Abstract
The second messenger cAMP acts via protein kinase A (PKA) to induce apoptosis by mechanisms that are poorly understood. Here, we assessed a role for mitochondria and analyzed gene expression in cAMP/PKA-promoted apoptosis by comparing wild-type (WT) S49 lymphoma cells and the S49 variant, D− (cAMP-deathless), which lacks cAMP-promoted apoptosis but has wild-type levels of PKA activity and cAMP-promoted G1 growth arrest. Treatment of WT, but not D−, S49 cells with 8-CPT-cAMP (8-(4-chlorophenylthio)-adenosine-3′:5′-cyclic monophosphate) for 24 h induced loss of mitochondrial membrane potential, mitochondrial release of cytochrome c and SMAC, and increase in caspase-3 activity. Gene expression analysis (using Affymetrix 430 2.0 arrays) revealed that WT and D− cells incubated with 8-CPT-cAMP have similar, but non-identical, extents of cAMP-regulated gene expression at 2 h (~800 transcripts) and 6 h (~1000 transcripts) (|Fold| >2, p <0.06); by contrast, at 24 h, ~2500 and ~1100 transcripts were changed in WT and D− cells, respectively. Using an approach that combined regression analysis, clustering, and functional annotation to identify transcripts that showed differential expression between WT and D− cells, we found differences in cAMP-mediated regulation of mRNAs involved in transcriptional repression, apoptosis, the cell cycle, RNA splicing, Golgi, and lysosomes. The two cell lines differed in cAMP-response element-binding protein (CREB) phosphorylation and expression of the transcriptional inhibitor ICER (inducible cAMP early repressor) and in cAMP-regulated expression of genes in the inhibitor of apoptosis (IAP) and Bcl families. The findings indicate that cAMP/PKA-promoted apoptosis of lymphoid cells occurs via mitochondrial-mediated events and imply that such apoptosis involves gene networks in multiple biochemical pathways.
The ability of the second messenger cAMP to alter the balance between cell growth and apoptosis is cell type-dependent. cAMP stimulates growth and inhibits apoptosis in some cell types, such as neuronal cells (1), but it promotes growth arrest and apoptosis in other types of cells, such as poorly differentiated lymphoid cells (2). In addition, cAMP analogs can enhance the pro-apoptotic effects of glucocorticoids, for example, of glucocorticoid-resistant multiple myeloma and leukemia cells (2–5). However, the mechanisms that mediate cAMP-induced apoptosis are poorly understood.
Cyclic AMP-promoted apoptosis of lymphoid, in particular T-cell-derived lymphoma, cells depends on the principal effector of cAMP action, protein kinase A (PKA)3 (2–4, 6). Although both PKA and the exchange protein directly activated by cyclic AMP (Epac), another mediator of cAMP action, are found in lymphoid cells, Epac does not appear to be involved in cAMP-mediated control of the cell cycle (7) or apoptosis in these cells (2, 8). Overexpression of certain anti-apoptotic proteins, such as Bcl-2, can protect lymphoma cells from cAMP-mediated apoptosis; such protection appears to be distinct from effects of cAMP on cell cycle arrest (9). Thus, cAMP-promoted growth arrest and apoptosis appear to occur via distinct mechanisms.
S49 lymphoma cells are a unique model system to explore cell growth arrest and apoptosis by the cAMP/PKA pathway. S49 cells, which arose in a BALB/c mouse as a T-cell tumor, grow in suspension culture with a doubling time of 14–16 h and undergo G1 growth arrest and apoptosis in response to elevation of cAMP (10, 11). S49 clonal variants have been isolated that are resistant to killing by cAMP or agents that increase cAMP levels. One of these cAMP-resistant variants, D− (cAMP deathless) S49 cells, has normal PKA activity and cAMP-promoted growth arrest in G1 but lacks cAMP-promoted apoptosis (12).
Treatment of wild-type, but not kin− (which lack PKA), S49 cells with cAMP induces a coordinated up-regulation of cell cycle checkpoint genes (e.g. Gadd45a, p27, cyclin G2) and down-regulation of cyclin E1 and E2, thus defining these as genes and proteins likely to be involved in cAMP/PKA-mediated G1 arrest (11). However, the mechanisms underlying cAMP/PKA-promoted apoptosis are not known. Two major pathways lead to apoptosis: 1) an intrinsic pathway that results from mitochondrial release of pro-apoptotic proteins and subsequent activation of caspases and 2) an extrinsic pathway initiated by extracellular signals that activate death receptors, caspase-8, and downstream effector caspases (13, 14). The intrinsic pathway involves Bcl-2 family members that localize to mitochondria and modulate the release of cytochrome c and Smac (second mitochondria-derived activator of caspase) from the mitochondrial intermembrane space (15). Inhibitor of apoptosis proteins (IAP), which can blunt apoptosis, bind and inhibit caspase activity (16). However, once in the cytosol, Smac binds to specific IAPs and antagonizes their anti-apoptotic activity.
We undertook the current studies to answer several questions regarding cAMP/PKA-promoted apoptosis in WT and D− S49 cells, reasoning that comparative studies of these two closely related cell types would provide new insights into apoptosis produced by cAMP/PKA. Does such apoptosis occur via the intrinsic or extrinsic pathway? Which gene expression changes characterize cAMP/PKA-promoted apoptosis? What mechanisms trigger and mediate cAMP-promoted apoptosis?
EXPERIMENTAL PROCEDURES
Cell Culture
WT and D− S49 cells were grown in suspension cultures with Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated horse serum and 10 mM HEPES in a humidified atmosphere containing 10% CO2 at 37 °C. Cells were incubated with 100 μM 8-CPT-cAMP for the indicated times. Cultures were initiated at a density of 2.5–5 × 105 cells/ml and carried out for the indicated times.
Apoptosis Assay and Analysis of DNA Content and Cell Cycle by Flow Cytometry
Apoptosis was evaluated by assessment of annexin V binding or propidium iodide staining, as described previously (9).
Detection of Mitochondrial Membrane Potential (MMP) ΔΨm
MMP was assessed by using the fluorescent probe DiOC6 (3,3′-dihexyloxacarbocyanine iodide, Molecular Probes). Cells were treated with 8-CPT-cAMP for the indicated times and then incubated with 40 nM DiOC6 for 15 min in a standard cell culture incubator. Cells were then removed from the incubator, centrifuged, and resuspended in PBS containing 1% fetal bovine serum. MMP was then measured at room temperature using fluorescence-activated cell sorter analysis with an excitation and an emission wavelength pair of 485 and 530 nm. As a positive control, cells were treated with 50 μM of the protonophore carbonyl cyanide m-chlorophenylhydrazone (Molecular Probes) for 5 min and then stained with DiOC6 and analyzed by fluorescence-activated cell sorter.
Caspase Activity Assay
Caspase-3 and caspase-8 activity were measured by a colorimetric assay (R&D Systems) according to the manufacturer’s instructions. Briefly, S49 cells were treated with 8-CPT-cAMP for 24 h and with anti-Fas for 16 h. Cell lysates were prepared and incubated with p-nitroaniline (pNA)-conjugated substrates for caspase-3 (DEVD-pNA) and caspase-8 (IETD-pNA). Absorbance was read at 405 nm using a spectrophotometer.
