Abstract
Polycationic nanocarriers attract increasing attention to the field of siRNA delivery. We investigated the self-assembly of siRNA vs pDNA with polycations, which are broadly used for nonviral gene and siRNA delivery. Although polyethyleneimine (PEI) was routinely adopted as siRNA carrier based on its efficacy in delivering pDNA, it has not been investigated yet why PEI efficiently delivers pDNA to cells but is controversially discussed in terms of efficacy for siRNA delivery. We are the first to investigate the self-assembly of PEI/siRNA vs PEI/pDNA and the steps of complexation and aggregation through different levels of hierarchy on the atomic and molecular scale with the novel synergistic use of molecular modeling, molecular dynamics simulation, isothermal titration calorimetry, and other characterization techniques. We are also the fist to elucidate atomic interactions, size, shape, stoichiometry, and association dynamics for polyplexes containing siRNA vs pDNA. Our investigation highlights differences in the hierarchical mechanism of formation of related polycation–siRNA and polycation–pDNA complexes. The results of fluorescence quenching assays indicated a biphasic behavior of siRNA binding with polycations where molecular reorganization of the siRNA within the polycations occurred at lower N/P ratios (nitrogen/phosphorus). Our results, for the first time, emphasize a biphasic behavior in siRNA complexation and the importance of low N/P ratios, which allow for excellent siRNA delivery efficiency. Our investigation highlights the formulation of siRNA complexes from a thermodynamic point of view and opens new perspectives to advance the rational design of new siRNA delivery systems.
Keywords: siRNA delivery, DNA delivery, polyethyleneimine, molecular modeling, isothermal titration calorimetry, RT-PCR, supramolecular complexation
Nanomedicine is the engineering, manufacturing, and application of nanotechnology for medical applications, among others, for drug or nucleic acids delivery.1,2 One of the most important applications of nanomedicine is gene delivery, a powerful approach for the treatment of cancer and genetic diseases. Compared with viral counterparts and liposomes, polymeric gene delivery systems have the advantages of lower toxicity and immunogenicity by design and allow for industrial production involving good manufacturing practice.3 A wide range of polymeric vectors were designed and developed based on the complexation of nucleic acids via electrostatic interaction between the negatively charged phosphates along the nucleic acid backbone and the positive charges on the cationic polymers.4 The cationic polymer poly(ethyleneimine) (PEI) is one of the best studied vectors for nonviral gene delivery. Starting in the 1990s, the polymeric nonviral vector PEI has been developed to achieve successful delivery of nucleic acids such as plasmid DNA, antisense oligonucleotides, and ribozymes, and lately has been adopted for siRNA delivery.2 Since the discovery of gene silencing by introduction of double-stranded RNA,5 RNA interference is widely used in functional genomics and drug development.6,7 Although the delivery of siRNA faces many of the same barriers and intracellular steps as delivery of plasmid DNA, the delivery of siRNA appears more difficult than DNA delivery, and the design of high affinity, good protection agents is a key point in the development of nanocarriers for siRNA delivery systems. To investigate why PEI efficiently delivers pDNA to cells but is controversially discussed in terms of efficacy for siRNA delivery, in this study, we used isothermal titration calorimetry (ITC) to investigate the complexation behavior of siRNA and DNA with polycations. These thermodynamic parameters also allow for the study of the hierarchical aggregation phenomena that result from the biomolecular interactions between nucleic acids and cationic polymers. Because the knowledge on structure conformation of cationic polymers and genetic materials is limited, molecular dynamics (MD) simulation was used to investigate the local mechanism of binding between pDNA or siRNA molecules and cationic polymers, providing detailed insight into the structural conformations and binding behavior.8–10 This synergetic use of MD simulation and ITC provides a complete description not only of the local binding between polymers and nucleic acids but also of the hierarchical aggregation steps that occur during polyplex formation. Additionally, the complexation of DNA and siRNA was also studied using heparin assays and dye quenching assays, and subsequently in vitro transfection experiments were conducted with both siRNA and pDNA. Our investigations are focused on the study of binding mechanisms, the different location of plasmid DNA and siRNA within complexes of cationic polymers, their different structural conformations and biophysical parameters, and the size and surface charge of the final polyplexes. By investigating these parameters and correlating them with functional studies including knockdown of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene expression measured by RT-PCR, we try to find distinguishing features of siRNA complexation and to explain why the principle of DNA transfection cannot generally be directly applied to siRNA transfection.11
