Abstract
Daphnia pulex, the crustacean with the first sequenced genome, is an important organism that has been widely used in ecological and toxicological research. MicroRNAs (miRNAs) are 21–25 nucleotide small non-coding RNAs that are involved in a myriad of physiological processes. In this research, we predicted 75 D. pulex miRNAs by sequence homology and secondary structure identification from the full genome sequence. Fourteen predicted miRNAs were selected for quantitative real time polymerase chain reaction (RT-PCR) validation. Out of these, eight (mir-8, mir-9, mir-12, mir-92, mir-100, mir-133, mir-153 and mir-283) were successfully amplified and validated. Next, expression levels were quantified at three different life stages (days 4, 8 and 12 of age) using U6 spliceosomal RNA as a reference gene. The expression of mir-8, mir-9, mir-12, mir-92 and mir-100 significantly differed across time suggesting these microRNAs might play a critical role during D. pulex development. This is the first study to identify and validate miRNAs in D. pulex, which is an important first step in further studies that evaluate their roles in development and response to environmental and ecological stimuli.
Introduction
Daphnia pulex is an ecologically important organism in freshwater ecosystems that has been studied for decades [1] and is the first crustacean for which a full genome sequence is available. D. pulex has a short life cycle and the phenotypic plasticity displayed by these organisms makes it an ideal model species for ecological, toxicological, and evolutionary research [2]. D. pulex can reproduce by either clonal (parthenogenetic) or sexual reproduction and often undergoes cyclical parthenogenesis. Usually, a healthy population will be genetically identical females that produce diploid parthenogenetic eggs. Haploid sexual eggs can be produced when females encounter certain environmental cues such as starvation or highly crowed conditions [3], [4]. Diploid juvenile D.pulex usually take 6–10 days and go through 5 or 6 juvenile instars to reach sexual maturity [4]. After reaching maturity, the growth rate of D. pulex decreases dramatically as energy is focused mostly on breeding rather than somatic development [4], [5]. Interestingly, D. pulex populations that are genetically identical can still show plasticity in certain phenotypes, e.g. predator induced helmet and “neckteeth” formation [6]. Thus clonal lines with phenotypically differential individuals enable us to examine whether there is an epigenetic influence on phenotype [7]. All these unique biological attributes, paired with the newly sequenced D. pulex genome, provide an unparalleled opportunity to study the epigenetic signatures in D. pulex and to examine how these signatures are changed and/or inherited across environmental conditions.
Micro-RNAs (miRNAs) are short, non-coding endogenous RNAs that are approximately 20–25 nucleotides in length. Recent research has revealed that miRNAs are involved in a variety of aspects of animal development including muscle development, aging and body size regulation [8]–[10]. During miRNA biogenesis, miRNA genes need to form a hairpin loop of ∼70 nucleotide in length [11]. The conservation of miRNA sequences across taxa, together with the secondary hairpin structure provide an opportunity to predict conserved miRNAs in species for which miRNAs have not been previously described. However, in silico prediction does not necessarily mean those predicted miRNAs exist functionally in an organism. Therefore, in vivo validation of predicted miRNAs is needed. Several approaches are available for this validation including real-time polymerase chain reaction (RT-PCR) and cloning and deep sequencing.
According to miRBase (Release 19), one of the largest miRNA databases [12], no D. pulex miRNAs have been experimentally verified. The objectives of this research were to 1) predict conserved D. pulex miRNAs by sequence homology and hairpin structure identification; 2) further validate predicted miRNAs by end point PCR and RT-PCR; 3) select stable reference genes for D. pulex miRNA expression; and 4) examine the expression level of validated miRNAs during D. pulex development. This research provides a foundation for future research on the role of D. pulex miRNAs during development.
Materials and Methods
In silico prediction of D. pulex miRNAs
Candidate miRNA loci were identified by conducting a nucleotide BLAST (BLAST-2.2.25, e-value 0.1)[13] using all animal mature miRNA sequences available (miRBase Release 19 http://www.mirbase.org/) against the D. pulex genome (DOE Joint Genome Institute http://www.jgi.doe.gov/). Only miRNA loci with at least 18 nucleotides with no more than 2 mismatches were considered for further analyses. In addition, if several miRNAs from the same miRNA family (e.g. has-mir-9 and mmu-mir-9 were considered from mir-9 family) matched the same D. pulex locus, the miRNA sequence with the highest identity was chosen.
