Abstract
Purpose
SCLC is an aggressive malignancy affecting nearly 30,000 people annually in the United States. We have previously identified elevated PARP1 levels in SCLC and demonstrated in vitro sensitivity to the PARP inhibitors AZD 2281 and AG014699. Here, we evaluate activity of a novel, potent PARP inhibitor, BMN 673, and identify markers of response as a basis for developing predictive markers for clinical application.
Experimental Design
Inhibition of SCLC proliferation by BMN 673 was assayed in vitro and effects on tumor growth were measured in SCLC xenograft models. Protein expression and pathway activation was assessed by reverse phase protein array and western blot analysis. PARP inhibition was confirmed using a PAR ELISA.
Results
We demonstrate striking, single agent activity of BMN 673 in SCLC cell lines and xenografts, with single agent BMN 673 exhibiting in vivo activity similar to cisplatin. Sensitivity to BMN 673 was associated with elevated baseline expression levels of several DNA repair proteins, whereas greater drug resistance was observed in SCLC models with baseline activation of the PI3K/mTOR pathway. Furthermore, we developed and confirmed these data with a novel “DNA repair score” consisting of a group of 17 DNA repair proteins.
Conclusions
Elevated expression of multiple DNA repair proteins, as well as a corresponding “DNA repair protein score,” predict response to BMN 673 in in vitro SCLC models. These observations complement recent work in which PI3K inhibition sensitizes breast cancer models to PARP inhibition, suggesting cooperation between DNA repair and PI3K pathways.
Keywords: PARP1, small cell lung cancer, DNA repair
Introduction
Small cell lung cancer (SCLC) is the most aggressive form of lung cancer, with a 5 year survival rate of only 6% (1). Approximately 30,000 new cases are diagnosed annually in the United States (representing 13% of lung cancers), making SCLC more common than many other types of cancer (1, 2). Most patients with SCLC respond to chemotherapy and radiation initially. However, relapse is nearly universal and a majority of patients do not respond to further systemic therapy. The development of new, active, and potentially targeted drugs for the treatment of small cell lung cancer represents a major unmet medical need. Unlike non-small cell lung cancers (NSCLC), there are currently no targeted therapies with demonstrated benefit for patients with this disease. Recently, using an integrated proteomic profiling approach, we identified PARP1 as a potential therapeutic target for SCLC (3), demonstrating in vitro activity of two PARP inhibitors, AZD 2281 and AG014699, in SCLC cell lines.
In the current study, we investigate a novel, highly potent PARP inhibitor, BMN 673, which has recently entered Phase I testing in solid tumors and hematologic malignancies. Striking activity has been observed in the dose escalation phase in BRCA-mutated breast and ovarian cancer, molecularly-defined populations that have previously exhibited susceptibility to PARP inhibition in other clinical trials (DeBono et al., ASCO 2013). Based on our preclinical data, a SCLC cohort is now being enrolled to the dose expansion phase of this study to investigate clinical activity in SCLC (NCT01286987). However, unlike other cancers where BRCA mutations predict greater sensitivity to PARP inhibitors, there are no validated biomarkers to select SCLC patients who are most likely to benefit from this class of drugs. Here, we provide both preclinical in vitro and in vivo sensitivity data of SCLC to BMN 673 and discover potential tumor biomarkers of PARP inhibitor response in SCLC.
Methods
Cell Lines
SCLC cell lines NCI-H524, NCI-H2081, NCI-H2107, NCI-H1930, NCI-H209, NCI-H69, NCI-H1048, NCI-H1092 and NCI-H446, DMS79 and COR-L279, obtained from ATCC or ECACC, were grown in suggested media supplemented with FBS and pen/strep. DNA fingerprinting was used to confirm the identity of each cell line at the time of total protein lysate preparation, as described previously (3). Mutation data for the cell lines was provided by the Minna lab or from previously published sources(4).