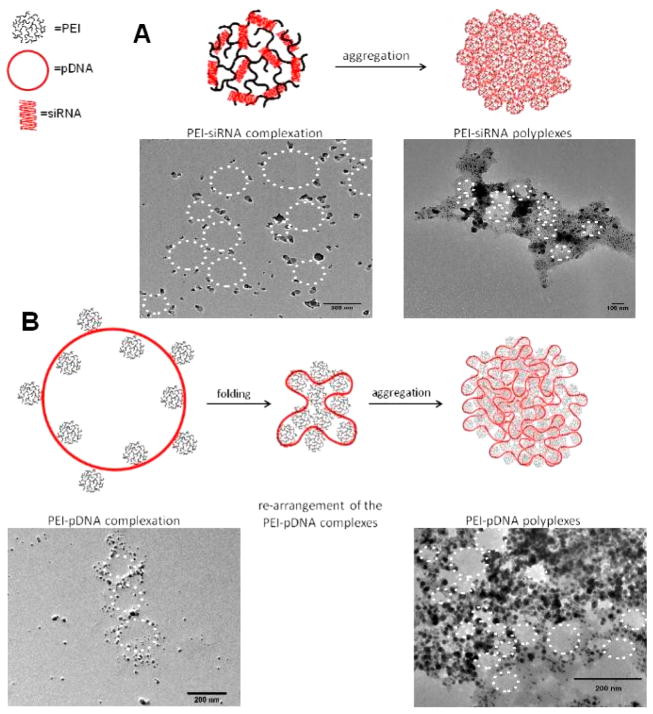
Our study of the complexation mechanism between nucleic acids and polycationic nanocarriers describes the very different nature of polycation–siRNA and polycation–DNA hierarchical aggregation. We chose PEI, the most widely used polycation for nucleic acid delivery, and an amphiphilic PEI-based triblock copolymer to elucidate the complex formation mechanism with siRNA vs pDNA and to investigate differences between hydrophilic and amphiphilic polycations. We demonstrate that siRNA complexation can be ideally schematized into two “rigid” steps, namely, (i) polycation–siRNA primary complexation, followed by the (ii) hierarchical association of multiple nanocomplexes into larger polyplexes (Figure 1A). DNA condensation, however, seems to be more extensive, involving multiple and more complex hierarchical steps. In fact, after the immediate binding of multiple PEI molecules to pDNA, the large primary complexes most probably undergo strong rearrangement and folding in solution. The aggregation of multiple primary complexes into polyplexes which follows the initial electrostatic interactions (Figure 1B) is thus less ordered and more chaotic than in the case of siRNA. In this hierarchical framework, siRNA aggregation results in a more uniform and stable complex formation, at low N/P ratios, which leads to increased siRNA delivery efficiency. Interestingly, with the following study of the relationship between nucleic acids/polycations aggregation mechanism and in vitro siRNA delivery efficiency, which is performed by RT-PCR and confocal laser scanning microscopy, the polycationic nanocarrier-based siRNA delivery systems showed the best knockdown effect with siRNA at N/P = 2, although higher N/P ratios were believed to be necessary until now by most of the researchers in the area of polycationic nanocarrier-based siRNA delivery. Our results show that self-assembled siRNA nanocarriers need to be characterized in more detail to optimize their delivery efficacy and that rules that apply for pDNA polyplexes cannot easily be transferred to siRNA.
Figure 1.
Model for different hierarchical aggregation mechanism. (A) PEI/siRNA. (B) PEI/pDNA. The synergic use of MD simulations, ITC, and dye quenching assays provides us a complete description not only of the local binding between polymers and nucleic acids but also of the hierarchical aggregation steps that occur during polyplex formation. TEM: During reduction of the silver cations into silver nanoparticles on the negatively charged sugar–phosphate backbone of the nucleic acids, siRNA and DNA were stained with Ag (black) and then condensed with polycations at low and high N/P ratios.
RESULTS AND DISCUSSION
Molecular Dynamics Simulation Study of PEI25 kDa Binding with DNA and siRNA
With the use of MD simulations, we aimed to compare the behavior of PEI25 kDa while binding DNA vs siRNA according to a 1:1 complexation model. All of the thermodynamic energies obtained from MD simulations were normalized per charge (and expressed in kcal mol−1) to allow for the comparison between the different nucleic acids (Figure 2). Interestingly, the binding entropy (ΔS) related to the siRNA and DNA complexation with PEI25 kDa was practically the same, while the enthalpy (ΔH) of siRNA/PEI25 kDa binding (−6.6 kcal mol−1) was higher than that of DNA/PEI25 kDa (ΔH = −5.7 kcal mol−1). As a consequence, the normalized free energy of binding of PEI25 kDa with siRNA (ΔG = −5.5 kcal mol−1) was more favorable than that of DNA complexation (ΔG =−4.8 kcal mol−1), indicating that PEI25 kDa polymers are slightly more strongly attracted by siRNA than by DNA. This can be explained with a more consistent curvature and a higher local flexibility of siRNA with respect to DNA, which facilitates the uniform binding between the negative charges present on the nucleic acid with the positive ones of the polymer. The models in Figure 2 were designed and simulated to study the possible presence of differences in the binding of PEI25 kDa with DNA and siRNA. While Dicer substrate interfering RNA (DsiRNA) molecules are double strands of 25/27mer, the plasmid DNA used in the experiments presented in this work contains about 4400 base pairs. The DNA model used for simulations is just a portion of the complete plasmid, and the simulation is thus representative of the local interactions between the polymers and the DNA double strands. Under physiological conditions, plasmid DNA exists usually as an elongated helix as B-form, while RNA exists as a more compact and curved double helix, which is known as A-form.12 That makes RNA locally more flexible in the case of local roll and tilt deformations13 and more adaptable14 in the case of binding with a charged spherical polymer rather than DNA.9 For PEI25 kDa, not all of the charged surface groups are sterically available to bind a single strand of nucleic acid because a large part of the charged amines is back folded. During the binding between PEI25 kDa and DNA/siRNA, parts of