Immunohistochemistry
S49 cells were treated with 8-CPT-cAMP for 24 h, incubated with 100 nM Mitotracker Red CMXRos (Molecular Probes) for 30 min, and placed on poly-D-lysine-coated coverslips. The cells were fixed for 15 min with 3.7% formaldehyde, Dulbecco’s modified Eagle’s medium, washed twice with PBS, 0.1% bovine serum albumin, permeabilized with 0.1% Triton X-100, PBS for 10 min, washed twice with PBS, 0.1% Tween 20 and finally with PBS, 1% bovine serum albumin, 0.05% Tween for 2 h. The cells were incubated for 2 h with an anti-cytochrome c antibody (Santa Cruz Biotechnology) diluted (1:100) in PBS containing 1% bovine serum albumin, 0.05% Tween and washed three times with PBS, 0.1% Tween. The cells were then incubated with a Cy2-conjugated secondary antibody anti-rabbit IgG (1:250), washed five times with PBS, 0.1% Tween, incubated with 4′,6-diamidino-2-phenylindole (1:5000) for 20 min, washed for 10 min with PBS, and mounted in gelvatol for microscopy. All incubation and wash steps were conducted at room temperature.
Image Deconvolution Analysis
Deconvolution images were obtained as described previously (17). Images were captured with a DeltaVision deconvolution microscope system (Applied Precision, Issaquah, WA). The system includes a Photometrics charge-coupled device mounted on a Nikon TE-200 inverted epifluorescence microscope. In general, between 30 and 60 optical sections spaced by ~0.1 μm were taken. Exposure times were set such that the camera response was in the linear range for each fluorophore. The data sets were deconvolved and analyzed by using SOFTWORX software (Applied Precision) on a Silicon Graphics Octane work station.
Sample Preparation for Expression Array Analysis
Total cellular RNA was isolated from cells by using RNeasy mini-columns (Qiagen, Santa Clara, CA) and reverse-transcribed with SuperScript II reverse transcriptase and an oligo(dT) primer containing a T7 RNA polymerase promoter. Single-stranded cDNA was converted into double-stranded cDNA and then purified (MessageAMP aRNA kit, Ambion, Austin, TX). Biotinylated cRNA was generated from double-stranded cDNA by in vitro transcription (MessageAMP aRNA Kit, Ambion). After a further round of purification, in vitro transcription reactions yielded 30–70 μg of biotinylated cRNA, which was fragmented to ~100-bp fragments before hybridization. Each Affymetrix 430 A chip was hybridized to 10 μg of fragmented cRNA. Arrays were hybridized and scanned with a GeneArray scanner (Hewlett-Packard/Affymetrix). For each array, the CEL files were generated with the Affymetrix Microarray Suite 5.0 and analyzed with GC RMA (18). Data are available at GEO (http://www.ncbi.nlm.nih.gov/projects/geo/) under the accession number GSE9727.
Regression Analysis and Hierarchical Ordered Partitioning and Collapsing Hybrid (HOPACH) Clustering of Transcripts
Log base 2 expression intensity estimates for all 22,690 probe sets on the Affymetrix chip were modeled using regression methods with the R statistical program utilizing the LIMMA package from the Bioconductor project. For each probe set, a polynomial regression model was fit to the 26 expression values with separate intercepts, and linear and quadratic time terms at 0, 2, 6, and 24 h after treatment, plus linear and quadratic time-by-cell line (WT versus D−) interaction terms. We used the p value from the moderated F statistic (LIMMA interaction p value) for the simultaneous contribution of the linear and quadratic interaction coefficients to test for time-dependent differential expression between WT and D− cells over 24 h of treatment. We found 2,572 probe sets with a LIMMA interaction p < 0.05. To functionally annotate these differentially expressed transcripts, we clustered them with HOPACH (19) and annotated clusters with Gene Ontology (GO) terms using the MAPPFinder program (20). The raw output was filtered to show the most non-redundant GO terms with the GO slim program.4 The top five GO terms ranked by Z-score are shown for each cluster. Complete unfiltered GO results are available as supplemental Table 1.
Real-time Quantitative Reverse Transcriptase-PCR (Real-time-PCR)
We used total RNA, prepared as described above, to generate cDNA templates with reverse transcriptase SuperScript II (Invitrogen) and quantified cDNA amplicons by incorporation of SYBR Green (Qiagen) into double-stranded DNA. Samples were compared by the relative (comparative) Ct method. -Fold induction or repression was measured relative to controls and calculated after adjusting for GAPDH using 2−ΔΔCt, where ΔCt = Ct interested gene − Ct GAPDH and ΔΔCt = ΔCt treatment − ΔCt control.
Immunoblot Analysis
For CREB and ICER (inducible cAMP early repressor) immunoblots, cells were centrifuged at various time points after treatments, washed twice in ice-cold PBS, and lysed (4 × 106 cells/aliquot) in 0.2 ml of SDS sample buffer (2% SDS, 10% glycerol, 0.01% bromphenol blue, 62.5 mM Tris-HCl, pH 6.8, 2.5% β-mercapto-ethanol), incubated for 30 min on ice, sonicated three times for 10 s, and centrifuged (5 min at 4 °C, 14,000 rpm). For cytochrome c and SMAC immunoblots, 2 × 107 cells were lysed in 0.5 ml of isotonic buffer containing 250 mM sucrose, 20 mM Hepes-KOH (pH 7.5), 10 mM KCl, 1.5 mM MgCl2, 1 mM Na-EDTA, 1 mM Na-EGTA, 1 mM dithi-othreitol, and a mixture of protease inhibitors, incubated for 15 min on ice, ground in a glass Dounce homogenizer with a tight pestle (b-type) about 30–50 times, and then centrifuged at 800 × g for 10 min. The supernatant was further centrifuged at 100,000 × g for 1 h in a Beckman Optima TLX ultracentrifuge. Lysates were treated and assayed on 12% NuPAGE bis-Tris gels according to the manufacturer’s instructions. Filters were blocked with 3% nonfat dry milk in PBS for 30 min and probed with primary antibodies overnight at 4 °C. After five washes in PBS containing 0.1% (v/v) Tween 20, filters were incubated with horseradish peroxidase-conjugated secondary antibodies at a final dilution of 1/5,000 and then washed five times. Proteins were visualized by ECL (Amersham Biosciences) using a UVP BioImaging system. Filters were stripped in Restore™ (Pierce) and reprobed with GAPDH antibody or Creb antibody for normalization.
Statistical Analyses
For apoptosis, mitochondrial membrane depolarization, caspase-3 and -8 activity, and real-time PCR analysis, all determinations were performed in duplicate or triplicate; each experiment was repeated at least three times, and values are expressed as mean ± S.E. For microarray analysis, we computed permutation t test statistics with the multtest package in R (from the Bioconductor project) and -fold calculations with the log2 GC RMA signal values. Four microarrays (n = 4) were used to hybridize baseline (0 h) experimental replicates; three microarrays were used for each time of treatment (2, 6, and 24 h). Because we had three replicates per time point, the permutation tests resulted in the smallest p values of either 0.02 or 0.057; therefore, we used a p < 0.06 and an absolute -fold change < 2 as the criteria for significance. Regression analysis was conducted as described above. For determination of protein level kinetics, statistical significance was determined by one-way analysis of variance followed by Bonferroni’s post hoc test for all possible pairwise comparisons. Comparison between two groups was based on an unpaired Student’s t test. A probability value of p < 0.05 was considered to be statistically significant; each experiment was repeated at least three times, and error bars represent the means ± S.E. in Figs. 1–3, 5, and 6.