Proliferation assays
For BMN 673 and cisplatin, cells were seeded in 96 well plates (at two different densities according to growth rate. After 24hr, BMN 673 or cisplatin was added at increasing concentrations in duplicate. After 10-11 days (for BMN 673) or 4 days (cisplatin), proliferation was assayed using Cell Titer Glo. For AZD 2281 (olaparib), cells were seeded in a 6-well plate (30,000 cells/well). After 24 hr, AZD 2281 was added at increasing concentrations (0, 0.16, 0.63, 2.5 and10 μM). After 14 days, the cells were counted. Assays were repeated at least twice at different cell densities.
Reverse Phase Protein Array
Protein lysate was collected from subconfluent cultures after 24-hr in full-serum media. Cells were lysed in a buffer containing 1% Triton X-100, 50 mM HEPES [pH 7.4], 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 100 mM NaF, 10 mM NaPPi, 10% glycerol, 1 mM PMSF, 1 mM Na3VO4, and 10 μg/mL aprotinin. Samples were quantified and RPPAs printed from lysates as previously described (3, 5). Immunostaining was performed with an automated autostainer (Dako, Carpinteria, CA). Each array was incubated with a primary antibody, and signal was detected using a catalyzed signal amplification system (DakoCytomation California, Inc., Carpinteria, CA). Primary antibodies were extensively validated via Western blots, where band quality and correlation of protein levels in previous RPPA experiments were determined, as previously described. An R package developed in-house were used to measure spot intensity. Protein levels were quantified by a SuperCurve method http://bioinformatics.mdanderson.org/Software/supercurve/ as described previously(6-8). Briefly, a fitted curve (“supercurve”) was plotted with the signal intensities on the Y-axis and the relative log2 concentration of each protein on the X-axis using the non-parametric, monotone increasing B-spline model. During the process, the raw spot intensity data were adjusted to correct spatial bias before model fitting. A QC metric was returned for each slide to help determine the quality of the slide: if the score is less than 0.8 on a 0-1 scale, the slide was dropped. In most cases, the staining was repeated to obtain a high quality score. If more than one slide was stained for an antibody, the slide with the highest QC score was used for analysis. Protein measurements were corrected for loading as described using median centering across antibodies. Statistical analyses were performed using R statistical software (version 2.10.0).
Animal model
Female Balb/c nude mice were obtained from Shanghai BK Laboratory Animal Center. All animal studies were conducted under an institutionally approved protocol. Human SCLC NCI-H1048 or NCI-H209 tumor cells (4×106 cells in 0.2ml in 1:1 mixture of media and matrigel) were injected s.c. in the flank. When tumors reached ~130mm3 average volume, animals (8 per group) were treated with vehicle (Q1D ×28, po), cisplatin (6 mg/kg, Q6D ×2 ip) or BMN 673 (0.33 mg/kg, Q1D ×28 po). Tumor volume and animal weight were measured every 2-3 days. Disease control was determined by comparing the change in drug-treated tumor volume (ΔT) versus vehicle-treated tumors (ΔC) using the ratio (ΔT/ΔC) at the day 21 for NCI-H1048 and day 26 for NCI-H209 (last day of tumor measurements in vehicle treated animals)(9).
Poly ADP-Ribose (PAR) Assay
To evaluate the effect of BMN 673 on PARP1 activity in vivo, lysates were prepared from xenografts 2 hours after drug delivery on day 3 of treatment. Poly ADP-Ribose (PAR) levels were assayed by ELISA per the manufacturer’s instructions (Trevigen, Inc., Gaithersburg, MD).
