trial showed that transcriptionally silent viruses can be reactivated in vivo under conditions that do not globally activate T cells. However, this study did not ascertain if there was an actual reduction in the viral reservoir, or whether the viral reservoir was uniformly responsive to the treatment.
Treatment with SAHA and VPA has been shown to increase the acetylation of a repressive nucleosome, Nuc1, which overlaps with the HIV transcription start site [58,59]. SAHA also dissociates HDAC1 from the viral promoter, which likely plays a role in the induction of Nuc1 acetylation [58]. Remodeling of Nuc1 could then lead to increased accessibility of host transcription factors such as NF-κB to the viral LTR. In addition to remodeling repressive chromatin architecture, some studies have shown that HDAC inhibitors may also increase P-TEFb availability for viral transcription. A study by the Peterlin laboratory showed that upon SAHA treatment, P-TEFb is released from the 7SK RNP complex and Cyclin T1 is recruited to the viral LTR [56]. This initial finding has also been confirmed and extended in a recent study, which suggested that HDAC inhibitors specific for different classes of HDACs can reactivate latent viruses independently of their effects on histone architecture, by instead increasing the cellular pool of free P-TEFb that is released from the 7SK RNP [57]. Interestingly, this recent study failed to find a correlation between the dose-dependent increase in viral reactivation by SAHA or ST-80 and an increase in cognate substrate acetylation by the HDAC inhibitors. SAHA also stimulated transcription from an unintegrated episomal HIV reporter plasmid, which is unlikely to accumulate histones and a chromatin configuration similar to latent integrated viruses. Furthermore, while SAHA and ST-80 dissociated P-TEFb from the 7SK RNP complex, the HDAC inhibitors MS-275 and TBSA were unable to do so in a Jurkat T-cell line [57]. As discussed below, the addition of BET inhibitors also results in dissociation of P-TEFb from the 7SK RNP complex, suggesting a common and overlapping mechanism of action for both classes of inhibitors. Minimal levels of P-TEFb in resting CD4+ T cells likely dampen the effects of HDAC and BET inhibitors on reactivating latent virus in these cells and may explain, in part, the heterogeneous response to the inhibitors seen in primary cells, as we discuss in detail below.
BET inhibitors mobilize P-TEFb for viral reactivation from latency
Several studies have demonstrated the effect of JQ1, a BET inhibitor, in reactivating latent HIV in primary CD4+ T cells, clonal and primary cell models of HIV latency [60–63]. In contrast to HDAC inhibitors, which modify the chromatin architecture, JQ1 binds to the conserved bromodomain fold found in all BET proteins and inhibits the recognition of acetylated lysine residues associated with actively transcribed chromatin [64]. JQ1 is a model, first-generation ligand designed and developed for its ability to bind the bromodomain-binding pocket as inferred from molecular modeling [64]. JQ1 was initially synthesized to target the activity of dysregulated bromodomain-containing proteins in human cancers, such as squamous cell carcinoma. However, its efficacy in reactivating latent HIV indicates that such inhibitors have a multifunctional utility.
The mechanism by which JQ1 induces HIV-1 gene expression and reactivates latent virus seems to involve an increase in P-TEFb availability, likely by freeing it up from either the 7SK RNP complex or from the BRD4 complex associated with euchromatin. BRD4 binds to P-TEFb through a conserved motif in its CTD termed the P-TEFb-interacting domain. When the P-TEFb-interacting domain is overexpressed as a peptide, it robustly represses Tat-mediated viral LTR transactivation and viral reactivation in a clonal cell line model of HIV latency [65]. Recent studies have further expanded the role of BRD4 as a suppressor of HIV transcription, albeit in a fashion that is different from conventional restriction factors [66]. Whereas the latter are host proteins that have evolved as an innate mechanism of defense against virus infection, proteins such as BRD4 appear to indirectly repress HIV infection by competing for a common and limited set of host proteins [63]. Indeed, the Brass group identified BRD4 as a host cofactor that represses HIV gene expression through two siRNA screens [63]. In their study, siRNA depletion of BRD4 or repression of BRD4 function with JQ1 resulted in comparable increases in HIV gene expression in multiple cell lines [63]. These treatments also enhanced RNAPII processivity in the chronically infected cell line U1 derived from promonocytes [63]. Another protein belonging to the BET family, BRD2, has also been shown to inhibit HIV transcription in a Tat-independent manner [62].