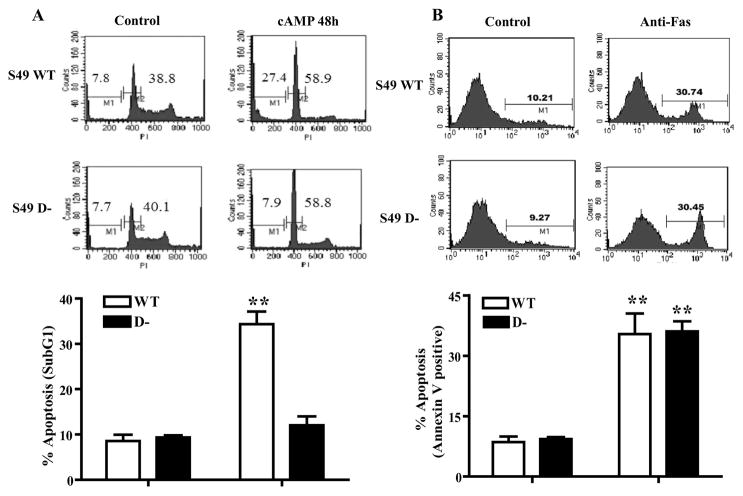
FIGURE 1. WT and D− S49 lymphoma cells reveal different patterns of cAMP-promoted apoptosis.
WT and D− S49 cells treated with CPT-cAMP for 48 h or anti-Fas antibody for 16 h and then G1 arrest and apoptosis were assessed. A, upper panels, representative histograms of S49 cells stained with propidium iodide (PI) to analyze G1 phase (M2) and Sub-G1 levels of DNA for apoptosis (M1) by flow cytometry. Lower panel, data from multiple experimental replicates are represented as percentage of apoptosis (SubG1), (n = 3), **, p < 0.001 when compared with control. B, upper panels, representative histograms of S49 cells stained with annexin V to analyze apoptosis (M1) by flow cytometry. Lower panel, data from multiple experimental replicates are represented, indicating percentage of apoptosis (annexin V-positive), (n = 3). **, p < 0.001 when compared with control.
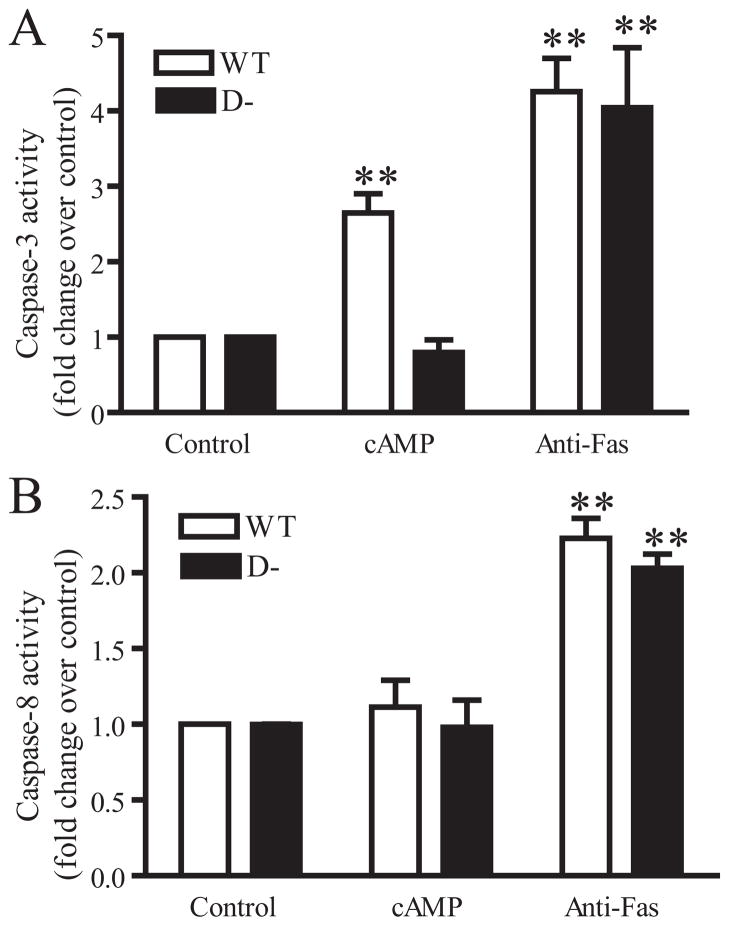
FIGURE 3. 8-CPT-cAMP increases caspase-3 activity in WT but not D− S49 cells, whereas caspase-8 activity is unaltered in either cell type by 8-CPT-cAMP.
WT and D− S49 cells were treated with 8-CPT-cAMP for 24 h or anti-Fas antibody for 16 h. Caspase activities were assayed according to “Experimental Procedures.” A, caspase-3 activity is expressed as -fold change in treated versus untreated cells (n = 3). B, caspase-8 activity, expressed as -fold change, in treated versus untreated cells (n = 3).
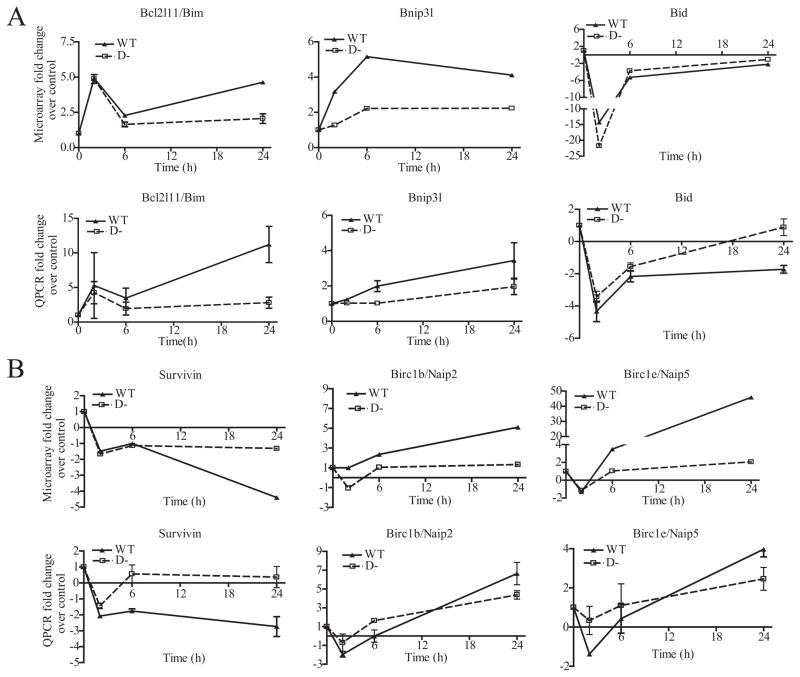
FIGURE 5. cAMP-PKA regulated pro- and anti-apoptotic genes in the Bcl2 and IAP families in WT and D− S49 cells.
A, differentially expressed genes in the Bcl-2 family: microarray data (upper panels) and real-time quantitative reverse transcriptase-PCR (QPCR) data (lower panels). Values were normalized to baseline expression (0 h). B, differentially expressed genes in the IAP family: microarray data (upper panels) and QPCR data (n = 3, lower panels).