Results
The novel PARP inhibitor BMN 673 is a highly effective inhibitor of SCLC proliferation
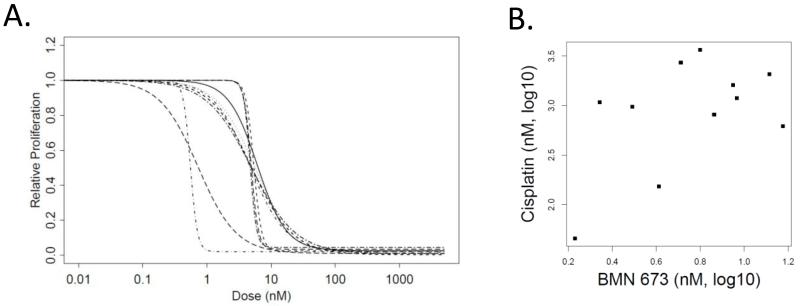
We found that BMN 673 exhibited strong, single-agent activity in our panel of 11 SCLC cell lines. The doses of BMN 673 required to inhibit 50% of SCLC cell growth (IC50) ranged from 1.7-15nM (Table 1 and Figure 1A), all of which are within clinically achievable ranges (de Bono, et al, ASCO 2013). We then compared the efficacy of BMN 673 with that of AZD 2281 (olaparib), a PARP inhibitor we have previously characterized in preclinical models of SCLC (3) and determined that BMN 673 inhibited cell proliferation to a greater extent than AZD 2281, suggesting greater in vitro potency. For example, the SCLC cell lines H82, H187 and H524 had 34-, 142- and 218-fold lower IC50 values to BMN 673 than those of AZD 2281, respectively. However, the rank-order of cell line sensitivity was the same between BMN673 and AZD 2281 in this experiment and was consistent with the relative sensitivities reported previously by our group following AZD 2281 treatment (3). Because platinum-based chemotherapy is the standard frontline treatment for SCLC, and patients with platinum-sensitive ovarian and breast cancers (often BRCA mutated) have shown greater clinical response to PARP inhibitors (10-12), we next tested whether a correlation existed between sensitivity to BMN 673 and cisplatin in SCLC cell lines. As shown in Figure 1B, we observed a moderate correlation between the IC50 values for BMN 673 and cisplatin (r=0.51, p=0.10 by Pearson correlation).
Table 1. SCLC cell lines: mutational status and IC50 values.
BMN 673 and cisplatin IC50 values calculated from proliferation assays. Mutational status of SCLC cell lines tested for TP53, RB1 and myc amplification (mt = mutant, wt = wild type, unk = unknown) are shown.
| Cell Line | TP53 | RB1 | myc amp. |
BMN 673 IC50 (nM) |
Cisplatin IC50 (nM) |
|---|---|---|---|---|---|
| H209* | mt | mt | none | 1.7 | 45.5 |
| H1048 | mt | mt | none | 2.2 | 1087 |
| H524* | mt | mt | myc | 3.1 | 976 |
| H1930* | mt | wt | none | 4.1 | 153 |
| H69* | mt | mt | n-myc | 5.2 | 2723 |
| H2081 | wt | mt | none | 6.3 | 2669 |
| H2107* | mt | mt | l-myc | 7.3 | 811 |
| H1092* | mt | wt | l-myc | 8.9 | 1611 |
| DMS-79* | mt | mt | none | 9.3 | 1189 |
| H446* | mt | mt | myc | 13 | 2074 |
| COR-L279 | mt | mt | unk | 15 | 620 |
Protein profile included in proteomic analysis
Figure 1. BMN 673 potently inhibits growth of SCLC in vitro.
Proliferation assays using BMN 673 showed SCLC to be exquisitely sensitive to BMN 673 (A). Interestingly the IC50 values of SCLC to BMN 673 and cisplatin indicated a moderate correlation (B).