the positive surface charges of the polymer establish strong electrostatic interactions with the nucleic acid. At the binding interface, positive and negative charges neutralize each other. But moving away from the binding site on the polymer surface, there are several other positively charged surface groups that do not participate actively in the binding with the nucleic acid (“primary complexes” in Figure 2). These free charges can potentially lead to interparticle electrostatic attractions with other siRNA/DNA molecules, giving rise to hierarchical aggregation phenomena. In fact, primary complexes can aggregate further and reorganize into larger polyplexes.15 Therefore, there is a balance that needs to be considered between the amount of charges and the ability to use these charges. In this framework, it is evident that the pure binding between the polymer and the nucleic acid that is depicted by MD simulation constitutes only the first, and most immediate, step in a complex hierarchical aggregation phenomenon that involves different scales and types of interactions, from strong electrostatic to weaker hydrophobic intermolecular forces. This hierarchy emerges when binding data from MD simulations are compared with the thermodynamic values calculated based on ITC measurements. The consequent molecular complexes can potentially undergo further structural reorganization and can interact with other polyplexes in solution. This causes slower complexation as compared to siRNA, where a consistent structural rearrangement is not expected due to the limited length of the nucleic acids. Moreover, DNA molecules need more polycations to achieve complete condensation and to form stable polyplexes. This hypothesis was challenged with the following ITC results.
Figure 2.
Molecular modeling and MD simulations. (A) Equilibrated configurations of the MD simulations of nucleic acids/PEI25 kDa polyplexes. (B) Simulated ΔG energies and the contributing potentials of the binding between branched PEI25 kDa and DNA or DsiRNA normalized to energy per charged surface amine expressed in kcal mol−1.
Isothermal Titration Calorimetry
The atomic binding results of local interactions from MD simulations are complemented by results from isothermal titration calorimetry experiments, which provide reliable thermodynamic interpretation16 of the aggregation of multiple polycation/nucleic acid nanocomplexes into higher-scale polyplexes. The ITC results are supported by the data from MD modeling and showed the same tendency of the binding behavior between polycations and different nucleic acids: the affinity between polycations and siRNA is higher than that between polycations and plasmid DNA, and the formation of hierarchical polycation/siRNA polyplexes is much easier and more stable than the complexation with plasmid DNA (Table 1). Even if the interaction between PEI25 kDa and DNA or siRNA is locally very similar, the flexible PEI25 kDa/DNA nanocomplexes can undergo structural rearrangement (folding), resulting in less uniform aggregation of multiple nanocomplexes into larger polyplexes (Figure 1B). Moreover, ITC indicates also that DNA molecules need a larger excess of polycations than siRNA to achieve complete condensation and to form stable polyplexes (Table 1). If on an atomic level the pure polymer–nucleic acid molecular recognition is controlled by electrostatic forces, on a higher-scale level, the interpolyplex interactions are also consistently characterized by hydrophobic forces, as is evidenced by data from ITC (Table 1). Hydrophobic aggregation is assumed to be typically an entropy-driven assembly phenomenon, accompanied by a lower favorable enthalpy (Table 1).17 This is particularly evident in the case of DNA. In fact, if PEI is modified with hydrophobic poly(caprolactone) segments, DNA/PEI25k-PCL1500-PEG2k nanocomplexes aggregate more strongly due to increased hydrophobicity18 and condense DNA more effectively. Concerning modified PEI25 kDa, only about five PEI25k-PCL1500-PEG2k molecules are required to condense one DNA molecule (N-value or site), whereas 11 molecules of unmodified PEI25 kDa are needed for the same effect.
TABLE 1.
Thermodynamic Parameters for the Specific Binding between Polycations and DNA or siRNAa
| N (sites) | K (M−1) | ΔH (cal/mol) | ΔS (cal/mol/deg) | ΔG (cal/mol) | |
|---|---|---|---|---|---|
| hyPEI25k/DNA | 1.59 ± 0.02 | 1.29 × 105 ± 1.37 × 104 | −2569 ± 40.96 | 14.8 | −6979.4 |
| hyPEI25k/siRNA | 2.26 ± 0.03 | 2.23 × 106 ± 9.28 × 105 | −2172 ± 48.09 | 21.8 | −8668.4 |
| hyPEI25k-PCL1500-PEG2k/DNA | 0.683 ± 0.01 | 2.58 × 105 ± 3.20 × 104 | −2795 ± 57.88 | 15.4 | −7384.2 |
| hyPEI25k-PCL1500-PEG2k/siRNA | 2.23 ± 0.05 | 2.80 × 105 ± 7.40 × 104 | −2063 ± 53.96 | 18.0 | −7427.0 |
All binding parameters are reliable experimental thermodynamic data calculated based on ITC. The larger dissociation constant K of siRNA/polycation complexation reflects that the affinity between polycations with siRNA is higher than that with pDNA. Concerning modified PEI, only about five PEI25k-PCL1500-PEG2k molecules were required to condense one pDNA molecule (N-value or site), whereas 11 PEI molecules were needed.
