Abstract
Cyclin D1 plays a key regulatory role during the G1 phase of the cell cycle and its gene is amplified and over-expressed in many cancers. The cyclin D1b mRNA variant was established in human cells and recent functional analyses revealed that its protein product harbors unique activities in human cancer cells. By performing reverse transcription-polymerase chain reaction (RT-PCR) and rapid amplification of cDNA ends (RACE) experiments, we identified the cyclin D1b mRNA variant in mouse. Similar to its human counterpart, the mouse cyclin D1b transcript consists of exon 1, 2, 3, 4 and part of intron 4, and contains a long open reading frame (ORF). The predicted peptide from this ORF is 34-amino acid longer than the human cyclin D1b. The expression of this mouse mRNA variant was investigated. It appears to be expressed ubiquitously and differentially in various mouse cell lines and tissues and its level might be proportional to that of the canonical endogenous cyclin D1a mRNA.
Keywords: RT-PCR, RACE, Exon, Intron, DNA sequencing, Cell cycle
Introduction
Cyclin D1 is an important regulator of cell cycle progression [1]. It functions mainly by activating cyclin dependent kinase (CDK) 4 or 6, which leads to phosphorylation of substrates that are critical for modulating G1 to S phase transition [1, 2]. Its aberrant expression is well established in the promotion of human tumorigenesis [3–7]. Betticher et al. [8] first reported the identification of an alternatively spliced cyclin D1 mRNA (now termed cyclin D1b) in human cells. Unlike the canonical cyclin D1 mRNA (now termed cyclin D1a), which consists of five exons, the cyclin D1b mRNA is made up of exon 1, 2, 3, 4 and part of intron 4. Subsequent studies revealed that human cyclin D1b protein is indeed expressed in cells and may be able to mediate aberrant cellular proliferation of human cancer [9–12]. Our group has a history of investigation on cyclin D1 in the mouse and we recently reported new insights for the function of the mouse cyclin D1a protein in mammary gland and pancreatic cell tumorigenesis [13, 14]. In order to expand our studies, we investigated whether cyclin D1b mRNA variant is expressed in mouse cells. Here we report, to our best knowledge, the first identification of the cyclin D1b mRNA in mouse.
Materials and methods
Cell lines
Various mouse cell lines 67NR, 66cl4, 168FARN, 4T1 and 4TO7 were kindly provided by Dr. Fred Miller at the Karmanos Cancer Institute in Detroit, Michigan. These cells were established from a single mouse mammary tumor, described previously [15]. Cell clones NN2 and NH2 are described in our previous report [14]. They are both derived from the benign mouse mammary epithelial cell line NMuMG: NN2 is NMuMG transfected with the pcDNA3.1 empty control expression plasmid (Invitrogen, CA) with neomycin selection, while NH2 isNMuMG transfected with the pcDNA3.1 empty control expression plasmid with hygromycin selection. Cell clones H5, M4 and M8 cells are described in our previous report [16]. They are established from an elastase-c-myc transgenic mouse pancreatic tumor by transfecting the tumor cells with either pcDNA3.1 control plasmid with hygromycin selection (for H5) or pcDNA3.1 plasmid over-expressing the c-myc gene (for M4 and M8).