PARP blockade inhibits the growth of SCLC tumors
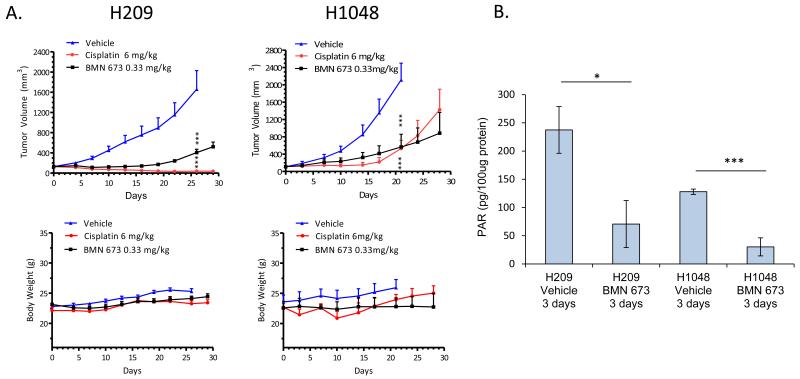
Using two SCLC xenograft models (NCI-H1048 and NCI-H209), we then tested the in vivo activity of daily BMN 673 (administered orally) as compared with either cisplatin (given intraperitoneally, i.p.) or vehicle control (given orally) in subcutaneous tumors grown in immunodeprived nu/nu mice. Tumors in animals receiving BMN 673 showed a significant reduction in growth as compared to vehicle-treated mice (p<0.0002 by t-test) (Figure 2A). Furthermore, growth delay observed with BMN 673 was similar to that observed with weekly cisplatin treatments (p<0.0002 for disease control in cisplatin vs. vehicle treated animals). Body weight was not significantly different between treatment arms, suggesting that the treatments were tolerated without significant toxicities (Figure 2). To confirm PARP targeting in vivo, we tested lysates from xenografts harvested 2-hr post-drug delivery on day 3 of treatment for the effect of BMN 673 on poly-ADP ribose (PAR) levels by ELISA (as readout of PARP activity). In both H1048 and H209 models, PAR production in tumors was significantly reduced following BMN 673 treatment (7.5 and 3.5 fold, respectively), as compared to vehicle-treated tumors (Figure 2B, p-values 0.003 and 0.036, respectively).
Figure 2. BMN 673 inhibits SCLC growth in vivo.
Animals bearing NCI-H209 and NCI-H1048 flank xenografts treated with BMN 673 showed delayed tumor growth without significant toxicity (A). Lysates prepared from xenografts harvested 2 hours post treatment on day 3 showed decreased poly-ADP ribose (PAR) levels (B) indicating strong inhibition of PARP activity in vivo by BMN 673. (Data presented as mean +/− SEM, *p<0.05, ***p<0.01)
Sensitivity to BMN 673 correlates to DNA repair protein expression and PI3K pathway activity
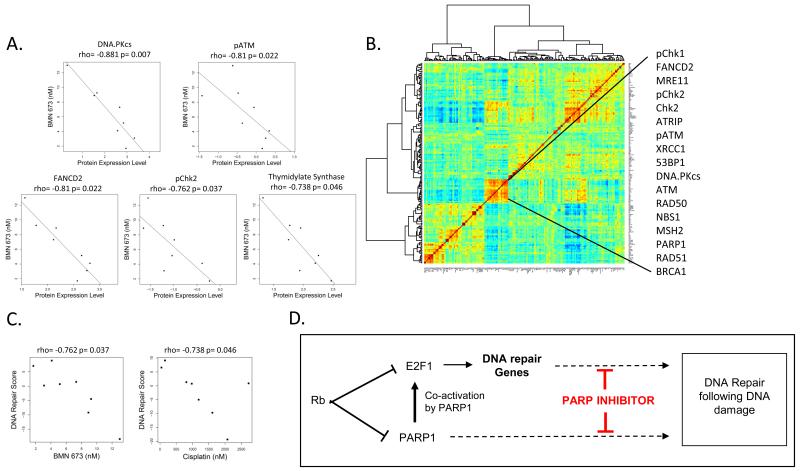
Although SCLC cell lines were in general highly sensitive to PARP inhibition in vitro, we did observe a 7-fold range of sensitivities. Therefore, to understand whether there are markers that might predict greater sensitivity or resistance in SCLC, we correlated the IC50 value to baseline expression levels of proteins involved in key oncologic signaling pathways or processes. Specifically, 193 total and/or phosphorylated proteins were quantified by reverse phase protein array (RPPA) as previously described (3, 8). Expression of each protein was then correlated with BMN 673 IC50 value. As shown in Figure 3A, the IC50 to BMN 673 showed a very strong and highly significant inverse correlation to a number of DNA repair proteins. That is, cell lines expressing higher levels of DNA repair proteins, such as FANCD2 (Rho= −0.81, p=0.02), and pChk2 (Rho= −0.76, p=0.04), were more sensitive to BMN 673, (by Spearman rank correlation).