In contrast with its effect in cell lines and clonal cell line models of HIV latency, JQ1 treatment has a heterogeneous effect on reactivating integrated latent virus in primary CD4+ T cells obtained from patients receiving HAART, or in primary cell models of viral latency [60,62,63]. In a study by Banerjee et al., JQ1 treatment moderately increased viral replication in resting CD4+ T cells obtained from one out of three patients with actively suppressed viremia [60]. By contrast, Zhu et al. found that JQ1 treatment alone was insufficient to reactivate latent virus in resting CD4+ T cells obtained from 19 patients on HAART therapy. However, their study showed that JQ1 synergizes with PKC agonists such as prostratin or phytohemagglutinin to reactivate transcriptionally silent virus in resting CD4+ T cells obtained from seven out of the 19 patients [63]. A similar variable response to JQ1 treatment was observed in different primary cell line models of viral latency. While JQ1 reactivates latent virus in a primary cell model stably transduced with the antiapoptotic protein Bcl-2, it was ineffective in reactivating latent virus from a central memory CD4+ T-cell model of viral latency [62].
The variable responses of resting CD4+ T cells to JQ1 treatment may be the result of a varied distribution and cellular equilibrium of P-TEFb in these cells. P-TEFb is partitioned in the 7SK RNP complex or associated with BRD4, and an emerging picture of JQ1’s mode of action suggests that it alters the association of P-TEFb in both complexes. In Jurkat cells, JQ1 treatment results in the release of P-TEFb from the 7SK RNP complex, which is then presumably available for basal viral transcription [61,67]. However, the exact mechanism by which JQ1 or SAHA treatment releases P-TEFb from the 7SK RNP complex is not known. It is possible that it could be a consequence of the cellular stress induced by JQ1 or SAHA treatment or an outcome of the chromatin remodeling initiated directly by SAHA or indirectly by JQ1 through displacement of BRD4. Previously, extracellular stress signals like UV light or treatment with HMBA have been shown to dissociate P-TEFb from the 7SK RNP complex [68,69]. Additionally, JQ1 treatment dissociates BRD4 bound to the HIV LTR in Jurkat cell models of latency and increases the association of Tat with the viral LTR [61,67]. Concomitant with an increase in P-TEFb levels, HEXIM1 levels also increase, thereby reconstituting the 7SK RNP complex in 6 h [61].
Besides increasing the availability of free P-TEFb, JQ1 treatment also increases the association of key SEC components, such as AFF4, ELL2 and CDK9, with the viral LTR, which unsurprisingly results in increased phosphorylation of the RNAPII CTD [67]. The available data regarding the recruitment of RNAPII to the viral LTR upon JQ1 treatment is contradictory. Whereas the Peterlin laboratory determined that JQ1 increases the occupancy of RNAPII at the viral LTR [61], the Zhou laboratory determined that, unlike prostratin, JQ1 treatment does not increase RNAPII occupancy at the viral LTR [67]. This is reminiscent of treatment with SAHA, which also does not increase basal levels of RNAPII at the viral LTR [56]. Prostratin treatment recruits P-TEFb to the viral LTR in an NF-κB-dependent manner to stimulate viral transcription initiation in the absence of Tat [70,71], whereas JQ1 treatment mediates recruitment of P-TEFb and SEC to the viral LTR by Tat in an NF-κB-independent manner. Therefore, the difference between JQ1 and prostratin-mediated recruitment of RNAPII to the viral LTR suggests that they likely act at discrete and nonredundant steps during viral transcription, and this may also explain the synergy between PKC agonists and JQ1.
Synergy between PKC agonists & HDAC & BET inhibitors
PKC agonists stimulate the translocation of NF-κB from the cytoplasm to the nucleus, where NF-κB can initiate proviral transcription in the absence of Tat [25,70]. Multiple studies have noted that the combination of PKC agonists and HDAC or BET inhibitors reactivate latent virus more potently from resting CD4+ T cells than individual application of these compounds [57,58,63]. Two important inferences can be drawn from this observation. First, it is quite evident that for successful viral reactivation, multiple blocks to viral transcription will likely need to be overcome. Second, because resting CD4+ T cells have limiting amounts of P-TEFb, a Tat-independent mechanism of initiating viral transcription will need to be initially induced, for example through the use of PKC agonists. Once Tat levels surpass the minimal threshold levels, a feed-forward loop will be initiated, the effect of which can be potentiated by the use of HDAC or BET inhibitors.
The addition of a PKC agonist such as a bryo-statin to resting CD4+ T cells latently infected with a replication-defective HIV virus along with SAHA results in a more potent activation of latent HIV than that achieved by SAHA alone [57]. Similarly, treatment of peripheral blood mononuclear cells derived from patients on HAART with prostratin and SAHA or VPA synergistically reactivated latent virus in 17 out of 25 samples, as opposed to reactivation of only three samples using HDAC inhibitors alone [58].