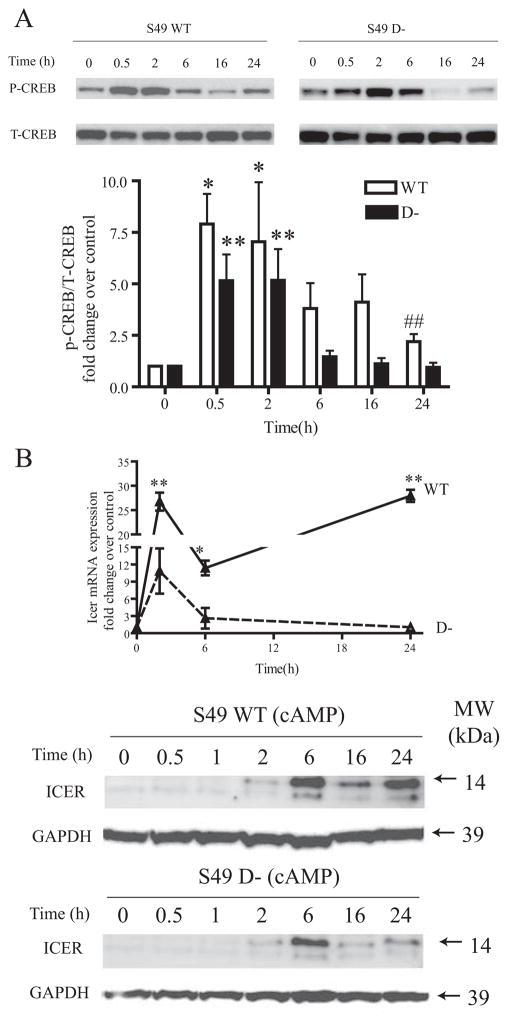
FIGURE 6. P-CREB and ICER induction by 8-CPT-cAMP treatment of WT and D− S49 cells.
WT and D−, S49 cells were treated with 8-CPT-cAMP for the indicated times. A, upper panel, representative immunoblot of P-CREB and total CREB (T-CREB). Lower panel, immunoblots with quantification of results from multiple (n = 6) replicates. Data are expressed as the ratio of phospho-CREB and total-Creb increase relative to control. Both lines displayed significant (p < 0.05) increases in protein levels by one-way analysis of variance over the treatment time course but did not show a significant interaction by two-way analysis of variance. Pairwise comparisons were also made: *, p < 0.05, **, p < 0.01 when compared with control (0 h) for each cell line; ##, p < 0.01 D− versus WT protein expression at the 24-h time. B, mRNA and protein expression levels of Icer. Upper panel, real-time PCR analysis of Icer mRNA expression after 8-CPT-cAMP treatment of WT and D− S49 cells, with data expressed as -fold change relative to control, unstimulated cells (n = 3); lower panel, representative immunoblot of ICER protein expression in WT and D− S49 cells treated with 8-CPT-cAMP for the indicated times (n = 3). Error bars indicate S.E. MW, molecular size markers; GADPH, glyceraldehyde-3-phosphate dehydrogenase.
RESULTS
PKA-mediated Apoptosis in Wild-type (WT) and Deathless (D−) S49 Lymphoma Cells
D− S49 cells are similar to WT S49 cells in that they have normal PKA activity and cAMP/PKA-induced G1 arrest, but akin to kin− S49 cells, D− cells are resistant to the apoptosis produced in WT S49 cells by elevated cAMP (12) (shown as sub-G1 cells in Fig. 1A). Thus, D− S49 cells provide a unique model system to identify mechanisms and pathways involved in cAMP/PKA-promoted apoptosis. The phenotype of D− cells is specific to cAMP/PKA-induced apoptosis since D− cells undergo apoptosis with similar kinetics to WT S49 cells in response to several inducers of apoptosis, e.g. treatment with dexamethasone (10), UV light (data not shown), and anti-Fas antibody (Fig. 1B).
cAMP/PKA Acts via a Mitochondrial Pathway to Induce Apoptosis in S49 Cells
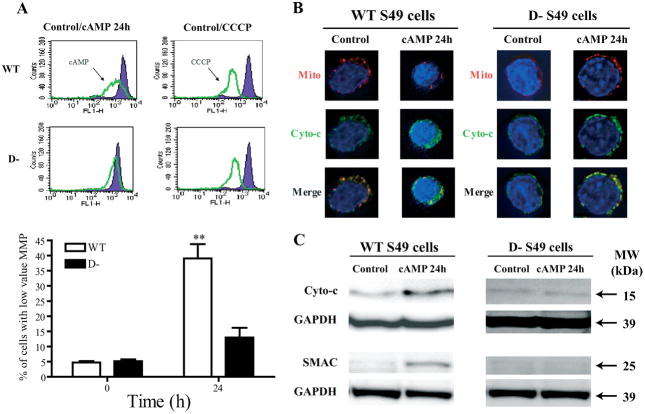
Previous data indicate that Bcl2 family members likely play an important role in cAMP-mediated apoptosis in WT S49 cells (2, 9). Since the action of such Bcl2 family members generally involves the mitochondria (21), we hypothesized that cAMP may act via a mitochondrial pathway to induce apoptosis. Indeed, treatment of WT cells with cAMP resulted in a prominent loss of mitochondrial membrane potential (Fig. 2A), whereas D− cells displayed a much smaller decrease in mitochondrial membrane potential in response to treatment with cAMP than did WT cells (Fig. 2A) despite having a similar loss of mitochondrial membrane potential as do WT cells in response to carbonyl cyanide m-chlorophenylhydrazone, an uncoupler of oxidative phosphorylation and disruptor of mitochondrial membrane potential. We corroborated the results for loss of mitochondrial membrane potential by using immunocytochemistry, which showed that cAMP induced a loss of cytochrome c from mitochondria in WT, but not D−, S49 cells (Fig. 2B). Apoptosis induced by cAMP can require transcriptional up-regulation of smac/diablo (22); we found that cAMP induced a release of SMAC into the cytosol in WT, but not D−, S49 cells (Fig. 2C).
FIGURE 2. cAMP induces loss of MMP and release of cytochrome c and SMAC from mitochondria in WT S49 cells.
A, upper panels, representative flow cytometry histograms of CPT-cAMP-induced alterations in MMP assayed with DiOC6 staining of WT and D− cells. Lower panel, data from multiple experimental replicates indicate percentage of cells with low MMP values (< 103 FL-1 signal), (n = 3). Error bars indicate S.E. **, p < 0.01 when compared with baseline. CCCP, carbonyl cyanide m-chlorophenylhydrazone. B, immunocytochemistry of mitochondria (Mito) and cytochrome c (Cyto-c) shows cAMP-mediated release of cytochrome c from the mitochondria and reduced mitochondrial staining in WT but not D− cells. C, representative immunoblot of cytochrome c and SMAC in cell lysates prepared according to “Experimental Procedures.” GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MW, molecular size markers.
cAMP-mediated Apoptosis Is Fas-independent
Cyclic AMP/PKA activation increases caspase-3 activity in WT but not D− S49 cells but does not increase caspase-8 activity in either cell type (Fig. 3). Caspase-3 activity can increase via either the mitochondria-dependent (intrinsic) apoptotic pathway or the death receptor/caspase-8-dependent (extrinsic) pathway (23, 24). To distinguish the effects of cAMP/PKA on those two pathways, we treated WT and D− S49 cells with anti-Fas antibody. Incubation of WT or D− S49 cells with anti-Fas antibody increased caspase-3 and -8 activities (Fig. 3). Together with the data above, these results show that cAMP/PKA acts via a Fas-independent, mitochondrial-mediated apoptotic pathway in WT S49 cells and that the apoptotic protective mutation in D− cells is upstream of caspase-3 activity.