Figure 3. SCLC IC50 to BMN 673 is inversely correlated to DNA repair protein expression.
IC50 to BMN correlated to expression of DNA repair proteins (A) demonstrates a strong inverse correlation, as cell lines with higher expression of DNA repair proteins by RPPA are more sensitive to PARP inhibition. DNA repair cluster within the correlation matrix, made up of a group of DNA repair proteins whose expression is highly correlated (B). The correlation to DNA repair is also observed when the “DNA repair score” is applied to the RPPA dataset (C). Proposed model of PARP inhibitor activity in SCLC (D).
We observed that a large number of DNA repair proteins are coordinately regulated in lung cancer cell lines, and developed a “DNA repair score” as a tool to measure DNA repair protein expression within a given sample. Specifically, in an unsupervised analysis, the expression of >190 total and/or phosphorylated proteins across 34 SCLC and 74 NSCLC cell lines (measured by RPPA, as above) were correlated with each other. This led to the identification of a DNA repair cluster within the correlation matrix, consisting of a group of 17 DNA repair proteins whose expression was highly correlated (Figure 3B). The score was derived by taking the average expression levels of these 17 DNA repair proteins. Proteins included in the DNA repair score are highlighted in the protein correlation matrix in Figure 3B (individual correlations for the 17 components of the DNA repair score are shown in figure 3A or supplemental Figure 1). Interestingly, an additional target of the transcription factor E2F1 – thymidylate synthase, which has previously been associated with resistance to the chemotherapeutic pemetrexed (13)– was one of the top five DNA repair proteins to correlated to BMN 673 IC50.
Pearson correlation between individual DNA repair proteins in the score range from r-values of 0.118-0.692 (median r-value=0.438), with corresponding p-values <0.003 for all markers included in the score. Consistent with expression of individual DNA repair markers, the DNA repair score shows a strong inverse correlation to BMN 673 IC50 (r= −0.762, p=0.037) (Figure 3C), further supporting the finding that cell lines with the overall highest expression of DNA repair makers are the most sensitive to BMN 673. Although several of the individual DNA repair proteins making up the score do not reach statistical significance for their association with BMN 673 sensitivity, all but one protein shows a similar trend to markers in Figure 3A, with higher expression levels associated with greater sensitivity. The one exception is PARP1 itself, which is expressed at relatively high levels across the entire SCLC panel, in contrast to the other proteins in the DNA repair signature which have a greater range of expression between most sensitive and most resistant cell lines. Therefore, while we believe that globally higher PARP expression in subsets of lung cancer is associated with greater sensitivity to PARP inhibitors (ex., greater sensitivity of SCLC as compared to NSCLC (3)), within cell lines that have high baseline PARP expression, other DNA repair markers show a greater dynamic range and are thus better predictors of relative sensitivity among PARP-overexpressing cell lines. Although our results indicate that the correlation between BMN 673 and cisplatin IC50s were of borderline statistical significance (r=0.52, p=0.10) (Figure 1C), we hypothesize that this is likely due to the relatively small number of cell lines tested. Interestingly, we also observed a strong correlation between the DNA repair score and in vitro sensitivity to cisplatin (r=−0.738, p=0.046), suggesting that the DNA repair score also predicts greater sensitivity to cisplatin (Figure 3C).