Genomic DNA extraction, reverse transcription-polymerase chain reaction (RT-PCR), and DNA sequencing
Genomic DNAs were extracted by adapting a procedure described previously [17]. The amount of genomic DNA was determined by UV absorption at 260 nm. 200 ng was used in each reaction of PCR amplification. Total RNA was isolated from exponentially growing cells using the RNeasy® Isolation Kit (Qiagen, Valencia, CA). The extracted RNAs (2 µg of each sample) were reverse-transcribed with the TaqMan® reverse transcription kit, following the manufacturer’s instructions (Roche, Applied Biosystems, Foster City, CA). The reactions were primed with both oligo (dT) 15 primer and random hexamer. The resulting cDNA preparations were subjected to PCR amplifications with HotMaster® Mix 2.5× (Eppendorf, Westbury, NY). Each PCR cycle included an initial denaturation step at 95°C for 2 min, a denaturation step at 95°C for 30 sec, a primer-annealing step at 60°C for 45–120 s, and an extension step at 72°C for 45 s. PCR were performed with an Eppendorf AG Mastercycler (Hamburg, Germany). Primers used to amplify mouse cyclin D1b or D1a cDNA are D1-L228 (5′ GCG CCA TGG AAC ACC AGC TC 3′), D1-L723 (5′ CAC GAT TTC ATC GAA CAC TT 3′), D1-R1064 (5′ CCA CTT CCC CCT CCT CCT CA 3′), and D1INTRON4-R246 (5′ AGA TAT GGT CCT ATG CTG GCT G 3′). The PCR products were analyzed by electrophoresis on 1% agarose gel containing ethidium bromide, and photographed over UV light. DNA sequencing service was provided by the Wayne State University DNA sequencing core facility with the Applied Biosystems ABI Prism 3700 sequencer (Applied Biosystems, CA). Sequencing reactions were performed using the ABI BigDye® Terminator v3.1.
Rapid amplification of cDNA ends (RACE)
5′ RACE and 3′ RACE were performed with the First-Choice® RLM-RACE kit from Ambion (Applied Biosystems, CA), following the manufacturer’s instructions. For 5′ RACE analysis, we used primer D1-R678 (5′ TGA GCT TGT TCA CCA GAA GC 3′); for 3′ RACE, we used the cyclin D1b specific primer D1INTRON4-L224 (5′ CAG CCA GCA TAG GAC CAT ATC T 3′), which is the reversed orientation version of D1INTRON4-R246.
Results and discussion
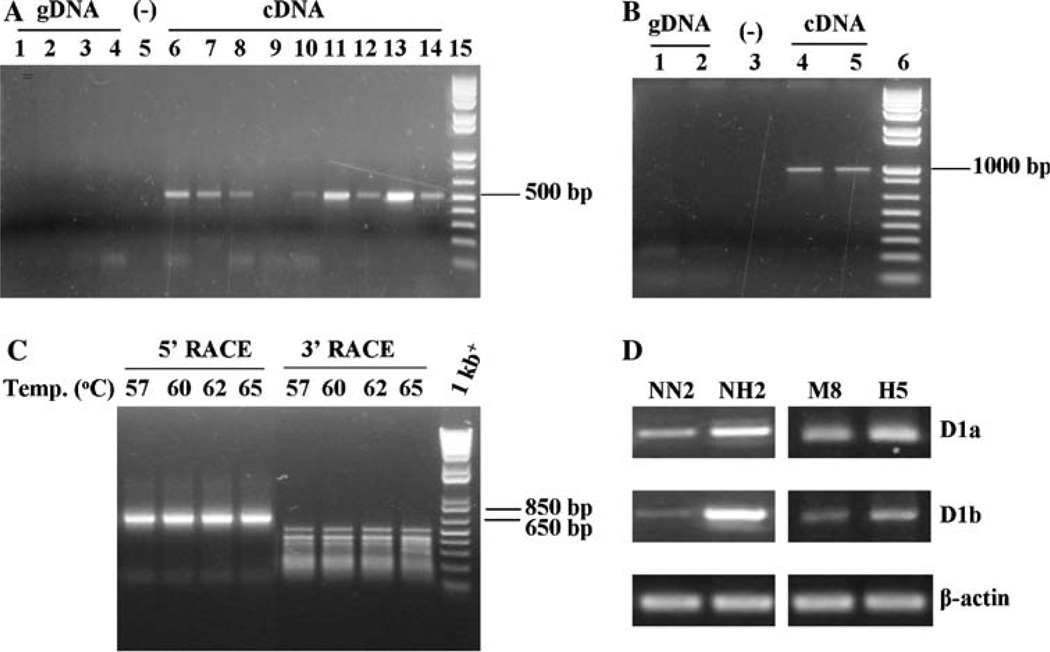
Human cyclin D1b mRNA consists of exon 1, 2, 3, 4 and intron 4 [8–12]. We speculated that mouse cyclin D1b mRNA might exist in a similar form. Therefore, we started PCR amplification with the primer pair of D1-L723 and D1INTRON4-R246, which has the left-hand primer in exon 3 and the right-hand primer in intron 4 and is expected to produce a PCR product of 498 bp. As shown in Fig. 1a, an approximately 500 bp fragment was indeed amplified from a variety of cDNA samples. Direct sequencing of this PCR fragment confirmed our speculation that it comprised part of mouse cyclin D1 exon 3, the whole exon 4, and part of intron 4. In comparison, there is no amplification from 4 independent genomic DNA samples (Fig. 1a) (this pair of primers is separated by a genomic fragment of approximately 4 kb, which is too big to be amplified in the PCR reaction). As a control, mouse p53 and beta actin genomic DNA fragments were successfully amplified from these four genomic DNA samples (data not shown).