The BET inhibitor JQ1 also synergizes with prostratin to reactivate latent virus from resting CD4+ T cells obtained from patients on HAART. In one study, treatment of cells with prostratin and JQ1 reactivated virus from seven out of 19 patient samples; interestingly, JQ1 treatment by itself did not reactivate virus [63].
We have previously shown that treatment of uninfected resting CD4+ T cells with prostratin dramatically increases Cyclin T1 levels and P-TEFb availability. Interestingly, it also increases 7SK RNA and HEXIM1 levels [50]. Therefore, it is likely that upon treatment with a PKC agonist, the increased levels of P-TEFb mean that it is assembled into the 7SK RNP complex, from which it can subsequently be extracted by SAHA. In addition to increasing P-TEFb availability, prostratin also activates NF-κB, which recruits P-TEFb to the NF-κB binding sites in the viral LTR and stimulates Tat-independent viral transcription, resulting in the production of basal levels of Tat [70,71]. Once Tat is synthesized, it can extract P-TEFb from the 7SK RNP complex or compete with BRD4 for binding to P-TEFb [48,72]. However, HDAC or BET inhibitors mitigate this competition by releasing P-TEFb from the 7SK RNP complex and/or inhibiting BRD4 function, thus increasing the availability of free P-TEFb and potentiating the effect of Tat.
HDAC & BET inhibitors: effects on T-cell activation
An important concern with using small-molecule inhibitors to reverse latency is their potential for inducing global T-cell activation, which would make them unacceptable for treatment. Advantages of using SAHA for viral reactivation over cytokines such as IL-2 are that SAHA does not induce T-cell activation and that it is an FDA-approved drug. Transcriptional profiling of a clonal Jurkat cell line used as a model of HIV latency showed that JQ1 downregulated genes involved in T-cell activation to a greater extent than SAHA [60]. Peripheral blood mono-nuclear cells treated with SAHA do not affect the ERK1/2 pathway involved in T-cell activation, in contrast with phorbol 12-myristate 13-acetate, which upregulates this pathway [56]. Similarly, transcriptional profiling of resting CD4+ T cells obtained from healthy blood donors and treated with clinically comparable doses of SAHA has also shown that SAHA treatment has minimal effects on global gene transcription. In this study, only 4.1% of the total number of genes profiled were responsive to treatment with SAHA, and the observed fold changes of these genes were very low, as opposed to the robust fold changes seen in cancer cells treated with SAHA [73]. While SAHA has been shown to downregulate CXCR4 expression in leukemia cells [74], it is not clear if SAHA is involved in regulating the expression of HIV host receptor proteins. However, a HDAC inhibitor called ITF2357 has been shown to downregulate CXCR4 expression in uninfected CD4+ T cells through undefined mechanisms [75].
Conclusion
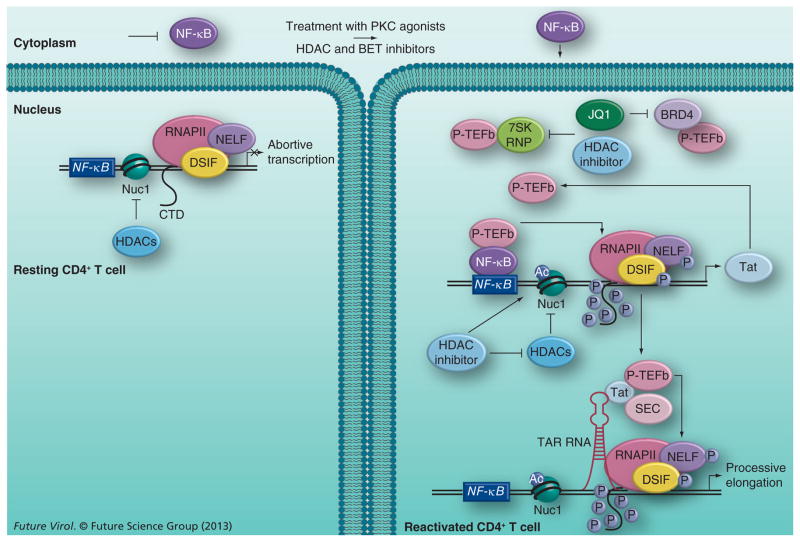
Studies of reactivation of latent HIV by HDAC and BET inhibitors clearly indicate that successful reactivation requires the removal of blocks to both RNAPII transcription initiation and elongation of the integrated provirus. While HDAC and BET inhibitors target different proteins involved directly or indirectly in maintaining viral latency, a commonality between their modes of action is likely to be the upregulation of P-TEFb. A speculative model incorporating the effects of HDAC and BET inhibitors on P-TEFb and HIV latency is shown in Figure 1. Whereas HDAC inhibitors appear to mobilize P-TEFb for viral transcription by dissociating it from the 7SK RNA complex, BET inhibitors likely augment cellular P-TEFb levels by two different mechanisms. In the first mechanism, recruitment of P-TEFb by BRD4 to regions of actively transcribed chromatin competitively inhibits viral protein Tat interaction with P-TEFb. BET inhibitors such as JQ1 block the recognition of acetylated histone markers by BRD4, and dissociate the BRD4–PTEFb complex from euchromatin. Tat targets the released P-TEFb to the SEC complex assembled at the viral LTR, thus stimulating processive transcription. In the second mechanism, BET inhibitors also appear to extract the P-TEFb sequestered in the 7SK RNP complex by undefined mechanisms. The resultant increase in P-TEFb levels upon HDAC or BET inhibitor treatment is accompanied with an upregulation of RNAPII-mediated transcription, and subsequent reactivation of the latent virus. In resting CD4+ T cells, however, inadequate levels of P-TEFb are likely to diminish the effect of HDAC and BET inhibitors, suggesting that in order for these inhibitors to be maximally effective, P-TEFb levels will need to be upregulated. This may explain the observed synergy between HDAC and BET inhibitors and PKC agonists, which are known to increase P-TEFb levels.