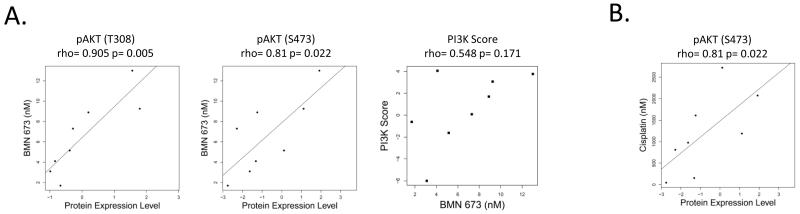
By contrast, we observed that cell lines with greater activity of the PI3K pathway were relatively more resistant to PARP inhibition and possibly to cisplatin treatment (Figure 4). Specifically, analysis of protein expression in the SCLC cell line panel revealed that cell lines with the greatest relative resistance to BMN 673 had the highest baseline expression of pAkt (T308) and pAkt (S473) (correlation between IC50 and pAkt expression levels were r=0.91 and 0.81, corresponding p-values=0.005 and 0.02, for T308 and S473, respectively). Strikingly, pAkt (S473) was also the top marker for cisplatin resistance in our SCLC panel (rho=0.81, p=0.02, Figure 4B). We next tested a previously published PI3K protein score consisting of several phosphorylated proteins within the PI3K pathway in the cell line panel (14). We observed higher PI3K scores in cell lines with higher BMN 673 IC50 levels, although this did not reach statistical significance (r=0.60, p=0.12). To confirm that the correlations described above are not due to an off target effect of BMN 673, we performed a similar analysis using the drug sensitivities we previously published for SCLC cell lines with an alternative PARP inhibitor, AZD 2281 (3). Although this analysis was limited by a small number of cell lines (n=5), the SCLC cell line most resistant to AZD 2281 (IC50 8.8uM) had the highest PI3K score and lowest DNA repair score, as compared to four relatively more sensitive cell lines (IC50 ≤2). Consistent with the BMN 673 results, we observed a trend towards higher DNA repair score (Pearson correlation, r=−0.86, p=0.06) and lower PI3K score (r=0.85, p=0.07) and greater sensitivity to AZD 2281 (by Pearson correlation).
Figure 4. PI3K pathway activity correlates to PARP inhibitor resistance.
SCLC cell lines with more active PI3K signaling are less sensitive to BMN 673 (A). Similarly, higher pAkt correlates to decreased sensitivity to cisplatin (B).
Discussion
In this study, we tested the anticancer activity of the novel PARP inhibitor BMN 673 in SCLC cell lines and animal models and used proteomic profiling data to explore potential biomarkers of BMN 673 response in SCLC. We found that BMN 673 is highly active in a panel of 11 SCLC cell lines and demonstrated significant single agent activity in a tumor xenograft model of SCLC. In fact, tumor growth inhibition in the SCLC models tested was similar to what was observed with cisplatin, which has been the backbone of standard of care chemotherapy for this disease for over 30 years.
Based on these and other preclinical studies (3), BMN 673 and other PARP inhibitors have entered clinical testing in patients with SCLC (NCT01286987, NCT01638546 at www.clinicaltrials.gov). Unlike breast and ovarian cancers where BRCA1/2 mutations have been strongly associated with response to PARP inhibitors due to the induction of synthetic lethality (15), there are no predictive biomarkers to suggest which SCLC patients may achieve benefit from this group of targeted drugs. Therefore, markers that could predict benefit would have important clinical application for development and implementation of PARP inhibitors and should be tested in ongoing clinical trials.
We examined biomarkers that were associated with sensitivity to PARP inhibition in order to develop markers that could eventually be applied to patient selection. In this analysis, elevated expression of 17 DNA repair proteins including FANCD2 and pChk2, as well as higher levels of expression of a novel “DNA repair protein score” were both associated with greater response to BMN 673. Conversely, cell lines with relatively higher expression of pAkt and a “PI3K score” were more resistant to the drug in vitro. Data from our xenograft models confirms this finding. We demonstrated that the NCI-H1048 xenograft model was somewhat less responsive to BMN 673 treatment than the NCI-H209 model (Figure 2). Interestingly, a previous study determined that the NCI-H1048 cell line, but not NCI-H209, carries a mutation in PIK3CA, resulting in elevated PI3K activity (16). This is in agreement with our above finding that cell lines with higher PI3K pathway activity were more resistant to BMN 673. Despite similar IC50 values in vitro, resistance associated with PI3K pathway is observed in vivo which may be due to different effects associated with the tumor environment.