Gene Expression Profiling of WT and D− S49 Cells Reveals That cAMP/PKA-mediated Apoptosis Is Associated with the Regulation of Lysosomal, Golgi, RNA Splicing, and Mitotic Genes
At zero time (basal conditions) prior to treatment with 8-CPT-cAMP, we identified 69 and 92 transcripts that are significantly (|Fold| > 2, p < 0.06) up- or down-regulated, respectively, in D− cells when compared with WT (supplemental Table 2). Gene Ontology analysis of these transcripts indicated that when compared with WT cells, D− had an up-regulated expression of genes encoding proteins compartmentalized in the lamellipodium (n = 3, p < 0.05) and involved in immune cell activation (n = 5, p < 0.05), lymphocyte differentiation (n = 3, p < 0.05), and immune response (n = 9). By contrast, transcripts involved in the negative regulation of transport (n = 3, p < 0.05), response to biotic stimulus (n = 15, p < 0.05), and integrin-mediated signaling (n = 3, p < 0.05) were down-regulated in D− cells when compared with WT cells under basal conditions.
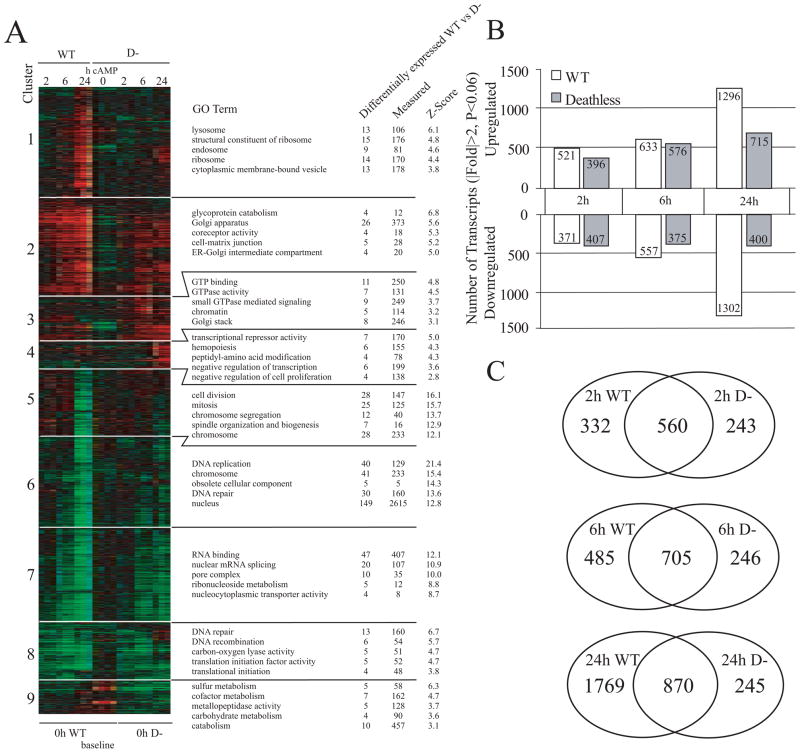
We used regression analysis (see “Experimental Procedures”) to identify transcripts that are differentially regulated between cAMP-treated WT and D− cells in an effort to identify key differences in gene regulation between the two cell types. We found ~2,500 transcripts that showed a significant difference (p < 0.05) in the kinetics of their regulation between WT and D− cells over the 24-h treatment period with 8-CPT-cAMP (Fig. 4A). Clustering and annotation of these transcripts indicated that several of these differentially regulated genes could be classified into functional groups. When compared with D− cells, WT cells had increased expression of lysosomal, ribosomal, and Golgi-related transcripts as a function of time of incubation with CPT-cAMP (Fig. 4A, clusters 1 and 2). Both the lysosomal and the Golgi compartments are known to undergo changes during apoptosis (25, 26). We also found several clusters of nuclei-related transcripts involved in mitosis and RNA processing and splicing that were down-regulated to a greater extent in WT versus D− cells 24 h after CPT-cAMP treatment (Fig. 4A, clusters 6 and 7). Alternative splicing has been shown to be an important mediator of apoptosis (27). In support of this, we found evidence of alternative splicing of several apoptotic mediators, as evidenced by the highly divergent signals of certain microarray probe sets that are specific to different transcript isoforms for a single gene (e.g. smac, bnip3l, data not shown, and crem/icer, see Fig. 6B).
FIGURE 4. Genome wide transcriptional analysis of WT and D− S49 cells treated with 8-CPT-cAMP for 24 h.
A, HOPACH cluster of 2,572 probe sets with interaction p < 0.05 reveals global differences in the kinetics of transcriptional regulation between WT and D− cells at 2, 6, and 24 h after treatment with 8-CPT-cAMP. Ratios were generated using baseline expression values as defined underneath the cluster figure. Rows represent individual transcripts, and columns represent experimental replicates. Red, green, and black colors indicate genes whose expression is up-regulated, down-regulated, or unchanged, respectively. Each main cluster was annotated by GO analysis to biologically describe the major differences in the regulation of transcripts between WT and D− cells over the course of 8-CPT-cAMP treatment. Differentially expressed WT vs D− indicates the number of transcripts in the defined cluster, Measured indicates the number of transcripts on the array that belong to the stated GO term, and Z-Score calculations are described in (20). A Z-score of 2 or greater is roughly equivalent to a p < 0.05. B, number of transcripts either up-regulated or down-regulated in WT and D− S49 cells at the defined time points and statistical cutoffs after 8-CPT-cAMP treatment. C, Venn diagrams indicating the numbers of transcripts regulated (|Fold| <2, p < 0.06) in common between the WT and D− cells at the defined time points. All common transcripts were regulated in the same directions between the two cell types (data not shown).
By contrast with the results observed after a 24-h incubation with 8-CPT-cAMP, a time at which WT cells had more than twice as many transcripts change >2-fold relative to D− cells, incubation with the cAMP analog for 2 and 6 h yielded more similar numbers of treatment-related changes and substantial overlap in identity of transcripts between WT and D− cells (Fig. 4, B and C). The latter results imply that the majority of gene expression changes reflective of apoptosis require >6 h of elevated cAMP levels and that the apoptotic resistance in D− cells is associated with a global blunting of gene expression at the 24-h time point. Washout experiments have revealed that >6 h of treatment with CPT-cAMP is required to irreversibly commit WT S49 cells to apoptosis (2). Because most of the gene expression changes in the two cell lines were similar at the 2-and 6-h time points (Fig. 4C), the apoptotic-resistant phenotype of D− cells may result from a relatively small subset of genes, although apoptosis and its consequences likely involve many more transcripts, as reflected by the much larger differences in gene expression observed between WT and D− treated with CPT-cAMP for 24 h.
WT and D− Cells Have Large Differences in Transcript Regulation 24 h after 8-CPT-cAMP Treatment
Transcriptional or posttranscriptional regulation of particular transcription factors is a likely explanation for the large differences in the number of transcripts between WT and D− cells treated with CPT-cAMP for 24 h. We found a cluster of seven transcriptional repressors that were up-regulated in the D− (but not WT) cells at 24 h, and thus, that may contribute to the differences in global mRNA regulation between the two cell lines (included in Fig. 4A, cluster 4, and in Table 1). Of note, down-regulation of one of these repressors, CCCTC-binding factor, has been shown to be required for stress-induced apoptotic response of human hematopoietic myeloid cells (28).
TABLE 1.