Fluorescence Quenching Assay
The dye quenching assay is another method to investigate the binding behavior of nucleic acids by polycations: the fluorescence of labeled siRNA molecules will be quenched by each other due to close spatial proximity in complexes where many siRNA molecules are compacted. Although we have used different polycations to condense siRNA, each curve has a minimum of fluorescence at N/P = 1 (N/P = nitrogen/phosphorus). After this minimum, the fluorescence increases again with increasing of N/P ratio (Figure 4A). Interestingly, an equilibrium in ITC is also reached at remarkably lower N/P ratios for siRNA than for DNA, highlighting the noteworthiness of this low N/P ratio. The endothermic peaks of siRNA binding isotherm curves close to N/P = 1 (Figure 3), together with the dye quenching assay (Figure 4A), reveal a special condensation phenomenon of siRNA: siRNA molecules “escape” from saturated “primary multimolecular nanocomplexes” at N/P = 1 and reorganize into more stable nanocomplexes with a lower energy level (N/P = 2) (Figure 4B). This trend was already observed with siRNA19 and oligonucleotides.20 Moreover, the particle size distribution (polydispersity index, PDI) measurements indicate that siRNA can be condensed into more ordered and uniform polyplexes with the lowest PDI at N/P = 2 (Figure S2). Additionally, heparin assays confirmed that siRNA polyplexes at N/P = 2 are particularly stable against competing polyanions (Figure S1). Therefore, we assumed that lower N/P ratios (N/P= 2 in the case of PEI) are especially effective for siRNA delivery.
Figure 4.
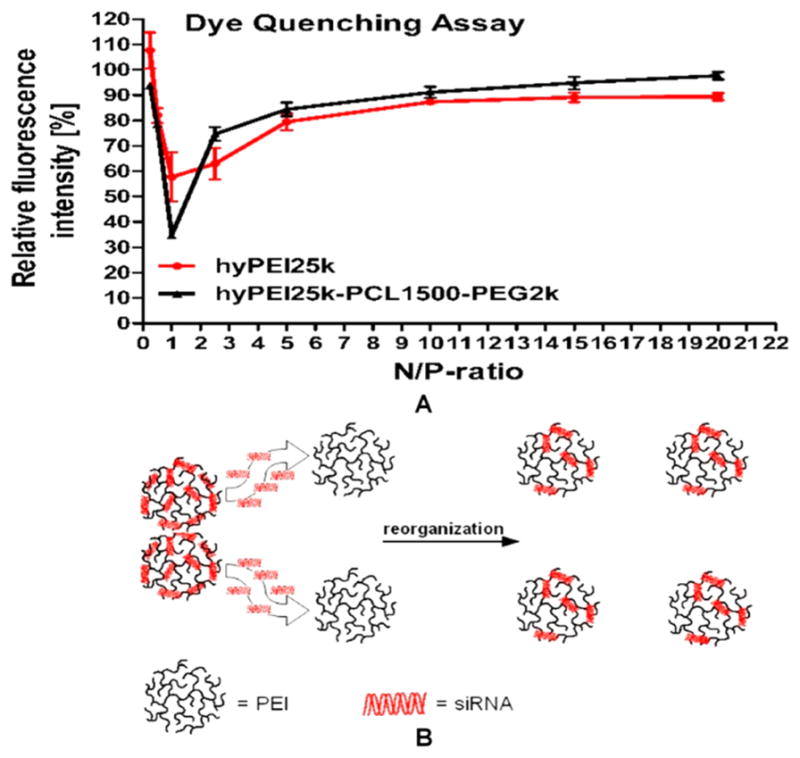
Dye quenching assay. (A) The fluorescence of Tye563-labeled siRNA molecules is quenched by each other in a “multimolecular complex” due to close spatial proximity. Each curve had a minimum of fluorescence at N/P = 1, after which the fluorescence increased again due to a decreased number of siRNA molecules per polyplex, resulting in less proximity of the labeled siRNA and thus in lower quenching. This special phenomenon of short nucleic acids condensation can be understood as a reorganization of the polyplexes. (B) Molecular reorganization of the siRNA within the polycations at lower N/P ratios.
Figure 3.

Thermodynamic interpretation was provided during isothermal titration calorimetry. Standard binding isotherm curve of siRNA and pDNA with polycations. The siRNA reorganization from the saturated complex into aggregates is an endothermic process, reflected in an endothermic peak at N/P = 1 in the ITC measurements.