Figure 1. Speculative model of HIV reactivation from latency using histone deacetylase or bromodomain and extraterminal inhibitors.
In resting CD4+ T cells, viral transcription is inhibited due to sequestration of NF-κB (and NFAT, not shown) in the cytoplasm and limiting levels of P-TEFb. The action of HDACs maintains the repressive Nuc1 structure formed at the viral long terminal repeat by deacetylating the histone tails. Upon the addition of PKC agonists and HDAC or BET inhibitors, latent viral reactivation likely occurs in a biphasic process. PKC agonists activate NF-κB, which, upon translocation into the nucleus, binds to the NF-κB site in the viral long terminal repeat and recruits P-TEFb, in a Tat-independent manner, to activate RNAPII transcription of the viral protein Tat. To perpetuate viral transcription, the newly synthesized Tat protein recruits P-TEFb and SEC to the TAR RNA element formed at the 5′ end of viral transcripts. P-TEFb phosphorylates the CTD of RNAPII and the negative elongation factors associated with it to stimulate processive transcription of the viral genome. HDAC and BET inhibitors augment the effect of PKC agonists by freeing up P-TEFb from different cellular reserves and making it available for Tat transactivation. While both inhibitors free it from the 7SK RNP complex through undefined mechanisms, BET inhibitors also release P-TEFb from the BRD4 complex by inhibiting the activity of the latter. The HDAC inhibitors also increase acetylation of the Nuc1 nucleosome by inhibiting the activity of HDACs, which contributes to reversing viral latency.
Ac: Acetylated; BET: Bromodomain and extraterminal; CTD: C-terminal domain; HDAC: Histone deacetylase; P: Phosphorylated; SEC: Super elongation complex.
Future perspective
Despite the promise that HDAC and BET inhibitors hold for contributing to a functional cure for HIV, several challenges remain. Studies with the these inhibitors have made it increasingly clear that more than one host cofactor or pathway involved in establishing and maintaining viral latency will need to be targeted to diminish the viral reservoir. We anticipate that the availability of primary CD4+ T-cell models of in vitro latency will facilitate the discovery of additional small-molecule inhibitors with increased potency. However even if the latent viral pool is successfully reactivated, it is uncertain whether the reactivated virus pool can be eliminated by either virus-mediated cytopathic effects, an intensified HAART regime or through the action of host CD8+ T cells. A recent study demonstrated that reactivation of latent virus from resting CD4+ T cells derived from patients on HAART was not sufficient to kill the infected CD4+ T cells. Furthermore, CD8+ T cells isolated from the patients were unable to eliminate autologous CD4+ T cells infected with HIV, unless they were prestimulated with an HIV-specific antigen [76]. This suggests that the host immune system, particularly the CD8+ T cells, will need to be primed either through vaccines or gene therapy before treatment with a combination of latency-reversing agents.
Executive summary.
Viral latency: to integrate or not to integrate: pre- & post-integration latency
Postintegration latency is the clinically relevant form of viral latency, thought to occur when an infected activated CD4+ T cell survives virus-mediated cytopathic effects and transitions to a resting memory state carrying an integrated transcriptionally silent provirus.
Viral latency: resting CD4+ T cells: the realm of HIV latency
Multiple restrictions operate at the transcriptional and post-transcriptional steps of the HIV life cycle establish and maintain viral latency in resting CD4+ T cells.
Transcription of the integrated provirus is inhibited by the limited availability of host transcription factors such as NF-κB and P-TEFb. Repressive chromatin architecture around the viral long terminal repeat (LTR) domain, maintained by the activity of histone deacetylases (HDACs), further restricts productive viral transcription.
