SUMMARY
The transition from transcription initiation into elongation is controlled by transcription factors, which recruit P-TEFb to promoters to phosphorylate RNA polymerase II. A fraction of P-TEFb is recruited as part of the inhibitory 7SK small nuclear ribonucleoprotein (snRNP), which inactivates the kinase and prevents elongation. It is unclear, however, how P-TEFb is captured from the promoter-bound 7SK snRNP to activate elongation. Here, we describe a novel mechanism by which transcription factors mediate the enzymatic release of P-TEFb from the 7SK snRNP at promoters to trigger activation in a gene-specific manner. We demonstrate that Tat recruits PPM1G/PP2Cγ to locally disassemble P-TEFb from the 7SK snRNP at the HIV promoter via dephosphorylation of the kinase T-loop. Similar to Tat, NF-κB recruits PPM1G in a stimulus-dependent manner to activate elongation at inflammatory-responsive genes. Recruitment of PPM1G to promoter-assembled 7SK snRNP provides a new paradigm for rapid gene activation through transcriptional pause release.
INTRODUCTION
The transition from transcription initiation into productive elongation is a key, rate-limiting step essential for gene activation (Core and Lis, 2008; Smith and Shilatifard, 2013; Zhou et al., 2012). This major restriction point occurs when Pol II pauses a few nucleotides downstream the transcription start site (TSS) by the action of negative elongation factors such as Negative ELongation Factor (NELF) and the 5,6-Dichloro-1-beta-D-ribofuranosylbenzimidazole Sensitivity-Inducing Factor (DSIF) (Marshall and Price, 1992; Yamaguchi et al., 1999). Conversion into the elongating form requires the recruitment of the P-TEFb kinase (Cdk9 and cyclin T1/2) by transcription factors, which hyperphosphorylates the C-terminal domain (CTD) of Pol II in addition to NELF and DSIF at promoters (Mancebo et al., 1997; Yamaguchi et al., 1999; Zhou et al., 2012). Several transcription factors such as c-Myc, p53, NF-κB, and HIV Tat utilize P-TEFb to activate the transition into elongation at their target genes (Barboric et al., 2001; Gomes et al., 2006; Lis et al., 2000; Mancebo et al., 1997; Rahl et al., 2010; Wei et al., 1998).
While unengaged in transcription, P-TEFb is held in a catalytically inactive state assembled into the 7SK snRNP through the kinase inhibitor Hexim1 and 7SK RNA. While P-TEFb and Hexim1 are reversibly bound, the La related protein-7 (Larp7) and the methylphosphate capping enzyme (MePCE) are constitutively bound to the 7SK snRNP by directly binding and stabilizing 7SK RNA (Chen et al., 2004; Jeronimo et al., 2007; Krueger et al., 2008). Two pools of P-TEFb (free and 7SK-bound) exist in a reversible equilibrium to accommodate the transcriptional demands of the cell (Peterlin and Price, 2006; Zhou et al., 2012). Previous works have shown that bulk nuclear levels of free P-TEFb increase upon disruption of the 7SK snRNP during activation of signaling cascades in response to environmental cues (Chen et al., 2008; Kim et al., 2011). However, it appears to be an inefficient mechanism for released P-TEFb to first diffuse around in the nucleoplasm before being attracted to promoters by transcription factors (Ji et al., 2013). We have previously shown that the 7SK snRNP is recruited to the HIV promoter where the viral transcription factor Tat stimulates its disassembly to transfer P-TEFb to the TAR stem-loop formed at the 5′-end of viral nascent transcripts (D’Orso and Frankel, 2010). Extending this discovery, recent work has demonstrated that the inhibitory 7SK snRNP complex occupies cellular promoters to directly prevent the transcriptional pause release (Ji et al., 2013). Thus, the presence of the 7SK snRNP at gene promoters appears to play a critical role in gene activation. To that end, we sought to characterize how transcription factors mediate the release of P-TEFb from 7SK snRNP to stimulate elongation in a gene-specific manner.
Here we report the unexpected findings that Tat and NF-κB recruit the PPM1G/PP2Cγ phosphatase to their target promoters to locally activate the P-TEFb kinase. PPM1G dephosphorylates the T-loop of Cdk9, releasing P-TEFb from the 7SK snRNP, and activating the switch from initiation into elongation. This enzymatic step occurs at the promoter, which increases the local concentration of the kinase before it is captured by transcription factors to activate elongation. Thus, recruitment of PPM1G to the promoter-assembled 7SK snRNP by transcription factors provides a new paradigm for rapid gene activation through transcriptional pause release.