In Vitro Uptake and Gene Knockdown Effect
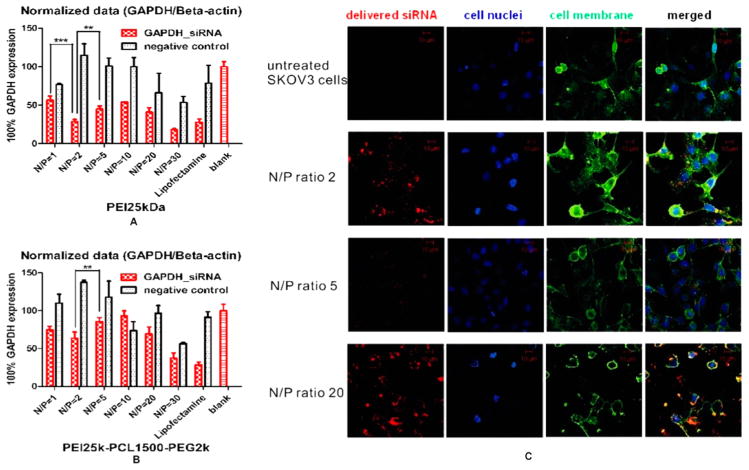
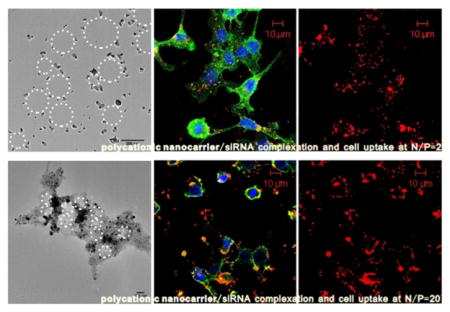
Interestingly, with the following study of the relationship between nucleic acids/polycations aggregation mechanism and in vitro siRNA delivery efficiency, which is performed by RT-PCR (Figure 5A, B) and confocal laser scanning microscopy (Figure 5C), we found that not only PEI25 kDa but also the PCL-PEG-modified copolymer hyPEI25k-PCL1500-PEG2k showed the best intracellular delivery and knockdown effect with siRNA at N/P = 2, although higher N/P ratios were believed to be necessary until now by most of the researchers in the area of polycationic nanocarrier-based siRNA delivery.21–24 The belief of increased transfection efficiency with increased N/P ratio can be explained by this exact behavior for the delivery of pDNA, which is true up to a point, where toxicity of free polymer decreases the transfection efficacy of pDNA (Figure S3). In the case of siRNA delivery with PEI25 kDa, by increasing the N/P ratio, the hGAPDH gene expression decreased from N/P 1 (53.26% knockdown) to N/P 2 (72.29% knockdown) and increased again with the increasing of N/P ratios. The knockdown effect in the graph is better at N/P = 30 than at N/P = 2, but the negative control at N/P = 30 is also very low, which indicates that at higher N/P ratio the knockdown effect is caused by not only gene silencing but also the cytotoxicity of the polycations. The confocal laser scanning microscopy (CLSM) micrographs reflected the same tendency: although the siRNA could be delivered effectively into the cytosol at N/P = 20, a disturbed cell morphology with partially dilapidated cellular membranes was observed, which indicated a high cytotoxicity of these polycationic delivery agents at high N/P ratios. On the other hand, the siRNA delivery efficiency at N/P = 2 was as good as at N/P = 20, but with a vital cell morphology (Figure 5C), as a result of more uniform and stable complex formation and lower cytotoxicity.
Figure 5. In vitro.
cell uptake and knockdown at different N/P ratios. (A) Knockdown effect of siRNA/PEI25 kDa polyplexes using RT-PCR. (B) Knockdown effect of siRNA/PEI25k-PCL1500-PEG2k polyplexes using RT-PCR: polycations showed the best knockdown effect with siRNA at N/P = 2. In the case of PEI25 kDa, by increasing the N/P ratio, the GAPDH gene expression decreased from N/P = 1 (53.26% knockdown) to N/P = 2 (72.29% knockdown) and increased again. The knockdown effect at N/P = 20 and 30 seems better than at N/P = 2, but the lower negative control bar indicated toxicity at higher N/P ratio. (C) Confocal laser scanning microscopy (CLSM) showed the cell uptake at different N/P ratios: the uptake efficiency at both N/P = 2 and N/P = 20 was good, but N/P = 20 was too toxic, causing a less vital cell morphology (siRNA was labeled with AF647 dyes; nuclei were stained with DAPI and cell membranes were labeled with FITC-wheat germ agglutinin).
CONCLUSION
In our research we investigated the different complexation and aggregation mechanism between polycationic nanocarriers and DNA or siRNA on the atomic and molecular scale. The novel synergic use of MD simulations, ITC, and dye quenching assay provided an exceptionally clear depiction of the different hierarchical aspects that control the formation of polyplexes. It is well accepted that the positively charged surface of poly(ethyleneimine) nanocomplexes induces not only increased cellular uptake through charge-mediated interactions25 (Figure 5C) but also disadvantageous higher cytotoxicity (especially true for high N/P ratios). While researchers seek to balance toxicity and transfection efficiency, our investigation highlights the need to address the actual assembly of polyelectrolyte complexes and to optimize the formulation of siRNA complexes from a thermodynamic point of view. Our study based on poly(ethyleneimine) as a model polycationic nanocarrier directs attention to lower N/P ratios, which emerge as an unnoticed “blind spot” in polycationic siRNA delivery. All our results emphasized one point: lower N/P ratios are especially effective for polycationic nanocarrier-based siRNA delivery. This could have broad implications for the delivery of siRNA, as less toxic and yet efficient delivery systems have been the bottleneck for the translation of this promising approach into the clinical arena. We recommend to the scientific community working in the area of polycationic siRNA delivery to study the actual assembly of self-assembled nanocarriers and thus to consider low N/P ratios, which could be particularly important for siRNA delivery but have been disregarded in previous studies.