The biomarkers identified here as potential predictive markers—in particular high levels of DNA repair proteins—are consistent with our previous data showing that SCLC were generally more sensitive to PARP inhibitors and had higher expression of DNA repair proteins, compared to NSCLC. We have previously proposed that sensitivity to PARP inhibitors may be due to loss of RB1, leading to dysinhibition of E2F1 and overexpression of its targets (including multiple DNA repair proteins, thymidylate synthase, and others) (Figure 3D) (3). In this analysis, additional E2F1 targets including thymidylate synthase (TS) and IGF1R were also correlated with response to BMN 673 (TS: r= −0.73, p=0.05, IGF1R: r=−0.76, p=0.03)), although to a lesser degree than DNA repair proteins. These findings support our proposed hypothesis that PARP inhibitors work in SCLC by indirectly decreasing the transcription of several key DNA repair proteins, in addition to direct effects on DNA repair response. However, the underlying cause of these differences in basal protein expression warrants further investigation. We also observed an association between these BMN673 predictive markers and markers of response to cisplatin. This preclinical finding in our SCLC models is consistent with clinical data from previous PARP inhibitor studies in breast and ovarian cancer patients that have shown a positive correlation between response to PARP inhibitors and platinum-sensitivity (10).
Finally, the observation that PI3K pathway activation is associated with greater resistance to BMN 673 is a novel finding and raises the question of whether the combination of PARP inhibitors with PI3K inhibitors might potentiate their effect or sensitize subsets of lung cancers to PARP inhibitors, particularly in light of recent publications in breast cancer demonstrating sensitization of breast tumors to PARP inhibition following the use of PI3K pathway inhibitors (17, 18).
Supplementary Material
Translational Relevance.
Small cell lung cancer (SCLC) is a highly lethal disease for which chemotherapy has not changed in more than 25 years. Although SCLC patients initially respond to chemotherapy and radiation, relapse is inevitable in the majority of patients. There is an urgent need, therefore, for specific, targeted therapeutics in SCLC. We have recently identified PARP1 as a potential target in SCLC. In the current study, we demonstrate remarkable sensitivity of SCLC xenograft models to PARP inhibition and identify DNA repair enzymes as potential predictive biomarkers, as well as a possible role for co-targeting the PI3K pathway to further sensitize SCLC to PARP inhibitors. Clinical trials of BMN 673 and other PARP inhibitors have been initiated for patients with SCLC which will allow clinical evaluation of the utility of the identified biomarkers.
Acknowledgments
We would like to acknowledge Drs. Emily Roarty, Len Post, and Shanghai Chempartner for their contributions to this project and for their scientific review and input on the manuscript.
Grant Support information: Cancer Center Support Grant (CA016672); UTSW and MDACC Lung SPORE 5 P50 CA070907; Cancer Clinical Investigator Team Leadership Award awarded by the National Cancer Institute though a supplement to P30CA016672 (LB); The Sidney Kimmel Scholar Award (LB); National Lung Cancer Research Partnership, made possible by the North Carolina Lung Cancer Partnership (LB); and the Lung Cancer Research Foundation (LB); LUNGevity Foundation (LB).
Footnotes
Conflicts of Interest: Y Feng and Y Shen are employees of BioMarin Pharmaceutical Inc.