Fig. 1.
(a) RT-PCR amplification with the cyclin D1 primer pair D1-L723 and D1INTRON4-R246. The left hand primer anneals to exon 3 sequence, and the right hand primer anneals to sequence in intron 4. The expected size of the PCR product is 498 bp. When genomic DNA (gDNA) was used as template, 200 ng from each sample was used in PCR. Lane 1–4, gDNA from mouse pancreatic tumor tissue, tail tissue, NH2 cells and M4 cells, respectively; lane 5, no template; lane 6–10, cDNA from cells 67NR, 66cl4, 168FARN, 4T1, and 4TO7, respectively; lane 11–14, cDNA from NH2, NN2, M4 and M8 cells, respectively; lane 15, 1kb+ DNA ladder from Invitrogen. (b) RT-PCR amplification with the cyclin D1 primer pair D1-L228 and D1INTRON4-R246. The left hand primer is a few base pairs upstream of the cyclin D1 protein translation start codon ATG in exon 1, and the right hand primer is in intron 4. The expected size of the PCR product is 974 bp. Lane 1–2, gDNA from NH2 and M4 cells, respectively; lane 3, no template; lane 4–5, cDNA from NH2 and M4 cells, respectively; lane 6, 1kb+ DNA ladder. (c) Rapid amplification of cDNA ends (RACE). 5′ RACE and 3′ RACE were performed as described in Materials and Methods. The template used in the PCR amplification is cDNA prepared from NH2 cells. In order to improve the chance of getting more specific amplification, we reproduced the reaction at 4 separate annealing temperatures: 57, 60, 62, and 65°C. (d) Comparison of cyclin D1a and cyclin D1b mRNA levels in NN2 and NH2 cells, and M8and H5 cells by RT-PCR. Cyclin D1a specific primer pair is D1-L723 and D1-R1064; cyclin D1b specific primer pair is D1-L723 and D1INTRON4-R246 (Fig. 2a). PCR amplification of β-actin served as cDNA template loading control. PCR products were separated by agarose gel electrophoresis (1% agarose in 0.5× TBE buffer). Each experiment was repeated a minimum of three times
Next, we carried out PCR amplification with primer pair of D1-L228 and D1INTRON4-R246, which has the left-hand primer a few base pair upstream of the cyclin D1 protein translation start codon ATG in exon 1 and the right-hand primer in intron 4 (Fig. 2a). The expected PCR fragment is 974 bp and contains a long open reading frame (ORF). As demonstrated in Fig. 1b, an approximately 1,000 bp fragment was amplified from two cDNA samples. This fragment was also sequenced directly and the results confirmed the existence of a cyclin D1 mRNA variant and identified it as the mouse cyclin D1b sequence that is similar to human cyclin D1b mRNA (part of exon 1, the whole of exons 2, 3 and 4, and part of intron 4). As a comparison, there was no amplification from two genomic DNA samples (Fig. 1b).
Fig. 2.