Pre-miRNA hairpin structure identification
During animal miRNA biogenesis, miRNA genes are initially transcribed into long primary miRNAs (pri-miRNAs) and then processed into ∼70 nucleotide precursor miRNAs (pre-miRNAs) that later form hairpin structures [11]. Identification of these pre-miRNA hairpin structures plays a critical role in miRNA computational prediction. We used a custom Perl script (File S1) to obtain the 200 nucleotide flanking sequence which center surrounded the candidate D. pulex miRNA loci (identified from BLAST above) as potential pre-miRNAs. These potential pre-miRNAs were then analyzed by Mfold [14] using default settings. For each potential pre-miRNA, Mfold outputs several structures with different minimum folding energy (MFE). We inspected structures that had the lowest MFE and only those that fulfilled the following criteria were considered to be authentic hairpin structures: 1) pre-miRNAs could form an appropriate hairpin structure with the potential mature miRNA located on one of the hairpin arms; 2) less than six mismatches were observed between the potential mature miRNA and its opposite strand; and 3) no breaks occurred between the mature miRNA and its opposite strand.
In vivo validation of D. pulex miRNAs
Animal culture and sample preparation
D. pulex were obtained from Dr. John Colbourne's lab at Indiana University (Bloomington, IN, USA). Organisms were cultured in hard water (NaHCO3 0.192 g/L, CaSO4·2H2O 0.120 g/L, MgSO4 0.120 g/L, KCl 0.008 g/L) and maintained in an environmental chamber at 25°C on a 16/8 light/dark cycle. Water was changed twice a week and D. pulex were fed YCT (yeast, cereal leaf, trout chow) mix (Aquatic Research Organisms Inc., Hampton, NH, USA) after every water change. D. pulex were collected from a single brood at 4, 8 and 12 days of age. At each time point, 3 samples of 10 individuals each were collected and flash frozen in liquid nitrogen. miRNAs (and small RNAs) were extracted using PureLink miRNA isolation kit (Invitrogen, Carlsbad, CA, USA). Total miRNA was quantified using a Qubit Fluorometer (Invitrogen, Carlsbad, CA, USA) and miRNA extracts were stored at −80°C until PCR analysis.
End-point PCR and RT-PCR validation
A subset of predicted miRNAs (n = 14) were randomly chosen for validation. Specific primers (Table S2) were designed following the method described by Chen et al. (2005) [15]. The protocol for miRNA validation was adapted from Varkonyi-Gasic et al. (2007) [16]. Briefly, miRNAs were first reverse-transcribed using 2 pmol miRNA specific stem-loop primers, a mix of 0.5 mM dNTP, and 1 µg of RNA template heated at 65°C for 5 min. A 1X first-strand buffer containing 5 mM DTT, 2 units of RNase OUT and 2.5 units of SuperScript III were then added and the mix incubated on ice for another 2 min. The final mix was incubated at 16°C for 30 min followed by 60 cycles at 30°C for 30 s, 42°C for 30 s and 50°C for 1 s and then incubated at 85°C for 5 min to inactivate the reverse transcriptase. cDNA was quantitated using a Qubit Fluorometer and stored at −80°C. The subsequent end-point PCRs were performed by mixing 0.5 mM dNTP, 0.2 µM forward primer, 0.2 µM universal reverse primer, 1 unit of Advantage 2 Polymerase mix (Clontech, Mountain View, CA, USA), 1 µl cDNA and nuclease-free water into a 20 µl volume reaction. PCR conditions were 94°C for 2 min, followed by 30–40 cycles at 94°C for 15 s and 60°C for 1 min. PCR products were visualized by electrophoresis on a 4% agarose gel.