MATERIALS AND METHODS
Materials
Hyperbranched polyethylenimine (hy-PEI) 25 kDa was obtained from BASF. Poly(ethylene glycol) monomethyl ether (mPEG) (5 kDa) and ε-caprolactone were purchased from Fluka (Taufkirchen, Germany). Beetle luciferin, heparin sodium salt, and all other chemicals were obtained from Sigma–Aldrich (Steinheim, Germany). Luciferase-encoding plasmid (pCMV-Luc) (lot no. PF461-090623) was amplified by The Plasmid Factory (Bielefeld, Germany). Negative control sequence, hGAPDH-DsiRNA, and TYE546-DsiRNA were obtained from Integrated DNA Technologies (IDT, Leuven, Belgium).
Molecular Modeling and MD Simulations
The binding of nucleic acid and PEI25 kDa was modeled according to a reported validated strategy.8,26 The MD simulations were conducted according to previous studies.8,26–28 Briefly, each of the molecular dynamics runs was carried out using the sander and pmemd.cuda modules within the AMBER 11 suite of programs.
Isothermal Titration Calorimetry
ITC was carried out with an iTC200 micro titration calorimeter (Microcal, Inc., Northampton, MA, USA) according to our earlier report.29 Bidistilled water was degassed at 293 K under vacuum for 10 min and equilibrated to room temperature before use. Measurements were performed at 298 K. The baseline (dilution energy) was recorded by titrating redundant amounts of polymer into water. Afterward, to modify the baseline, a redundant of 3 μL of polymer, 3.0 mM nitrogen of PEI was added and titrated into the stirred cell containing plasmid DNA (0.1 mM base pairs) or siRNA (0.05 mM base pairs). To prevent a systematic error from syringe filling, an initial injection of 1.5 μL was necessary and the following injections were constantly maintained at 3 μL at intervals of 180 s until the plasmid DNA or siRNA was saturated with polymer. Energy differences caused by the condensation of polymer and nucleic acids during each polymer injection were detected from the integral of the calorimetric signal, after subtraction of the baseline. ITC data were analyzed with ORIGIN software (Microcal, Inc.). After integration and correction of peaks with a single-site-binding assumption, the thermodynamic parameters enthalpy (ΔH), entropy (ΔS), and the dissociation constant K of binding were calculated.
Dye Quenching Assay
Dye quenching assays were conducted according to a previous study by Merkel et al.19 Briefly, 60 pmol of Tye563-labeled DsiRNA (IDT, Leuven, Belgium) was complexed with hyPEI25k or hyPEI25k-PCL1500-PEG2k at different N/P ratios. Remaining fluorescence of the polyplex solutions (200 μL) was determined in opaque FluoroNunc 96 well plates (Nunc, Thermo Fisher Scientific, Langenselbold, Germany) using a fluorescence plate reader (SAFIRE II, Tecan Group Ltd., Männedorf, Switzerland) at 549 nm excitation and 563 nm emission wavelengths. Experiments were performed in replicates of three, and the results are given as mean relative fluorescence intensity values ± SD. For normalization purposes, free polymer in buffer represents 0% fluorescence, while free siRNA in buffer represents 100% fluorescence.
In Vitro Cell Uptake and Knockdown Experiments
SKOV3 cells were seeded with 106 cells per well in six wells 24 h prior to transfection and transfected with 50 pmol of siRNA. The mRNA was isolated 24 h after transfection (PureLink RNA Mini Kit, Invitrogen GmbH, Germany) and reverse transcribed to cDNA (First Strand cDNA Synthesis kit, Fermentas, Germany). RT-PCR was performed using QuantiFast SYBR Green PCR kits (Qiagen, Germany) and the Rotor-Gene 3000 RT-PCR thermal cycler (Corbett Research, Sydney, Australia). For confocal laser scanning microscopy, cells were incubated with nanocomplexes containing AF647-labeled siRNA for 4 h and then fixed. Nuclei were stained with DAPI, and cell membranes were labeled with FITC-wheat germ agglutinin (Invitrogen, Karlsruhe, Germany).
Transmission Electron Microscopy
To determine the different location of plasmid-DNA and siRNA within complexes of the polycations, first, polyplexes of pDNA (0.1 mg/mL, 300 μL in 5% glucose) or siRNA (6.1 μM, 300 μL in 5% glucose) were prepared at N/P = 2 and N/P = 20 as described above. After complexation of the nucleic acids with polymer (15 min of incubation at 25 °C), 600 μL of polyplexes was metalized during incubation with 30 μL of AgNO3 (0.1 M) for 2 h at 25 °C. The concentration of plasmid DNA and siRNA was calculated and selected to obtain the same concentration of base pairs, respectively. Polyplexes for TEM analysis were prepared by drop-coating the solutions on carbon-coated Cu meshes (S160-3, Plano, Germany). TEM measurements were performed using a JEM-3010 microscope (Jeol Ltd., Tokyo, Japan), operated at 300 kV, equipped with a high-resolution CCD camera for image recording.