RESULTS
Novel 7SK RNA co-factors assemble with Tat into a ribonucleoprotein complex to control HIV transcription
To investigate the mechanisms by which transcription factors recruit P-TEFb from the 7SK snRNP during selective activation of transcriptional programs, we used HIV as a model system. Tat promotes the disassembly of the 7SK snRNP at the viral promoter and transfers P-TEFb to TAR, stimulating Pol II phosphorylation and transcription elongation (Figure 1A). Because Tat directly binds 7SK RNA in cells (Figure S1A) and complexes with the 7SK snRNP (D’Orso and Frankel, 2010; Krueger et al., 2010; Muniz et al., 2010; Sobhian et al., 2010; Faust et al., 2012), we hypothesized that Tat assembles a Tat-7SK ribonucleoprotein (RNP) complex to regulate the release of P-TEFb from the 7SK snRNP at the HIV promoter during the transition into elongation.
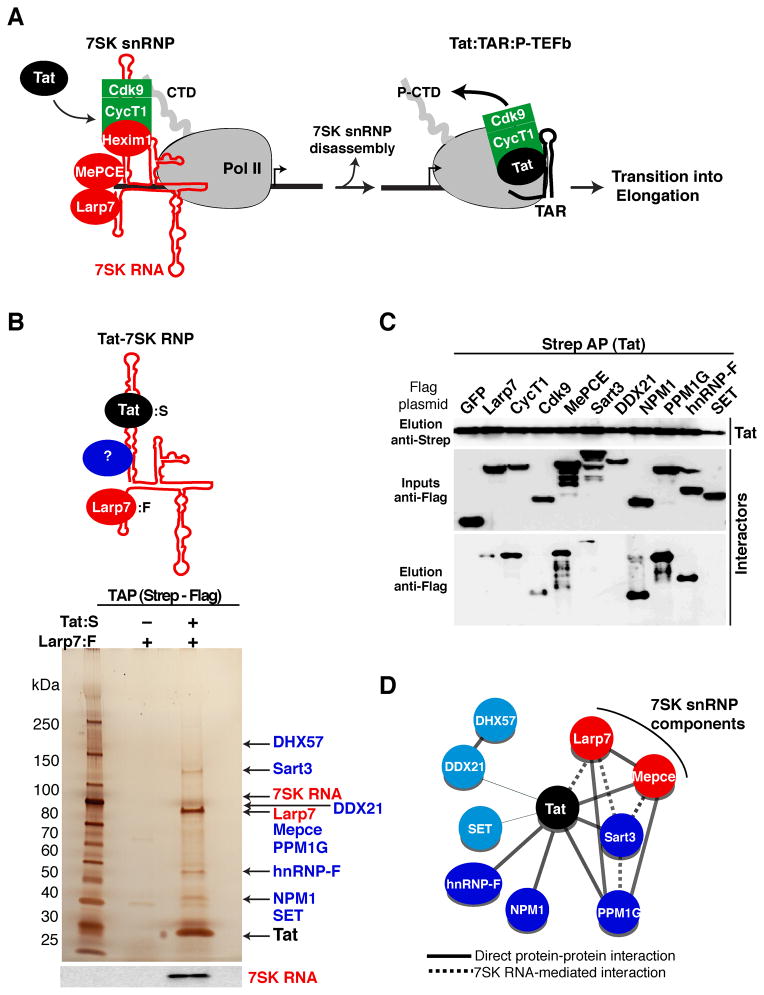
Figure 1. Identification of an HIV-host Tat-7SK protein-RNA complex implicated in transcriptional control.
(A) Model of HIV transcription activation by Tat. Tat promotes the disassembly of the promoter-bound 7SK snRNP to transfer P-TEFb (CycT1:Cdk9) to the nascent viral RNA (TAR), allowing for Pol II CTD hyperphosphorylation (P-CTD) and the transition into elongation.
(B) Tandem affinity purification (TAP) of the Tat-7SK ribonucleoprotein (RNP) complex by sequential Strep-tagged Tat (Tat:S) and Flag-tagged Larp7 (Larp7:F) purifications from 293T cells. High-confidence Tat-7SK RNP interactors were identified by mass spectrometry analysis (see also Table S1) and indicated by arrows on a silver-stained gel. The bottom panel shows a 7SK RNA Northern blot.