(a) Illustration of the locations of primers used in PCR amplifications and the components of mouse cyclin D1a and cyclin D1b cDNA. Coding exon sequences are represented by black rectangles; non-coding exon sequences are represented by empty rectangles; and cyclin D1b cDNA fragment derived from intron 4 are represented by a shadowed rectangle. (b) Comparison of the cyclin D1b peptide sequences derived from human and mouse intron 4, respectively
After the existence of the cyclin D1b mRNA in mouse was confirmed, we performed 5′ and 3′ RACE to try to identify the 5′- and 3′-end of the transcript. The cDNA template used was prepared from the NH2 cell line, which showed a high level of cyclin D1b transcript (Fig. 1a) and elevated expression of cyclin D1a as determined by RT-PCR and Western blot (data not shown). The right hand primer used in the 5′ RACE PCR amplification was D1-R678, which anneals to both cyclin D1a and cyclin D1b cDNA (Fig. 2a). Since cyclin D1b and cyclin D1a transcripts have a large stretch of 5′ sequence in common, it is quite challenging to select a primer specific for cyclin D1b. We tried two primers from intron 4 cyclin D1b specific sequences, but the PCR amplifications were not successful. By using primer D1-R678, we reasoned that if cyclin D1b shares the same transcription starting site with cyclin D1a, the one expected PCR fragment would be approximately 700 bp. On the other hand, if cyclin D1b starts down stream of the cyclin D1a transcription start site, its expected PCR fragment from 5′ RACE PCR amplification would be shorter than 700 bp, but longer than 450 bp, because as presented above, cyclin D1b cDNA was successfully RT-PCR amplified with the left hand primer, D1-L228. Additionally, if cyclin D1b transcription starts from upstream of the cyclin D1a start site, the PCR fragment would be longer than 700 bp. As shown in Fig. 1c, the 5′ RACE PCR amplification resulted in only one major specific band of approximately 700 bp (Fig. 1c). This band was cut out from the agarose gel, purified and sequenced. The sequencing data revealed that this fragment represents the transcription of the canonical cyclin D1a mRNA. There was no PCR fragment between 450 and 700 bp. Therefore, we can rule out the possibility that the cyclin D1b transcript starts from downstream of the cyclin D1a transcription start site. However, we remain uncertain about the possibility that the cyclin D1b transcript may start from a 5′ transcription site upstream of the cyclin D1a start site. On the other hand, the 3′ RACE PCR amplification produced quite a few bands with the longest one at approximately 550 bp (Fig. 1c), which we purified from the gel and sequenced. The sequence for the 550 bp product matches that for cyclin D1 intron 4 sequence, ending at around 818 base pair of the intron 4. Based on our RT-PCR and RACE experiments, we think it is quite likely that mouse cyclin D1b mRNA transcript consists of exon 1, 2, 3, 4 and approximately 818 base pair of intron 4 (Fig. 2a). This putative mouse cyclin D1b complete cDNA sequence was submitted to NCBI GenBank and the assigned accession number is EU600170.
We investigated further to compare the expression of cyclin D1b in various mouse tissues and cell lines. The general observation is that cyclin D1b mRNA is expressed ubiquitously and its level is in good correlation with that of the canonical cyclin D1a, i.e., when cyclin D1a is highly expressed, so is cyclin D1b; and when cyclin D1a expression is relatively low, so is cyclin D1b expression. For example, although PCR fragments were amplified successfully from cDNA samples prepared from mouse liver, kidney, heart, normal pancreas and pancreatic tumors, the level of cyclin D1b mRNA was in such low abundance in all non-tumor samples, which usually show very low levels of cyclin D1a expression, that a second round of PCR amplification was required to visualize the expected bands (data not shown). The general observation was also confirmed by the comparison of cyclin D1b levels in two pairs of cell lines: NN2/NH2 and M8/H5. As described in the Materials and Methods section, NN2 is NMuMG transfected with the neomycin pcDNA3.1 control plasmid, while NH2 is NMuMG transfected with the hygromycin control plasmid. In our studies, we found that hygromycin resistant NMuMG cell clones, such as NH2, have elevated expression of cyclin D1 at the mRNA and protein levels [14]. The H5 cells are pancreatic tumor cells transfected with hygromycin pcDNA3.1 control plasmid, and the M8 cells were pancreatic tumor cells transfected with hygromycin pcDNA3.1 plasmid over-expressing the mouse c-myc gene. It is documented in our previous report that elevated expression of c-myc would depress the level of the canonical cyclin D1a at mRNA and protein levels [16]. Consistent with these earlier observations, NH2 cells showed higher levels of cyclin D1a and cyclin D1b mRNA compared with NN2 cells, and so did H5 cells compared with M8 cells (Fig. 1d).