Statistics
All analytical assays were conducted in replicates of three or four. Results are given as mean values ± standard deviation. Two-way ANOVA and statistical evaluations were performed using Graph Pad Prism 4.03 (Graph Pad Software, La Jolla, CA, USA).
Supplementary Material
Acknowledgments
The authors wish to acknowledge Dr. Ayse Kilic and Dr. Holger Garn (Institute of Laboratory Medicine and Pathobiochemistry, Philipps Universität Marburg) for use of the Rotor-Gene real time cycler, Eva Mohr (IPTB) for expert technical support in the cell culture, Michael Hellwig (Center of Material Science, Philipps Universität Marburg) for TEM imaging, Prof. Dr. Wolfgang Parak and Yu Xiang (Department of Physics, Philipps-Universität Marburg) for CLSM imaging, and Dr. Dafeng Chu (Department of Pharmaceutics and Biopharmacy, Philipps Universität Marburg) for excellent discussions.
Footnotes
Conflict of Interest: The authors declare no competing financial interest.
Supporting Information Available: Results of binding and siRNA protection assays, dynamic light scattering and zeta potential analysis, in vitro transfection experiments with plasmid DNA, and in vitro cytotoxicity can be downloaded online. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES AND NOTES
- 1.Riehemann K, Schneider SW, Luger TA, Godin B, Ferrari M, Fuchs H. Nanomedicine--Challenge and Perspectives. Angew Chem, Int Ed. 2009;48:872–897. doi: 10.1002/anie.200802585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petros RA, DeSimone JM. Strategies in the Design of Nanoparticles for Therapeutic Applications. Nat Rev Drug Discovery. 2010;9:615–627. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- 3.Lollo CP, Banaszczyk MG, Chiou HC. Obstacles and Advances in Non-Viral Gene Delivery. Curr Opin Mol Ther. 2000;2:136–142. [PubMed] [Google Scholar]
- 4.Park TG, Jeong JH, Kim SW. Current Status of Polymeric Gene Delivery Systems. Adv Drug Delivery Rev. 2006;58:467–486. doi: 10.1016/j.addr.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and Specific Genetic Interference by Double-Stranded RNA in Caenorhabditis Elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 6.Ferrari M. Cancer Nanotechnology: Opportunities and Challenges. Nat Rev Cancer. 2005;5:161–171. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L, Gu FX, Chan JM, Wang AZ, Langer RS, Farokhzad OC. Nanoparticles in Medicine: Therapeutic Applications and Developments. Clin Pharmacol Ther. 2008;83:761–769. doi: 10.1038/sj.clpt.6100400. [DOI] [PubMed] [Google Scholar]
- 8.Pavan GM, Danani A, Pricl S, Smith DK. Modeling the Multivalent Recognition between Dendritic Molecules and DNA: Understanding How Ligand “Sacrifice” and Screening Can Enhance Binding. J Am Chem Soc. 2009;131:9686–9694. doi: 10.1021/ja901174k. [DOI] [PubMed] [Google Scholar]
- 9.Pavan GM, Kostiainen MA, Danani A. Computational Approach for Understanding the Interactions of UV-Degradable Dendrons with DNA and siRNA. J Phys Chem B. 2010;114:5686–5693. doi: 10.1021/jp911439q. [DOI] [PubMed] [Google Scholar]
- 10.Pavan GM, Danani A. Dendrimers and Dendrons for siRNA Binding: Computational Insights. J Drug Delivery Sci Technol. 2012;22:83–89. [Google Scholar]
- 11.Gary DJ, Puri N, Won YY. Polymer-Based siRNA Delivery: Perspectives on the Fundamental and Phenomenological Distinctions from Polymer-Based DNA Delivery. J Controlled Release. 2007;121:64–73. doi: 10.1016/j.jconrel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 12.Arnott S. Principles of Nucleic-Acid Structure - Saenger, W. Nature. 1984;312:174–174. [Google Scholar]
- 13.Noy A, Perez A, Lankas F, Javier Luque F, Orozco M. Relative Flexibility of DNA and RNA: A Molecular Dynamics Study. J Mol Biol. 2004;343:627–638. doi: 10.1016/j.jmb.2004.07.048. [DOI] [PubMed] [Google Scholar]
- 14.Pavan GM, Albertazzi L, Danani A. Ability to Adapt: Different Generations of PAMAM Dendrimers Show Different Behaviors in Binding siRNA. J Phys Chem B. 2010;114:2667–2675. doi: 10.1021/jp100271w. [DOI] [PubMed] [Google Scholar]