Transcriptional repressors from cluster 4
| Gene title | Gene symbol | -Fold 2-h WT | 2-h WT p value | -Fold 6-h WT | 6-h WT p value | -Fold 24-h WT | 24-h WT p value | -Fold 2-h D− | 2-h D−p value | -Fold 6-h D− | 6-h D−p value | -Fold 24-h D− | 24 h D− p value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CCR4-NOT transcription complex, subunit 8 | Cnot8 | 1.1 | 0.51 | −1.0 | 0.66 | −1.1 | 0.46 | 1.0 | 0.37 | −1.2 | 0.09 | 1.3 | 0.03 |
| MYST histone acetyltransferase monocytic leukemia 4 | Myst4 | −1.1 | 0.77 | 1.1 | 0.91 | 3.6 | 0.03 | 1.1 | 0.63 | 1.9 | 0.03 | 8.8 | 0.03 |
| RE1-silencing transcription factor | Rest | −1.5 | 0.11 | −1.2 | 0.23 | 1.0 | 1.00 | −1.6 | 0.17 | −1.3 | 0.14 | 2.2 | 0.03 |
| methyl-CpG binding domain protein 2 | Mbd2 | −1.1 | 0.17 | −1.0 | 1.00 | −1.1 | 0.23 | −1.1 | 0.34 | 1.2 | 0.06 | 1.2 | 0.06 |
| Nuclear factor of κ light polypeptide gene enhancer in B-cells inhibitor, ε | Nfkbie | 1.2 | 0.37 | −1.3 | 0.40 | 1.5 | 0.09 | 2.2 | 0.03 | 1.3 | 0.37 | 5.6 | 0.03 |
| CCCTC-binding factor | Ctcf | 1.2 | 0.06 | −1.1 | 0.51 | −1.2 | 0.11 | 1.1 | 0.40 | −1.0 | 0.83 | 1.2 | 0.09 |
| Dr1-associated protein 1 (negative cofactor 2 α) | Drap1 | 1.1 | 0.54 | −1.1 | 0.66 | −1.0 | 0.83 | 1.1 | 0.40 | 1.1 | 0.40 | 1.4 | 0.03 |
Extension of our analysis revealed that 70 transcription factors had interaction p values of < 0.05 (supplemental Table 3), highlighting potential targets for the transcriptional blunting effect seen at the 24-h time point in WT cells when compared with D− cells. The most significantly changed such transcript was peroxisome proliferator-activated receptor-γ (p < 0.000001), which was up-regulated >15-fold at 24 h in WT cells but without significant change in D− cells. Peroxisome proliferator-activated receptor-γ has been shown to inhibit proliferative response, decrease viability, and increase apoptosis in T-cells (29, 30). Stat1 and an interferon γ (IFN-γ)-activated transcript (ifi203) also displayed significantly different expression patterns between WT and D− cells incubated with CPT-cAMP, suggesting that the IFN-γ pathway is more active in WT than in D− S49 cells. Further analysis revealed that mRNA expression of IFN-γ and both of its receptors was up-regulated to a greater extent in WT than in D− cells (see Fig. 7). We tested whether recombinant IFN-γ acts synergistically with cAMP to promote killing in D− or whether a blocking antibody to IFN-γ could inhibit killing by cAMP in WT cells but found that neither treatment significantly affected cAMP-promoted apoptosis of WT or D− S49 cells (data not shown).
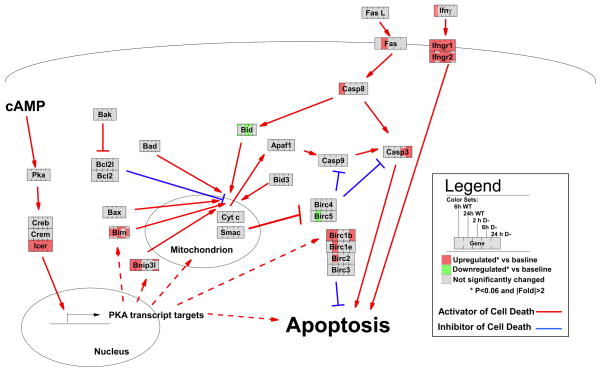
FIGURE 7. GenMAPP (48) pathway-based representation of the kinetics of gene expression induced by 8-CPT-cAMP in WT and D− S49 cells as measured by microarray data.
The pathway represents key transcripts and proteins studied and discussed in this study. Proteins are represented as boxes. Colors represent transcript -fold changes measured by microarray. Solid lines indicate known direct interactions between proteins, and dashed lines indicate potential direct interactions or indirect interactions. Incompletely shaded boxes (e.g. caspase-3 at the 6-h time point for D− cells) indicate the presence of multiple transcripts for the same gene with different expression profiles (i.e. putative alternative splicing).
cAMP-mediated Regulation of Bcl-2 and IAP Gene Families
WT and D− S49 cells displayed differential regulation of the Bcl-2 and IAP gene families, as shown by both microarray and real-time PCR analysis. For the Bcl-2 family, cAMP/PKA up-regulated the expression of the pro-apoptotic genes bim and bnip3l, whereas down-regulating bid (Fig. 5A). In addition, cAMP exhibited bidirectional regulation of pro-survival genes of the IAP family: down-regulation of survivin (birc5) and up-regulation of nAIP2 (birc1b-neuronal apoptosis inhibitory protein) and nAIP5 (birc1e). Of these pro- and anti-apoptotic genes, bim and Survivin showed the most pronounced differences in expression between WT and D− cells. Bim expression levels were higher and survivin levels were lower in WT cells when compared with D− cells, implicating these genes as potential regulators of apoptosis in S49 cells.
Sustained Expression of Icer May Contribute to cAMP-mediated Gene Regulation to Promote Cell Death
Cyclic AMP can regulate transcription factors by inducing their phosphorylation or changing their expression levels (31). PKA phosphorylates the cAMP-response element (CRE)-binding protein CREB at serine 133 and activates it to stimulate the transcription of many cAMP-responsive genes by binding to CRE sequences in their promoters. In S49 cells, cAMP-mediated apoptosis is dependent on PKA and phosphorylated CREB (P-CREB) (2). To test the hypothesis that P-CREB might contribute to differences in transcriptional patterns in WT and D− S49 cells, we assessed the time course of CREB phosphorylation and found a similar pattern between WT and D cells for the first 2 h of treatment of cells with CPT-cAMP (Fig. 6A). However, the responses diverged thereafter with WT cells showing a higher and more prolonged level of P-CREB than did D− cells.
Icer is an isoform of the Crem family of CRE modulators that is induced by Creb. We found that Icer mRNA and protein expression levels had a similar kinetic pattern with higher values in WT than in D− S49 cells at all time points (Fig. 6B). In WT cells, icer displayed a biphasic increase, peaking at 2 h and then again at 24 h. This biphasic response is absent in D− cells, which showed a smaller peak of icer than did WT cells at 2 h and a return to basal levels by 24 h. The differential regulation of CREB and ICER proteins likely contributes to the global differences in transcriptional regulation observed 24 h after cAMP treatment of WT and D− cells and thus may help regulate the pro-apoptotic responses to cAMP in WT S49 cells.