Viral latency: regulation of P-TEFb in resting CD4+ T cells: complexity of 7SK RNA & BRD4
P-TEFb, a key HIV host transcription cofactor, exists in at least two complexes in actively dividing cells: the 7SK RNP, in which its activity is inhibited, or the BRD4 complex, which targets P-TEFb to cellular promoters.
The activity of P-TEFb in resting CD4+ T cells is limited by the stringent regulation of its regulatory subunit Cyclin T1 and catalytic subunit CDK9, and not by its association with the 7SK RNP complex.
Viral reactivation from latency: involvement of P-TEFb in viral reactivation using HDAC inhibitors
HDAC inhibitors show variable effects in reactivating latent virus from resting CD4+ T cells, likely in part due to the low levels of P-TEFb in these cells.
These inhibitors remodel the nucleosomal architecture around the HIV LTR and use unknown mechanisms to dissociate P-TEFb from the 7SK RNP complex, which can be recruited by the viral protein Tat to the viral LTR for processive transcription.
Viral reactivation from latency: bromodomain & extraterminal inhibitors mobilize P-TEFb for viral reactivation
Bromodomain and extraterminal (BET) inhibitors prevent targeting of BRD4 to euchromatin and thus dissociate P-TEFb bound to BRD4. They also release P-TEFb from the 7SK RNP complex and increase the occupancy of super elongation complex components on the viral LTR.
Similar to HDAC inhibitors, primary resting CD4+ T cells show a heterogeneous reactivation response to BET inhibitor treatment.
Viral reactivation from latency: synergy between PKC agonists & HDAC & BET inhibitors
PKC agonists increase NF-κB and P-TEFb levels in resting CD4+ T cells, which initiates transcription of the viral protein Tat by recruiting P-TEFb to the viral LTR.
HDAC and BET inhibitors augment the effect of PKC agonists by freeing up additional P-TEFb from the 7SK RNP or BRD4 complexes.
Viral reactivation from latency: HDAC & BET inhibitors: effect on T-cell activation
Transcriptional profiling of a T-cell line has revealed that a HDAC inhibitor, suberoylanilide hydroxamic acid, and the BET inhibitor JQ1 downregulate genes involved in T-cell activation. Treatment of resting CD4+ T cells with suberoylanilide hydroxamic acid also causes minimal changes in global cellular transcription.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
Research in the laboratory of AP Rice was supported by NIH grants AI102483 and DA030166. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
▪ of interest
- 1.Arhel N, Kirchhoff F. Host proteins involved in HIV infection: new therapeutic targets. Biochim Biophys Acta. 2010;1802(3):313–321. doi: 10.1016/j.bbadis.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Dorr P, Westby M, Dobbs S, et al. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob Agents Chemother. 2005;49(11):4721–4732. doi: 10.1128/AAC.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carr A, Cooper DA. Adverse effects of antiretroviral therapy. Lancet. 2000;356(9239):1423–1430. doi: 10.1016/S0140-6736(00)02854-3. [DOI] [PubMed] [Google Scholar]
- 4.Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med. 1995;1(12):1284–1290. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- 5.Chomont N, El-Far M, Ancuta P, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15(8):893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chun TW, Carruth L, Finzi D, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387(6629):183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 7▪.Siliciano RF, Greene WC. HIV latency. Cold Spring Harb Perspect Med. 2011;1(1):a007096. doi: 10.1101/cshperspect.a007096. Comprehensive review on HIV latency. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davey RT, Jr, Bhat N, Yoder C, et al. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci USA. 1999;96(26):15109–15114. doi: 10.1073/pnas.96.26.15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Planelles V, Wolschendorf F, Kutsch O. Facts and fiction: cellular models for high throughput screening for HIV-1 reactivating drugs. Curr HIV Res. 2011;9(8):568–578. doi: 10.2174/157016211798998826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siliciano R. Detection of latently infected cells. Presented at: Strategies for an HIV Cure; Washington, DC, USA. 28–30 November 2012. [Google Scholar]
- 11.Siliciano R. HIV-1 eradication strategies: design, assessment, and clinical consequences. Presented at: 20th Conference on Retroviruses and Opportunistic Infections; Atlanta, GA, USA. 3–6 March 2013. [Google Scholar]
- 12.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. The challenge of finding a cure for HIV infection. Science. 2009;323(5919):1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 13.Van Praag RM, Prins JM, Roos MT, et al. OKT3 and IL-2 treatment for purging of the latent HIV-1 reservoir in vivo results in selective long-lasting CD4+ T cell depletion. J Clin Immunol. 2001;21(3):218–226. doi: 10.1023/a:1011091300321. [DOI] [PubMed] [Google Scholar]