RT-PCR was conducted on a StepOnePlus real-time PCR system (Applied Biosystems, Inc. Foster City, CA, USA). Each reaction contained 2 µM SYBR Green I master mix, 1 µM forward primer, 1 µM reverse primer and 2 µl RT product. Cycling parameters were 95°C for 5 min, followed by 35–45 cycles at 95°C for 5 s, 60°C for 10 s and 72°C for 1 s. A melting curve analysis was performed to check that no primer-dimers were present.
Expression of D.pulex miRNAs at different life stages
To better understand the role of miRNAs in D. pulex development, we further tested the expression of a validated set of miRNAs at different ages. Several widely used small RNA reference genes in other model species were selected as candidate reference genes. A nucleotide BLAST (BLAST-2.2.25, e-value 0.1) search was conducted using candidate reference gene sequences against the D. pulex genome. The BLAST hits on D. pulex genome were used for primer design using Primer 3 (v.0.4.0) [17]. RT-PCR was performed using miRNA samples at different life stages following the method already described. The stability of candidate reference genes was evaluated using geNorm [18] and NormFinder [19]. GeNorm is a widely used algorithm for selection of the most stable reference genes. M values calculated by geNorm represent the stability of each gene with smallest values indicative of highest expression stability. NormFinder is another acknowledged algorithm used for determining the stability of reference genes with smaller values representing higher expression stability. Fold-change differences during development were calculated based on expression during the first time point (day 4) using the ΔΔCt method[20] and normalized to the expression of U6. One-way ANOVA tests were performed to test for differential expression over time using SAS (SAS Statistical Institute, Cary, NC). For genes where expression was significantly different over time, pair wise differences between time points was assessed with Tukey's HSD tests. Type I error was set at alpha = 0.05.
Results and Discussion
In silico prediction of D. pulex miRNAs
From miRBase 16.0, 16,564 mature animal miRNA sequences were obtained to query against the D. pulex genome. Following our criteria described above, 1,171 candidate miRNA loci were identified in the D. pulex genome. After the secondary structure analysis by mFold, a total of 75 D. pulex miRNAs were predicted (Table S1). Two clustered groups of miRNAs, mir-12/mir-283 and mir-100/mir-125, were identified.
Choosing a stable D. pulex reference gene
Several widely used miRNA reference genes (U6 Spliceosomal RNA, RNU1A, RNU5A, SNORD25, and SCARNA17) were selected as candidate reference genes in our study. Out of these, only RNU1A and U6 had BLAST hits to the D. pulex genome. After several attempts to amplify RNU1A and U6 using sequence specific primers, only the U6 gene could be successfully amplified. This is not surprising since U6 is one of the most highly conserved spliceosomal RNAs [21]. Next, geNorm and NormFinder were used to test the expression stability of U6 during D. pulex development. U6 gene had the smallest M value (0.09) and the smallest stability value (0.15) (Table 1) further supporting its use as a stable reference gene for miRNA studies in D. pulex (genes with M values≤1.5 are considered stably expressed).
Table 1. M value and stability value of candidate reference genes calculated by geNorm and NormFinder.
| miRNA | Stability value | M value |
| mir8 | 0.40 | 0.36 |
| mir9 | 0.20 | 0.17 |
| mir12 | 0.41 | 0.12 |
| mir92 | 0.24 | 0.14 |
| mir100 | 0.52 | 0.13 |
| mir153 | 0.35 | 0.18 |
| mir283 | 0.30 | 0.38 |
| U6 | 0.15 | 0.09 |
Genes with M value≤1.5 are considered stably expressed genes. Stability values represent the combination of intra- and intergroup expression variation.
In vivo validation of D. pulex miRNAs
Out of the 14 miRNAs selected for validation, 8 (mir-8, mir-9, mir-12, mir-92, mir-100, mir-133, mir-153 and mir-283) were successfully amplified. This suggests that our computational prediction method is an efficient way to discover conserved miRNAs in D. pulex. Since only one pair of primers was tested for each miRNA, additional primers might need to be designed for further validation of the remaining 6 miRNAs (mir-1, mir-10, mir-34, mir-96, mir-124 and mir-137). Because mir-133 expression could only be detected at day 12, it was excluded from the expression stability test and expression changes test.