- 15.Merkel OM, Mintzer MA, Librizzi D, Samsonova O, Dicke T, Sproat B, Garn H, Barth PJ, Simanek EE, Kissel T. Triazine Dendrimers as Nonviral Vectors for in Vitro and in Vivo RNAi: the Effects of Peripheral Groups and Core Structure on Biological Activity. Mol Pharm. 2010;7:969–983. doi: 10.1021/mp100101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koch C, Heine A, Klebe G. Tracing the Detail: How Mutations Affect Binding Modes and Thermodynamic Signatures of Closely Related Aldose Reductase Inhibitors. J Mol Biol. 2011;406:700–712. doi: 10.1016/j.jmb.2010.11.058. [DOI] [PubMed] [Google Scholar]
- 17.Doni G, Kostiainen MA, Danani A, Pavan GM. Generation-Dependent Molecular Recognition Controls Self-Assembly in Supramolecular Dendron-Virus Complexes. Nano Lett. 2011;11:723–728. doi: 10.1021/nl103857e. [DOI] [PubMed] [Google Scholar]
- 18.Zheng M, Liu Y, Samsonova O, Endres T, Merkel O, Kissel T. Amphiphilic and Biodegradable hy-PEI-g-PCL-b-PEG Copolymers Efficiently Mediate Transgene Expression Depending on Their Graft Density. Int J Pharm. 2011;427:80–87. doi: 10.1016/j.ijpharm.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 19.Merkel OM, Beyerle A, Librizzi D, Pfestroff A, Behr TM, Sproat B, Barth PJ, Kissel T. Nonviral siRNA Delivery to the Lung: Investigation of PEG-PEI Polyplexes and Their in Vivo Performance. Mol Pharm. 2009;6:1246–1260. doi: 10.1021/mp900107v. [DOI] [PubMed] [Google Scholar]
- 20.Van Rompaey E, Engelborghs Y, Sanders N, De Smedt SC, Demeester J. Interactions between Oligonucleotides and Cationic Polymers Investigated by Fluorescence Correlation Spectroscopy. Pharm Res. 2001;18:928–936. doi: 10.1023/a:1010975908915. [DOI] [PubMed] [Google Scholar]
- 21.Park JW, Bae KH, Kim C, Park TG. Clustered Magnetite Nanocrystals Cross-Linked with PEI for Efficient siRNA Delivery. Biomacromolecules. 2011;12:457–465. doi: 10.1021/bm101244j. [DOI] [PubMed] [Google Scholar]
- 22.Wu Y, Wang WW, Chen YT, Huang KH, Shuai XT, Chen QK, Li XX, Lian GD. The Investigation of Polymer-siRNA Nanoparticle for Gene Therapy of Gastric Cancerin Vitro. Int J Nanomed. 2010;5:129–136. doi: 10.2147/ijn.s8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dimitrova M, Affolter C, Meyer F, Nguyen I, Richard DG, Schuster C, Bartenschlager R, Voegel JC, Ogier J, Baumert TF. Sustained Delivery of siRNAs Targeting Viral Infection by Cell-Degradable Multilayered Polyelectrolyte Films. Proc Natl Acad Sci U S A. 2008;105:16320–16325. doi: 10.1073/pnas.0800156105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Urban-Klein B, Werth S, Abuharbeid S, Czubayko F, Aigner A. RNAi-Mediated Gene-Targeting through Systemic Application of Polyethylenimine (PEI)-Complexed siRNA in Vivo. Gene Ther. 2005;12:461–466. doi: 10.1038/sj.gt.3302425. [DOI] [PubMed] [Google Scholar]
- 25.Godbey WT, Wu KK, Mikos AG. Size Matters: Molecular Weight Affects the Efficiency of Poly(ethylenimine) as a Gene Delivery Vehicle. J Biomed Mater Res. 1999;45:268–275. doi: 10.1002/(sici)1097-4636(19990605)45:3<268::aid-jbm15>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 26.Merkel OM, Zheng M, Mintzer MA, Pavan GM, Librizzi D, Maly M, Hoffken H, Danani A, Simanek EE, Kissel T. Molecular Modeling and in Vivo Imaging Can Identify Successful Flexible Triazine Dendrimer-Based siRNA Delivery Systems. J Controlled Release. 2011;153:23–33. doi: 10.1016/j.jconrel.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pavan GM, Mintzer MA, Simanek EE, Merkel OM, Kissel T, Danani A. Computational Insights into the Interactions between DNA and siRNA with “Rigid” and “Flexible” Triazine Dendrimers. Biomacromolecules. 2010;11:721–730. doi: 10.1021/bm901298t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen LB, Mortensen K, Pavan GM, Kasimova MR, Jensen DK, Gadzhyeva V, Nielsen HM, Foged C. Molecular Characterization of the Interaction between siRNA and PAMAM G7 Dendrimers by SAXS, ITC, and Molecular Dynamics Simulations. Biomacromolecules. 2010;11:3571–3477. doi: 10.1021/bm101033g. [DOI] [PubMed] [Google Scholar]
- 29.Steuber H, Czodrowski P, Sotriffer CA, Klebe G. Tracing Changes in Protonation: A Prerequisite to Factorize Thermodynamic Data of Inhibitor Binding to Aldose Reductase. J Mol Biol. 2007;373:1305–1320. doi: 10.1016/j.jmb.2007.08.063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.