- 14.Karn J, Stoltzfus CM. Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harb Perspect Med. 2012;2(2):a006916. doi: 10.1101/cshperspect.a006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15▪.Archin NM, Liberty AL, Kashuba AD, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487(7408):482–485. doi: 10.1038/nature11286. Established that histone deacetylase inhibitors could in principle reactivate latently infected CD4+ T cells obtained from a cohort of patients on HAART. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loetscher P, Moser B, Baggiolini M. Chemokines and their receptors in lymphocyte traffic and HIV infection. Adv Immunol. 2000;74:127–180. doi: 10.1016/s0065-2776(08)60910-4. [DOI] [PubMed] [Google Scholar]
- 17.Stevenson M. HIV-1 pathogenesis. Nat Med. 2003;9(7):853–860. doi: 10.1038/nm0703-853. [DOI] [PubMed] [Google Scholar]
- 18.Bukrinsky MI, Sharova N, Dempsey MP, et al. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc Natl Acad Sci USA. 1992;89(14):6580–6584. doi: 10.1073/pnas.89.14.6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coiras M, Lopez-Huertas MR, Perez-Olmeda M, Alcami J. Understanding HIV-1 latency provides clues for the eradication of long-term reservoirs. Nat Rev Microbiol. 2009;7(11):798–812. doi: 10.1038/nrmicro2223. [DOI] [PubMed] [Google Scholar]
- 20.Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61(2):213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 21.Gao WY, Cara A, Gallo RC, Lori F. Low levels of deoxynucleotides in peripheral blood lymphocytes: a strategy to inhibit human immunodeficiency virus type 1 replication. Proc Natl Acad Sci USA. 1993;90(19):8925–8928. doi: 10.1073/pnas.90.19.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiu YL, Soros VB, Kreisberg JF, Stopak K, Yonemoto W, Greene WC. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature. 2005;435(7038):108–114. doi: 10.1038/nature03493. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Y, Zhang H, Siliciano JD, Siliciano RF. Kinetics of human immunodeficiency virus type 1 decay following entry into resting CD4+ T cells. J Virol. 2005;79(4):2199–2210. doi: 10.1128/JVI.79.4.2199-2210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pierson TC, Zhou Y, Kieffer TL, Ruff CT, Buck C, Siliciano RF. Molecular characterization of preintegration latency in human immunodeficiency virus type 1 infection. J Virol. 2002;76(17):8518–8531. doi: 10.1128/JVI.76.17.8518-8531.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colin L, Van Lint C. Molecular control of HIV-1 postintegration latency: implications for the development of new therapeutic strategies. Retrovirology. 2009;6:111. doi: 10.1186/1742-4690-6-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keedy KS, Archin NM, Gates AT, Espeseth A, Hazuda DJ, Margolis DM. A limited group of class I histone deacetylases acts to repress human immunodeficiency virus type 1 expression. J Virol. 2009;83(10):4749–4756. doi: 10.1128/JVI.02585-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Archin NM, Keedy KS, Espeseth A, Dang H, Hazuda DJ, Margolis DM. Expression of latent human immunodeficiency type 1 is induced by novel and selective histone deacetylase inhibitors. AIDS. 2009;23(14):1799–1806. doi: 10.1097/QAD.0b013e32832ec1dc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 29.Verdin E. DNase I-hypersensitive sites are associated with both long terminal repeats and with the intragenic enhancer of integrated human immunodeficiency virus type 1. J Virol. 1991;65(12):6790–6799. doi: 10.1128/jvi.65.12.6790-6799.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karn J. The molecular biology of HIV latency: breaking and restoring the Tat-dependent transcriptional circuit. Curr Opin HIV AIDS. 2011;6(1):4–11. doi: 10.1097/COH.0b013e328340ffbb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23(3):297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 32.He N, Liu M, Hsu J, et al. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell. 2010;38(3):428–438. doi: 10.1016/j.molcel.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sobhian B, Laguette N, Yatim A, et al. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell. 2010;38(3):439–451. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rice AP, Herrmann CH. Regulation of TAK/P-TEFb in CD4+ T lymphocytes and macrophages. Curr HIV Res. 2003;1(4):395–404. doi: 10.2174/1570162033485159. [DOI] [PubMed] [Google Scholar]