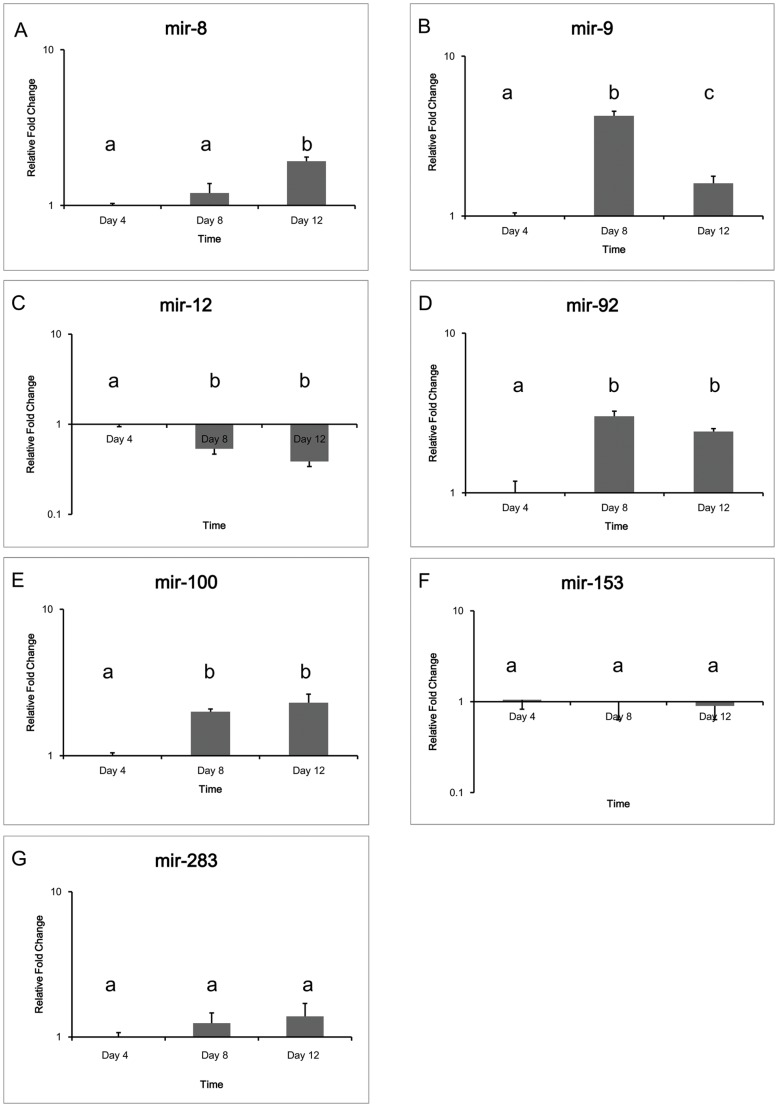
With exception of only two miRNAs (mir-153 and mir-283) all miRNAs changed in expression during the three time points monitored, with only one miRNA (mir-12) decreasing in expression at days 8 and 12 (Figure 1). The significant up-regulation of mir-8 at day 12 and of mir-9 at day 8 coincides with dramatic morphological changes including molting, somatic growth, brood chamber development and egg development in D. pulex [4], [5]. mir-8 is an important insulin signaling regulator that controls body size in Drosophila by suppressing its target gene (u-shaped, ush) [10], [22] (see Table S3). mir-9 is known to control the timing of neurogenesis [23], [24]. Thus, we propose that mir-8 and mir-9 might also play important roles in D. pulex somatic growth and neurogenesis, but further studies are needed to corroborate these findings. mir-12 is known to regulates the MCT1 and MCM6 genes in Wolbachia-infected mosquito cell line[25]. mir-92 is a novel marker for acute leukemia known to increase the proliferation of myeloid cells [26], [27]. mir-100 acts as a tumor suppressor in acute myeloid leukemia by regulating cell differentiation and survival [28], [29]. The roles of mir-92 and mir-100 in D. pulex development are unknown at this time. Interestingly, mir-12 and mir-283 are located within a 1 kb region and transcribe the same pri-miRNA, but had opposite expression patterns. This uncoordinated expression profile has also been identified in Drosophila [30] and provides evidence of post-translational regulation in these clustered miRNAs.