DISCUSSION
Cyclic AMP and PKA can stimulate proliferation (and be anti-apoptotic) or promote cell death via apoptosis depending on the cell type (32). For example, cAMP analogs promote apoptosis in lymphoid cells (2) but protect certain other cell types from apoptosis; the latter include neurons (1), hepatocytes (33), neutrophils (34), and intestinal epithelial cells (35). In the current study, we used S49 lymphoma cells as a unique model system to investigate how cAMP/PKA induces apoptosis by assessing mechanisms and gene expression patterns that relate to apoptosis in WT S49 cells and in D−, a mutant S49 cell line that is resistant to apoptosis. Our results show that cAMP/PKA-induced apoptosis in S49 cells involves activation of the intrinsic apoptotic pathway (i.e. mitochondrial membrane potential depolarization and mitochondrial release of cytochrome c and SMAC) and is FAS-independent. SMAC release was not observed in either D− cells treated with cAMP or kin− S49 cells that lack PKA activity.5 Our findings that implicate SMAC in cAMP/PKA-mediated apoptosis are consistent with data in other cell systems (22). SMAC can antagonize the IAP proteins that inhibit activated caspases and sensitize leukemic cells to apoptosis (36) (Fig. 7).
Our microarray analyses indicated that cAMP/PKA-promoted apoptosis likely involves the up-regulation of transcripts that are involved in lysosomal, ribosomal, and Golgi-related functions and a down-regulation of cell cycle, nuclear, RNA splicing, and translational initiation transcripts (Fig. 4A). Most striking was the global blunting of changes in gene expression in D− cells when compared with WT cells at 24 h, implying that the D− phenotype involves the delayed regulation of large groups of genes, perhaps as a result of differences in expression of particular transcription factors. Our data are the first to show that cAMP and PKA can down-regulate survivin (birc5) and up-regulate naip2 (birc1b) and naip5 (birc1e) (Fig. 7). Survivin is highly expressed in transformed cell lines and in human lung, colon, pancreas, prostate, and breast cancers and in ~50% of high grade non-Hodgkin lymphomas but undetectable in terminally differentiated adult tissues (37). Survivin suppresses activation and/or activity of caspase-3 (38), although others have found that it can bind caspase-9 and/or neutralize the proapoptotic protein, DIABLO/SMAC (39, 40). Thus, decreased expression of survivin may contribute to cAMP/PKA-mediated apoptosis in S49 cells. Our results also implicate Bim, a proapoptotic Bcl-2 family member, as the most differentially regulated between WT and D− cells; in addition, the expression pattern of Bim after CPT-cAMP suggests that it may help regulate apoptosis in S49 cells (Fig. 7) (2).
Differential levels of expression of icer in WT and D− S49 cells may also contribute to cAMP/PKA-promoted apoptosis in the WT S49 cells. Icer, a crem isoform that is associated with regulation of cell growth, is produced by an alternative start site in the crem gene locus (41). ICER is a powerful inhibitor of cAMP-induced transcription because it contains the DNA binding and leucine zipper domains but not the N-terminal transactivation domain of CREM. Icer can be rapidly induced by cAMP and then shuts off cAMP-inducible gene expression driven by CRE DNA consensus elements, including its own promoter that harbors two closely spaced CREs (42). Sustained elevation of ICER levels can elicit pathological consequences, such as death of neuronal cells and cardiomyocytes (43, 44). ICER-promoted apoptosis occurs in part through CREB-mediated transcription and down-regulation of bcl-2 (45), although we did not find a significant change in bcl-2 expression, as measured by microarray in either WT or D− cells. CPT-cAMP induces phosphorylation of CREB and increases icer expression in WT but not kin− S49 cells (2, 11). Other data indicate that phosphorylation of CREB appears to be necessary, but not always sufficient, for the activation of icer expression (46, 47).
In conclusion, the current data show that cAMP promotes apoptosis of WT S49 cells via a mitochondrial signaling pathway that depends on PKA activity and transcriptional regulation. cAMP/PKA regulates both pro-apoptotic and anti-apoptotic genes in S49 cells (summarized in Fig. 7). Our data implicate multiple genes and pathways as contributing to cAMP/PKA-promoted apoptosis and the anti-apoptotic phenotype of D− cells. It is conceivable that a limited subset of such genes and their gene products serve as “master regulators,” i.e. rate-limiting pro-/anti-apoptotic genes, whose altered expression and action in response to increases in cellular cAMP levels and PKA activation critically control the commitment to and events mediating apoptosis. Our future studies will focus on candidate genes identified in the current experiments that may serve as such regulators.
Supplementary Material
Footnotes
This work was supported by Grants from the Leukemia and Lymphoma Society, National Institutes of Health (GM61774), and National Institutes of Health sponsored Institutional Research and Academic Career Development Award (IRACDA) fellowship (GM06852).
The on-line version of this article (available at http://www.jbc.org) contains three supplemental tables.
The abbreviations used are: PKA, protein kinase A; WT, wild type; 8-CPT-cAMP, 8-(4-chlorophenylthio)-adenosine-3′:5′-cyclic monophosphate; CRE, cAMP-response element; CREB, CRE-binding protein; P-CREB, phosphorylated CREB; ICER, inducible cAMP early repressor; IAP, inhibitor of apoptosis; MMP, mitochondrial membrane potential; pNA, p-nitroaniline; PBS, phosphate-buffered saline; GO, Gene Ontology; real-time PCR, real-time quantitative reverse transcriptase-PCR; SMAC, second mitochondria-derived activator of caspase; IFN, interferon; HOPACH, hierarchical ordered partitioning and collapsing hybrid; bis-Tris, 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol.