Sanchez et al. [18] reported very recently that a mutated transcription factor up-regulates cyclin D1b in human cancer. There are also early interesting reports that a common polymorphism within human cyclin D1, A/G at nucleotide 870 at the exon 4-intron 4 splicing site, does not alter its encoded amino acid but may influence the ratio of transcript a and transcript b [19, 20]. Reported results have been inconsistent, however, with the A allele favoring production of transcript b. Because genomic DNA sequence at the mouse cyclin D1 exon 4-intron 4 junction sites is identical to that of human cyclin D1, we investigated the potential connection between the A/G polymorphism and the ratio of mouse cyclin D1a and cyclin D1b. The exon 4-intron 4 junction region of mouse cyclin D1 was PCR amplified by using genomic DNA templates prepared from three independent cell lines with high level of cyclin D1b mRNA—cell lines 67NR, NH2, and M4. The resulting PCR fragments were cloned into pCR2.1®TOPO® cloning vector (Invitrogen, CA, USA). Plasmid DNAs were isolated from one dozen clones for each group and subjected to DNA sequencing. It turned out that all 36 clones represented the G allele. Therefore, there might be no connection between the A/G polymorphism and the ratio of mouse cyclin D1a and cyclin D1b.
Amino acid sequences and functional domains are highly conserved between mouse and human cyclin D1 protein. Basically, the N-terminal 20 amino acids are important for phosphorylation of retinoblastoma protein (Rb), amino acids 50 to 165 forms the so-calledCyclinBox,which is required for the interaction with CDK4 or 6, and the C-terminal sequences are important for protein stability [21]. It was reported that the Cyclin Box and the C-terminal sequences are essential for cyclin D1’s oncogenic transformation capability [21]. Both human and mouse cyclin D1b transcripts would affect the crucial C-terminal sequences of cyclin D1 protein. Nevertheless, the human cyclin D1b protein is indeed expressed in cells and has been implicated inmediation of aberrant cellular proliferation of human cancer [9–12, 18]. The mouse D1b transcript contains a long ORF as well. It is noticed that the predicted peptide sequence derived fromthe mouse intron 4 is 34-amino acid longer than that from the human intron 4 (Fig. 2b). More studies are needed in order to answer the questions that whether the mouse D1b protein expresses in cells and what functions it may be able to perform.
Acknowledgement
This work was supported by NIH grant R01CA100864 (J.D.Liao).
Contributor Information
Jack Wu, Karmanos Cancer Institute, Wayne State University, Detroit, MI 48201, USA.
Si-hung Wu, Hormel Institute, University of Minnesota, Austin, MN 55912, USA.
Aliccia Bollig, Karmanos Cancer Institute, Wayne State University, Detroit, MI 48201, USA.
Archana Thakur, Karmanos Cancer Institute, Wayne State University, Detroit, MI 48201, USA.