REFERENCES
- 1.Herrmann LL, Zabramski JM. Nonaneurysmal subarachnoid hemorrhage: a review of clinical course and outcome in two hemorrhage patterns. J Neurosci Nurs. 2007;39:135–42. [PubMed] [Google Scholar]
- 2.Govindan R, Page N, Morgensztern D, Read W, Tierney R, Vlahiotis A, et al. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2006;24:4539–44. doi: 10.1200/JCO.2005.04.4859. [DOI] [PubMed] [Google Scholar]
- 3.Byers LA, Wang J, Nilsson MB, Fujimoto J, Saintigny P, Yordy J, et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov. 2012;2:798–811. doi: 10.1158/2159-8290.CD-12-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rudin CM, Hann CL, Peacock CD, Watkins DN. Novel systemic therapies for small cell lung cancer. J Natl Compr Canc Netw. 2008;6:315–22. doi: 10.6004/jnccn.2008.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Molecular cancer therapeutics. 2006;5:2512–21. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 6.He Y, Zhou Z, Hofstetter WL, Zhou Y, Hu W, Guo C, et al. Aberrant expression of proteins involved in signal transduction and DNA repair pathways in lung cancer and their association with clinical parameters. PloS one. 2012;7:e31087. doi: 10.1371/journal.pone.0031087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ihle NT, Byers LA, Kim ES, Saintigny P, Lee JJ, Blumenschein GR, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. Journal of the National Cancer Institute. 2012;104:228–39. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nanjundan M, Byers LA, Carey MS, Siwak DR, Raso MG, Diao L, et al. Proteomic profiling identifies pathways dysregulated in non-small cell lung cancer and an inverse association of AMPK and adhesion pathways with recurrence. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2010;5:1894–904. doi: 10.1097/JTO.0b013e3181f2a266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cascone T, Herynk MH, Xu L, Du Z, Kadara H, Nilsson MB, et al. Upregulated stromal EGFR and vascular remodeling in mouse xenograft models of angiogenesis inhibitor-resistant human lung adenocarcinoma. J Clin Invest. 2011;121:1313–28. doi: 10.1172/JCI42405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fong PC, Yap TA, Boss DS, Carden CP, Mergui-Roelvink M, Gourley C, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28:2512–9. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- 11.Byrski T, Huzarski T, Dent R, Gronwald J, Zuziak D, Cybulski C, et al. Response to neoadjuvant therapy with cisplatin in BRCA1-positive breast cancer patients. Breast Cancer Res Treat. 2009;115:359–63. doi: 10.1007/s10549-008-0128-9. [DOI] [PubMed] [Google Scholar]
- 12.Byrski T, Gronwald J, Huzarski T, Grzybowska E, Budryk M, Stawicka M, et al. Pathologic complete response rates in young women with BRCA1-positive breast cancers after neoadjuvant chemotherapy. J Clin Oncol. 2010;28:375–9. doi: 10.1200/JCO.2008.20.7019. [DOI] [PubMed] [Google Scholar]
- 13.Rossi A, Galetta D, Gridelli C. Biological prognostic and predictive factors in lung cancer. Oncology. 2009;77(Suppl 1):90–6. doi: 10.1159/000258500. [DOI] [PubMed] [Google Scholar]
- 14.Creighton CJ, Fu X, Hennessy BT, Casa AJ, Zhang Y, Gonzalez-Angulo AM, et al. Proteomic and transcriptomic profiling reveals a link between the PI3K pathway and lower estrogen-receptor (ER) levels and activity in ER+ breast cancer. Breast Cancer Res. 2010;12:R40. doi: 10.1186/bcr2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. The New England journal of medicine. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto H, Shigematsu H, Nomura M, Lockwood WW, Sato M, Okumura N, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res. 2008;68:6913–21. doi: 10.1158/0008-5472.CAN-07-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juvekar A, Burga LN, Hu H, Lunsford EP, Ibrahim YH, Balmana J, et al. Combining a PI3K Inhibitor with a PARP Inhibitor Provides an Effective Therapy for BRCA1-Related Breast Cancer. Cancer Discov. 2012;2:1048–63. doi: 10.1158/2159-8290.CD-11-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ibrahim YH, Garcia-Garcia C, Serra V, He L, Torres-Lockhart K, Prat A, et al. PI3K Inhibition Impairs BRCA1/2 Expression and Sensitizes BRCA-Proficient Triple-Negative Breast Cancer to PARP Inhibition. Cancer Discov. 2012;2:1036–47. doi: 10.1158/2159-8290.CD-11-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.