Figure 1. miRNA (mir-8 (A), mir-9(B), mir-12(C), mir-92(D), mir-100(E), mir-153(F), mir283(G)) expression changes during D. pulex development. Bars plotted represent means and standard errors.
Letters indicate Tukey's groupings (P<0.05) of gene expression level at three different life stages.
Conclusions
In this research, we predicted 75 conserved D. pulex miRNAs and successfully validated 8 miRNAs by RT-PCR. Using U6 as reference gene, we tested the expression of these miRNAs during different D. pulex life stages (days 4, 8, and 12). Significant changes in the expression of mir-8, mir-9, mir-12, mir-92 and mir-100 were observed, suggesting they play an important role during Daphnia development. As a next step, a specific designed D. pulex miRNA target prediction program will be developed to better understand the roles that miRNAs play in D. pulex development. This study is the first to report expression of miRNAs on D. pulex and will facilitate future epigenetic research on this species and daphnids in general.
Acknowledgments
The authors acknowledge Dr. John Colbourne's lab at Indiana University for providing D. pulex and Ashley Chin-Baarstad and Dr. Matthew Hale for their valuable comments.
Supporting Information
Predicted D. pulex miRNAs, genomic coordinates, and mature miRNA sequences.
(DOCX)
miRNAs and primer sequences.
(DOCX)
Biological functions associated with the differentially expressed miRNAs during D. pulex development.
(DOCX)
PERL code for pre-miRNA sequence analyze.
(TXT)
Funding Statement
This research is supported by Department of Forestry and Natural Resources, Purdue University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Korovchinsky N (1997) On the history of studies on cladoceran taxonomy and morphology, with emphasis on early work and causes of insufficient knowledge of the diversity of the group. Hydrobiologia 360: 1–11 10.1023/A:1003156802800 [DOI] [Google Scholar]
- 2. Shaw JR, Pfrender ME, Eads BD, Klaper R, Callaghan A, et al. (2008) Daphnia as an emerging model for toxicological genomics. Adv Exp Biol 02: 165–219 10.1016/S1872-2423(08)00005-7 [DOI] [Google Scholar]
- 3. Kleiven OT, Larsson P, Hobæk A, Hobaek A (1992) Sexual reproduction in Daphniamagna requires three stimuli. Oikos 65: 197–206. [Google Scholar]
- 4.Ebert D (2005) Ecology, epidemiology, and evolution of parasitism in Daphnia. National Library of Medicine (US), National Center for Biotechnology Information,Bethesda,MD.
- 5. Green J (1955) Growth, size and reproduction in Daphnia (Crustacea: cladocera). Proc Zool Soc Lon 126: 173–204. [Google Scholar]
- 6. Imai M, Naraki Y, Tochinai S, Miura T (2009) Elaborate regulations of the predator-induced polyphenism in the water flea Daphnia pulex: kairomone-sensitive periods and life-history tradeoffs. J Exp Zool A Ecol Genet Physiol 311: 788–795 10.1002/jez.565 [DOI] [PubMed] [Google Scholar]
- 7.Harris KDM, Bartlett NJ, Lloyd VK (2012) Daphnia as an emerging epigenetic model organism. Genet Res Int 2012: 147892 1-8. doi:10.1155/2012/147892. [DOI] [PMC free article] [PubMed]
- 8. Güller I, Russell AP (2010) MicroRNAs in skeletal muscle: their role and regulation in development, disease and function. The Journal of Physiology 588: 4075–4087 10.1113/jphysiol.2010.194175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen LH, Chiou GY, Chen YW, Li HY, Chiou SH (2010) MicroRNA and aging: a novel modulator in regulating the aging network. Ageing Res Rev 9 Suppl 1S59–66 10.1016/j.arr.2010.08.002 [DOI] [PubMed] [Google Scholar]
- 10. Hyun S, Lee JH, Jin H, Nam J, Namkoong B, et al. (2009) Conserved microRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell 139: 1096–1108 10.1016/j.cell.2009.11.020 [DOI] [PubMed] [Google Scholar]