D. Joshua Liao, Email: djliao@hi.umn.edu, Hormel Institute, University of Minnesota, Austin, MN 55912, USA.
References
- 1.Sherr CJ. D-type cyclins. Trends Biochem Sci. 1995;20:187–190. doi: 10.1016/s0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- 2.Sherr CJ. The Pezcoller lecture: cancer cell cycles revisited. Cancer Res. 2000;60:3689–3695. [PubMed] [Google Scholar]
- 3.Peters G. The D-type cyclins and their role in tumorigenesis. J Cell Sci Suppl. 1994;18:89–96. doi: 10.1242/jcs.1994.supplement_18.13. [DOI] [PubMed] [Google Scholar]
- 4.Arnold A. The cyclin D1/PRAD1 oncogene in human neoplasia. J Invest Med. 1995;43:543–549. [PubMed] [Google Scholar]
- 5.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 6.Palmero I, Peters G. Perturbation of cell cycle regulators in human cancer. Cancer Surv. 1996;27:351–367. [PubMed] [Google Scholar]
- 7.Diehl JA. Cycling to cancer with cyclin D1. Cancer Biol Ther. 2002;1:226–231. doi: 10.4161/cbt.72. [DOI] [PubMed] [Google Scholar]
- 8.Betticher DC, Thatcher N, Altermatt HJ, Hoban P, Ryder WD, Heighway J. Alternate splicing produces a novel cyclin D1 transcript. Oncogene. 1995;11:1005–1011. [PubMed] [Google Scholar]
- 9.Solomon DA, Wang Y, Fox SR, Lambeck TC, Giesting S, Lan Z, Senderowicz AM, Conti CJ, Knudsen ES. Cyclin D1 splice variants. Differential effects on localization, RB phosphorylation, and cellular transformation. J Biol Chem. 2003;278:30339–30347. doi: 10.1074/jbc.M303969200. [DOI] [PubMed] [Google Scholar]
- 10.Lu F, Gladden AB, Diehl JA. An alternatively spliced cyclin D1 isoform, cyclin D1b, is a nuclear oncogene. Cancer Res. 2003;63:7056–7061. [PubMed] [Google Scholar]
- 11.Burd CJ, Petre CE, Morey LM, Wang Y, Revelo MP, Haiman CA, Lu S, Fenoglio-Preiser CM, Li J, Knudsen ES, Wong J, Knudsen KE. Cyclin D1b variant influences prostate cancer growth through aberrant androgen receptor regulation. Proc Natl Acad Sci USA. 2006;103:2190–2195. doi: 10.1073/pnas.0506281103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knudsen KE. The cyclin D1b splice variant: an old oncogene learns new tricks. Cell Div. 2006;24:1–15. doi: 10.1186/1747-1028-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biliran H, Jr, Wang Y, Banerjee S, Xu H, Heng H, Thakur A, Bollig A, Sarkar FH, Liao JD. Over-expression of cyclin D1 promotes tumor cell growth and confers resistance to cisplatin-mediated apoptosis in an elastase-myc transgene-expressing pancreatic tumor cell line. Clin Cancer Res. 2005;11:6075–6086. doi: 10.1158/1078-0432.CCR-04-2419. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Thakur A, Sun Y, Wu J, Biliran H, Bollig A, Liao DJ. Synergistic effect of cyclin D1 and c-Myc leads to more aggressive and invasive mammary tumors in severe combined immunodeficient mice. Cancer Res. 2007;67:3698–3707. doi: 10.1158/0008-5472.CAN-06-4000. [DOI] [PubMed] [Google Scholar]
- 15.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52:1399–1405. [PubMed] [Google Scholar]
- 16.Biliran H, Jr, Banerjee S, Thakur A, Sarkar FH, Bollig A, Ahmed F, Wu J, Sun Y, Liao JD. c-Myc-induced chemosensitization is mediated by suppression of cyclin D1 expression and nuclear factor-kappa B activity in pancreatic cancer cells. Clin Cancer Res. 2007;13:2811–2821. doi: 10.1158/1078-0432.CCR-06-1844. [DOI] [PubMed] [Google Scholar]
- 17.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez G, Bittencourt D, Laud K, Barbier J, Delattre O, Auboeuf D, Dutertre M. Alteration of cyclin D1 transcript elongation by a mutated transcription factor up-regulates the oncogenic D1b splice isoform in cancer. Proc Natl Acad Sci USA. 2008 doi: 10.1073/pnas.0710748105. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howe D, Lynas C. The cyclin D1 alternative transcript [a] and [b] are expressed in normal and malignant lymphocytes and their relative levels are influenced by the polymorphism at codon 241. Haematologica. 2001;86:563–569. [PubMed] [Google Scholar]
- 20.Carrère N, Belaud-Rotureau MA, Dubus P, Parrens M, de Mascarel A, Merlio JP. The relative levels of cyclin D1a and D1b alternative transcripts in mantle cell lymphoma may depend more on sample origin than on CCND1 polymorphism. Haemotologica. 2005;90:854–856. [PubMed] [Google Scholar]
- 21.Zwicker J, Brüsselbach S, Jooss KU, Sewing A, Behn M, Lucibello FC, Müller R. Functional domains in cyclin D1: pRb-kinase activity is not essential for transformation. Oncogene. 1999;18:19–25. doi: 10.1038/sj.onc.1202286. [DOI] [PubMed] [Google Scholar]