- 35.Chiang K, Rice AP. MicroRNA-mediated restriction of HIV-1 in resting CD4+ T cells and monocytes. Viruses. 2012;4(9):1390–1409. doi: 10.3390/v4091390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiang K, Sung TL, Rice AP. Regulation of cyclin T1 and HIV-1 replication by MicroRNAsin resting CD4+ T lymphocytes. J Virol. 2012;86(6):3244–3252. doi: 10.1128/JVI.05065-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramakrishnan R, Dow EC, Rice AP. Characterization of CDK9 T-loop phosphorylation in resting and activated CD4+ T lymphocytes. J Leukoc Biol. 2009;86(6):1345–1350. doi: 10.1189/jlb.0509309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Budhiraja S, Ramakrishnan R, Rice AP. Phosphatase PPM1A negatively regulates P-TEFb function in resting CD4+ T cells and inhibits HIV-1 gene expression. Retrovirology. 2012;9:52. doi: 10.1186/1742-4690-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larochelle S, Amat R, Glover-Cutter K, et al. Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat Struct Mol Biol. 2012;19(11):1108–1115. doi: 10.1038/nsmb.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40▪.Budhiraja S, Famiglietti M, Bosque A, Planelles V, Rice AP. Cyclin T1 and CDK9 T-loop phosphorylation are downregulated during establishment of HIV-1 latency in primary resting memory CD4+ T cells. J Virol. 2013;87(2):1211–1220. doi: 10.1128/JVI.02413-12. Shows that sequestration of P-TEFb in the 7SK RNP complex is unlikely to be a latency determinant in resting memory CD4+ T cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Florence B, Faller DV. You bet-cha: a novel family of transcriptional regulators. Front Biosci. 2001;6:D1008–D1018. doi: 10.2741/florence. [DOI] [PubMed] [Google Scholar]
- 42.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein BRD4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci USA. 2003;100(15):8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein BRD4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19(4):523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 44.Yang Z, He N, Zhou Q. BRD4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol Cell Biol. 2008;28(3):967–976. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414(6861):317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 46.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414(6861):322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 47.Peterlin BM, Brogie JE, Price DH. 7SK snRNA: a noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip Rev RNA. 2012;3(1):92–103. doi: 10.1002/wrna.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48▪.Yang Z, Yik JH, Chen R, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein BRD4. Mol Cell. 2005;19(4):535–545. doi: 10.1016/j.molcel.2005.06.029. BRD4 was shown to competitively recruit P-TEFb from the 7SK RNP complex. [DOI] [PubMed] [Google Scholar]
- 49.Haaland RE, Herrmann CH, Rice AP. Increased association of 7SK snRNA with tat cofactor P-TEFb following activation of peripheral blood lymphocytes. AIDS. 2003;17(17):2429–2436. doi: 10.1097/00002030-200311210-00004. [DOI] [PubMed] [Google Scholar]
- 50▪.Sung TL, Rice AP. Effects of prostratin on Cyclin T1/P-TEFb function and the gene expression profile in primary resting CD4+ T cells. Retrovirology. 2006;3:66. doi: 10.1186/1742-4690-3-66. Demonstrates that prostratin treatment activates Cyclin T1 and HEXIM1 levels in uninfected resting CD4+ T cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shirakawa K, Chavez L, Hakre S, Calvanese V, Verdin E. Reactivation of latent HIV by histone deacetylase inhibitors. Trends Microbiol. 2013;21(6):277–285. doi: 10.1016/j.tim.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lehrman G, Hogue IB, Palmer S, et al. Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet. 2005;366(9485):549–555. doi: 10.1016/S0140-6736(05)67098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Routy JP, Tremblay CL, Angel JB, et al. Valproic acid in association with highly active antiretroviral therapy for reducing systemic HIV-1 reservoirs: results from a multicentre randomized clinical study. HIV Med. 2012;13(5):291–296. doi: 10.1111/j.1468-1293.2011.00975.x. [DOI] [PubMed] [Google Scholar]
- 54.Archin NM, Cheema M, Parker D, et al. Antiretroviral intensification and valproic acid lack sustained effect on residual HIV-1 viremia or resting CD4+ cell infection. PLoS ONE. 2010;5(2):e9390. doi: 10.1371/journal.pone.0009390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sagot-Lerolle N, Lamine A, Chaix ML, et al. Prolonged valproic acid treatment does not reduce the size of latent HIV reservoir. AIDS. 2008;22(10):1125–1129. doi: 10.1097/QAD.0b013e3282fd6ddc. [DOI] [PubMed] [Google Scholar]