N. Salomonis, A. C. Zambon, and B. R. Conklin, manuscript in preparation.
L. Zhang and P. A. Insel, unpublished results.
References
- 1.Wang X, Tang X, Li M, Marshall J, Mao Z. J Biol Chem. 2005;280:16705–16713. doi: 10.1074/jbc.M501819200. [DOI] [PubMed] [Google Scholar]
- 2.Zhang L, Insel PA. J Biol Chem. 2004;279:20858–20865. doi: 10.1074/jbc.M310643200. [DOI] [PubMed] [Google Scholar]
- 3.Krett NL, Zell JL, Halgren RG, Pillay S, Traynor AE, Rosen ST. Clin Cancer Res. 1997;3:1781–1787. [PubMed] [Google Scholar]
- 4.Halgren RG, Traynor AE, Pillay S, Zell JL, Heller KF, Krett NL, Rosen ST. Blood. 1998;92:2893–2898. [PubMed] [Google Scholar]
- 5.Ji Z, Mei FC, Johnson BH, Thompson EB, Cheng X. J Biol Chem. 2007 Sep 25;282:37370–37377. doi: 10.1074/jbc.M703697200. [DOI] [PubMed] [Google Scholar]
- 6.Yan L, Herrmann V, Hofer JK, Insel PA. Am J Physiol. 2000;279:C1665–C1674. doi: 10.1152/ajpcell.2000.279.5.C1665. [DOI] [PubMed] [Google Scholar]
- 7.Fuld S, Borland G, Yarwood SJ. Exp Cell Res. 2005;309:161–173. doi: 10.1016/j.yexcr.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 8.Tiwari S, Felekkis K, Moon EY, Flies A, Sherr DH, Lerner A. Blood. 2004;103:2661–2667. doi: 10.1182/blood-2003-06-2154. [DOI] [PubMed] [Google Scholar]
- 9.Zhang L, Insel PA. Am J Physiol. 2001;281:C1642–C1647. doi: 10.1152/ajpcell.2001.281.5.C1642. [DOI] [PubMed] [Google Scholar]
- 10.Albert DA. J Clin Investig. 1995;95:1490–1496. doi: 10.1172/JCI117820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zambon AC, Zhang L, Minovitsky S, Kanter JR, Prabhakar S, Salomonis N, Vranizan K, Dubchak I, Conklin BR, Insel PA. Proc Natl Acad Sci U S A. 2005;102:8561–8566. doi: 10.1073/pnas.0503363102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemaire I, Coffino P. Cell. 1977;11:149–155. doi: 10.1016/0092-8674(77)90325-7. [DOI] [PubMed] [Google Scholar]
- 13.Muller M, Wilder S, Bannasch D, Israeli D, Lehlbach K, Li-Weber M, Friedman SL, Galle PR, Stremmel W, Oren M, Krammer PH. J Exp Med. 1998;188:2033–2045. doi: 10.1084/jem.188.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joza N, Kroemer G, Penninger JM. Trends Genet. 2002;18:142–149. doi: 10.1016/s0168-9525(01)02618-x. [DOI] [PubMed] [Google Scholar]
- 15.Baell JB, Huang DC. Biochem Pharmacol. 2002;64:851–863. doi: 10.1016/s0006-2952(02)01148-6. [DOI] [PubMed] [Google Scholar]
- 16.Roy N, Deveraux QL, Takahashi R, Salvesen GS, Reed JC. EMBO J. 1997;16:6914–6925. doi: 10.1093/emboj/16.23.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swaney JS, Roth DM, Olson ER, Naugle JE, Meszaros JG, Insel PA. Proc Natl Acad Sci U S A. 2005;102:437–442. doi: 10.1073/pnas.0408704102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Z, Irizarry RA. Nat Biotechnol. 2004;22:656–658. doi: 10.1038/nbt0604-656b. author reply 658. [DOI] [PubMed] [Google Scholar]
- 19.van der Laan MJ, Pollard KS. Technical Report #93. Division of Biostatistics, University of California; Berkeley, CA: 2001. [Google Scholar]
- 20.Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim R, Emi M, Tanabe K, Toge T. Cancer. 2004;101:2491–2502. doi: 10.1002/cncr.20696. [DOI] [PubMed] [Google Scholar]
- 22.Martinez-Velazquez M, Melendez-Zajgla J, Maldonado V. Cell Signal. 2007;19:1212–1220. doi: 10.1016/j.cellsig.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Kim R, Tanabe K, Uchida Y, Emi M, Inoue H, Toge T. Cancer Chemother Pharmacol. 2002;50:343–352. doi: 10.1007/s00280-002-0522-7. [DOI] [PubMed] [Google Scholar]
- 24.Thorburn A. Cell Signal. 2004;16:139–144. doi: 10.1016/j.cellsig.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 25.He J, Tohyama Y, Yamamoto K, Kobayashi M, Shi Y, Takano T, Noda C, Tohyama K, Yamamura H. Genes Cells. 2005;10:23–35. doi: 10.1111/j.1365-2443.2004.00811.x. [DOI] [PubMed] [Google Scholar]
- 26.Machamer CE. Trends Cell Biol. 2003;13:279–281. doi: 10.1016/s0962-8924(03)00101-6. [DOI] [PubMed] [Google Scholar]
- 27.Schwerk C, Schulze-Osthoff K. Mol Cell. 2005;19:1–13. doi: 10.1016/j.molcel.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 28.Li T, Lu L. Exp Cell Res. 2007;313:3057–3065. doi: 10.1016/j.yexcr.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harris SG, Phipps RP. Eur J Immunol. 2001;31:1098–1105. doi: 10.1002/1521-4141(200104)31:4<1098::aid-immu1098>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 30.Harris SG, Phipps RP. Immunology. 2002;105:23–34. doi: 10.1046/j.0019-2805.2001.01340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Servillo G, Della Fazia MA, Sassone-Corsi P. Exp Cell Res. 2002;275:143–154. doi: 10.1006/excr.2002.5491. [DOI] [PubMed] [Google Scholar]
- 32.Muller Igaz L, Refojo D, Costas MA, Holsboer F, Arzt E. Biochim Biophys Acta. 2002;1542:139–148. doi: 10.1016/s0167-4889(01)00174-4. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Kim PK, Peng X, Loughran P, Vodovotz Y, Zhang B, Billiar TR. Apoptosis. 2006;11:441–451. doi: 10.1007/s10495-005-4293-6. [DOI] [PubMed] [Google Scholar]
- 34.Conran N, Almeida CB, Lanaro C, Ferreira RP, Traina F, Saad ST, Costa FF. Br J Haematol. 2007;139:148–158. doi: 10.1111/j.1365-2141.2007.06748.x. [DOI] [PubMed] [Google Scholar]
- 35.Nishihara H, Kizaka-Kondoh S, Insel PA, Eckmann L. Proc Natl Acad Sci U S A. 2003;100:8921–8926. doi: 10.1073/pnas.1533221100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verhagen AM, Vaux DL. Apoptosis. 2002;7:163–166. doi: 10.1023/a:1014318615955. [DOI] [PubMed] [Google Scholar]
- 37.Ambrosini G, Adida C, Altieri DC. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 38.Kobayashi K, Hatano M, Otaki M, Ogasawara T, Tokuhisa T. Proc Natl Acad Sci U S A. 1999;96:1457–1462. doi: 10.1073/pnas.96.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shinjyo T, Kuribara R, Inukai T, Hosoi H, Kinoshita T, Miyajima A, Houghton PJ, Look AT, Ozawa K, Inaba T. Mol Cell Biol. 2001;21:854–864. doi: 10.1128/MCB.21.3.854-864.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du C, Fang M, Li Y, Li L, Wang X. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 41.Molina CA, Foulkes NS, Lalli E, Sassone-Corsi P. Cell. 1993;75:875–886. doi: 10.1016/0092-8674(93)90532-u. [DOI] [PubMed] [Google Scholar]
- 42.Lamas M, Monaco L, Zazopoulos E, Lalli E, Tamai K, Penna L, Mazzucchelli C, Nantel F, Foulkes NS, Sassone-Corsi P. Philos Trans R Soc Lond B Biol Sci. 1996;351:561–567. doi: 10.1098/rstb.1996.0055. [DOI] [PubMed] [Google Scholar]
- 43.Jaworski J, Mioduszewska B, Sanchez-Capelo A, Figiel I, Habas A, Gozdz A, Proszynski T, Hetman M, Mallet J, Kaczmarek L. J Neurosci. 2003;23:4519–4526. doi: 10.1523/JNEUROSCI.23-11-04519.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding B, Abe J, Wei H, Xu H, Che W, Aizawa T, Liu W, Molina CA, Sadoshima J, Blaxall BC, Berk BC, Yan C. Proc Natl Acad Sci U S A. 2005;102:14771–14776. doi: 10.1073/pnas.0506489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomita H, Nazmy M, Kajimoto K, Yehia G, Molina CA, Sadoshima J. Circ Res. 2003;93:12–22. doi: 10.1161/01.RES.0000079794.57578.F1. [DOI] [PubMed] [Google Scholar]
- 46.Monaco L, Sassone-Corsi P. Oncogene. 1997;15:2493–2500. doi: 10.1038/sj.onc.1201636. [DOI] [PubMed] [Google Scholar]
- 47.Mioduszewska B, Jaworski J, Kaczmarek L. J Neurochem. 2003;87:1313–1320. doi: 10.1046/j.1471-4159.2003.02116.x. [DOI] [PubMed] [Google Scholar]
- 48.Salomonis N, Hanspers K, Zambon AC, Vranizan K, Lawlor SC, Dahlquist KD, Doniger SW, Stuart J, Conklin BR, Pico AR. BMC Bioinformatics. 2007;8:217. doi: 10.1186/1471-2105-8-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.