- 11. Lee Y, Kim M, Han J, Yeom KH, Lee S, et al. (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051–4060 10.1038/sj.emboj.7600385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kozomara A, Griffiths-Jones S (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res39: D152–157 10.1093/nar/gkq1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Altschul S (1990) Basic Local Alignment Search Tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 14. Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, et al. (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 2005 33(20): e179 10.1093/nar/gni178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 1–12 10.1186/1746-4811-3-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Misener S&, Krawetz SA, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa (NJ): Humana Press Inc. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 18. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes.Genome Biol. 3(7): 1–11 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research 64: 5245–5250 10.1158/0008-5472.CAN-04-0496 [DOI] [PubMed] [Google Scholar]
- 20. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 21. Brow D, Guthrie C (1988) Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature 333: 213–218 10.1038/334213a0 [DOI] [PubMed] [Google Scholar]
- 22. Kennell J, Gerin I, MacDougald O, Cadigan KM (2008) The microRNA miR-8 is a conserved negative regulator of Wnt signaling. Proceedings of the National Academy of Sciences of the United States of America 105: 15417–15422 10.1073/pnas.0807763105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Selcuklu SD, Donoghue MTA, Rehmet K, de Souza Gomes M, Fort A, et al. (2012) MicroRNA-9 inhibition of cell proliferation and identification of novel miR-9 targets by transcriptome profiling in breast cancer cells. J Biol Chem 287: 29516–29528 10.1074/jbc.M111.335943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Coolen M, Thieffry D, Drivenes Ø, Becker TS, Bally-Cuif L (2012) miR-9 controls the timing of neurogenesis through the direct inhibition of antagonistic factors. Developmental cell 22: 1052–1064 10.1016/j.devcel.2012.03.003 [DOI] [PubMed] [Google Scholar]
- 25. Osei-Amo S, Hussain M, O'Neill SL, Asgari S (2012) Wolbachia-induced aae-miR-12 miRNA negatively regulates the expression of MCT1 and MCM6 genes in Wolbachia-infected mosquito cell line. PloS one 7: e50049 10.1371/journal.pone.0050049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tanaka M, Oikawa K, Takanashi M, Kudo M, Ohyashiki J, et al. (2009) Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PloS one 4: e5532 10.1371/journal.pone.0005532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Manni I, Artuso S, Careccia S, Rizzo MG, Baserga R, et al. (2009) The microRNA miR-92 increases proliferation of myeloid cells and by targeting p63 modulates the abundance of its isoforms. FASEB J 23: 3957 10.1096/fj.09-131847 [DOI] [PubMed] [Google Scholar]
- 28. Zheng YS, Zhang H, Zhang XJ, Feng DD, Luo XQ, et al. (2012) MiR-100 regulates cell differentiation and survival by targeting RBSP3, a phosphatase-like tumor suppressor in acute myeloid leukemia. Oncogene 31: 80–92 10.1038/onc.2011.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Giangreco AA, Vaishnav A, Wagner D, Finelli A, Fleshner N, et al. (2013) Tumor suppressor microRNAs, miR-100 and -125b, are regulated by 1,25-dihydroxyvitamin D in primary prostate cells and in patient tissue. Cancer Pre Res (Phila) 6: 483–494 10.1158/1940-6207.CAPR-12-0253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryazansky SS, Gvozdev VA, Berezikov E (2011) Evidence for post-transcriptional regulation of clustered microRNAs in Drosophila. BMC Genomics 12–371. doi: doi:10.1186/1471-2164-12-371. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Predicted D. pulex miRNAs, genomic coordinates, and mature miRNA sequences.
(DOCX)
miRNAs and primer sequences.
(DOCX)
Biological functions associated with the differentially expressed miRNAs during D. pulex development.
(DOCX)
PERL code for pre-miRNA sequence analyze.
(TXT)