- 56▪.Contreras X, Schweneker M, Chen CS, et al. Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J Biol Chem. 2009;284(11):6782–6789. doi: 10.1074/jbc.M807898200. Suberoylanilide hydroxamic acid was shown to dissociate P-TEFb from the 7SK RNP complex in this study, thus establishing a role for P-TEFb in latent viral reactivation using histone deacetylase inhibitors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bartholomeeusen K, Fujinaga K, Xiang Y, Peterlin BM. HDAC inhibitors that release positive transcription elongation factor B (P-TEFb) from its inhibitory complex also activate HIV transcription. J Biol Chem. 2013;288(20):14400–14407. doi: 10.1074/jbc.M113.464834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reuse S, Calao M, Kabeya K, et al. Synergistic activation of HIV-1 expression by deacetylase inhibitors and prostratin: implications for treatment of latent infection. PLoS ONE. 2009;4(6):e6093. doi: 10.1371/journal.pone.0006093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Archin NM, Espeseth A, Parker D, Cheema M, Hazuda D, Margolis DM. Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res Hum Retroviruses. 2009;25(2):207–212. doi: 10.1089/aid.2008.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Banerjee C, Archin N, Michaels D, et al. BET bromodomain inhibition as a novel strategy for reactivation of HIV-1. J Leukoc Biol. 2012;92(6):1147–1154. doi: 10.1189/jlb.0312165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61▪.Bartholomeeusen K, Xiang Y, Fujinaga K, Peterlin BM. Bromodomain and extra-terminal (BET) bromodomain inhibition activate transcription via transient release of positive transcription elongation factor B (P-TEFb) from 7SK small nuclear ribonucleoprotein. J Biol Chem. 2012;287(43):36609–36616. doi: 10.1074/jbc.M112.410746. The effect of JQ1 was shown not to be restricted to releasing P-TEFb from the BRD4 complex, as it also frees up P-TEFb from the 7SK RNP complex for Tat transactivation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boehm D, Calvanese V, Dar RD, et al. BET bromodomain-targeting compounds reactivate HIV from latency via a Tat-independent mechanism. Cell Cycle. 2013;12(3):452–462. doi: 10.4161/cc.23309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu J, Gaiha GD, John SP, et al. Reactivation of latent HIV-1 by inhibition of BRD4. Cell Rep. 2012;2(4):807–816. doi: 10.1016/j.celrep.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64▪.Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. doi: 10.1038/nature09504. Details the design, development and synthesis of the first bromodomain and extraterminal inhibitor, JQ1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bisgrove DA, Mahmoudi T, Henklein P, Verdin E. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc Natl Acad Sci USA. 2007;104(34):13690–13695. doi: 10.1073/pnas.0705053104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Malim MH, Bieniasz PD. HIV restriction factors and mechanisms of evasion. Cold Spring Harb Perspect Med. 2012;2(5):a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Z, Guo J, Wu Y, Zhou Q. The BET bromodomain inhibitor JQ1 activates HIV latency through antagonizing BRD4 inhibition of Tat-transactivation. Nucleic Acids Res. 2013;41(1):277–287. doi: 10.1093/nar/gks976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Contreras X, Barboric M, Lenasi T, Peterlin BM. Hmba releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog. 2007;3(10):1459–1469. doi: 10.1371/journal.ppat.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen R, Liu M, Li H, et al. PP2B and PP1alpha cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev. 2008;22(10):1356–1368. doi: 10.1101/gad.1636008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell. 2001;8(2):327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- 71.Williams SA, Chen LF, Kwon H, et al. prostratin antagonizes HIV latency by activating NF-kappaB. J Biol Chem. 2004;279(40):42008–42017. doi: 10.1074/jbc.M402124200. [DOI] [PubMed] [Google Scholar]
- 72.Barboric M, Yik JH, Czudnochowski N, et al. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res. 2007;35(6):2003–2012. doi: 10.1093/nar/gkm063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Beliakova-Bethell N, Zhang JX, Singhania A, et al. Suberoylanilide hydroxamic acid induces limited changes in the transcriptome of primary CD4+ T cells. AIDS. 2013;27(1):29–37. doi: 10.1097/QAD.0b013e32835b3e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Crazzolara R, Johrer K, Johnstone RW, et al. Histone deacetylase inhibitors potently repress CXCR4 chemokine receptor expression and function in acute lymphoblastic leukaemia. Br J Haematol. 2002;119(4):965–969. doi: 10.1046/j.1365-2141.2002.03955.x. [DOI] [PubMed] [Google Scholar]
- 75.Matalon S, Palmer BE, Nold MF, et al. The histone deacetylase inhibitor ITF2357 decreases surface CXCR4 and CCR5 expression on CD4+ T-cells and monocytes and is superior to valproic acid for latent HIV-1 expression in vitro. J Acquir Immune Defic Syndr. 2010;54(1):1–9. doi: 10.1097/QAI.0b013e3181d3dca3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76▪.Shan L, Deng K, Shroff NS, et al. Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity. 2012;36(3):491–501. doi: 10.1016/j.immuni.2012.01.014. Shows that reactivation of latently infected cells by itself is not sufficient to cause virus or CD8+ T-cell-mediated death of the infected cells. [DOI] [PMC free article] [PubMed] [Google Scholar]