(C) Validation of the interactors identified. Tat:S was co-expressed into 293T cells along with the identified Flag-tagged factors. Strep AP followed by Western blot reveals proteins co-immunoprecipitating with Tat.
(D) A network representation of the protein-protein and protein-RNA interactions within the Tat-7SK RNP complex. The thickness of the edges corresponds to the semi-quantititative score of the interactions among nodes. Solid and dashed edges correspond to direct or RNA-mediated interactions (according to the results of Figures 1C and S1, and data not shown). Light blue nodes represent interactors identified by mass spectrometry but not validated by protein-protein interaction assays. See also Figure S1.
To identify Tat co-factors involved in the release of P-TEFb from the 7SK snRNP, we isolated the Tat-7SK RNP complex through tandem affinity purification (TAP) of Tat and Larp7 (Figure 1B). Larp7 was used to select for 7SK-associated fractions of Tat because it is constitutively bound to the RNA (Jeronimo et al., 2007; Krueger et al., 2008). Silver staining and Northern blot of the final eluate revealed the presence of Tat, Larp7, 7SK RNA, as well as several bands that were not present upon co-purification of Larp7 without Tat (Figure 1B) or a K41A non-functional Tat (data not shown). To reveal the identity of the additional bands, we subjected gel slices and an in-solution complex mixture to mass spectrometry analysis. In addition to Tat and Larp7, we identified eight high-confidence interactors (Table S1). Despite repeated attempts, however, P-TEFb and Hexim1 were not detected in the Tat-7SK RNP complex through Western blot and mass spectrometry (data not shown), suggesting they are released upon binding of Tat along with the co-factors identified.
The proteomics dataset was validated by protein-protein interaction assays by co-transfecting cells with Tat along with each of the identified interactors or negative (GFP) and positive (Larp7, CycT1 and Cdk9) controls. We observed that MePCE, Sart3, NPM1, PPM1G, and hnRNP-F co-purified with Tat, while SET and DDX21 did not under the conditions tested (Figure 1C). Among the interactors, only Larp7 demonstrates an RNase sensitive interaction pattern with Tat (data not shown), suggesting that Tat makes direct protein contacts in the absence of 7SK RNA. To identify whether the Tat interactors associate with the 7SK snRNP in cells independently of Tat, we performed RNA-ImmunoPrecipitation (RIP) assays followed by Northern blot, as well as RNA gel shift assays, and observed variable association with 7SK RNA, except for NPM1 and SET (Figures S1B–D). By compiling the protein-protein and protein-RNA interaction datasets, we defined a network of interactions within the Tat-7SK RNP complex (Figure 1D). We reasoned that factors associated with this RNP complex dictate the release of P-TEFb from the 7SK snRNP-assembled at the HIV promoter (Figure 1A). Here we focus on the PPM1G phosphatase because of the possible enzymatic disassembly of the snRNP during HIV transcription activation (see below).
Tat recruits PPM1G to dephosphorylate the T-loop of Cdk9 and release P-TEFb from the 7SK snRNP complex
Previous studies have demonstrated that phosphorylation of Thr186 (Phos-T186) at the activating T-loop of Cdk9 (P-Cdk9) promotes the assembly of P-TEFb with Hexim1 and 7SK RNA (Chen et al., 2004; Li et al., 2005). Because the Tat-7SK RNP lacks P-TEFb, but contains PPM1G (a member of the PPM/PP2C family of nuclear, metal-dependent Ser/Thr phosphatases (Allemand et al., 2007)), we hypothesized that Cdk9 dephosphorylation at Thr186 by PPM1G is a major determinant by which Tat induces the disassembly of P-TEFb from the 7SK snRNP. To test this, we purified wild-type PPM1G and a catalytically inactive D496A mutant (Allemand et al., 2007) and observed that PPM1G (but not D496A) dephosphorylates Cdk9 at Thr186 in vitro (Figure 2A). To study if Cdk9 dephosphorylation triggers P-TEFb release from the snRNP, we incubated 7SK-bound P-TEFb with PPM1G under dephosphorylation conditions, and subsequently purified P-TEFb to monitor its assembly into the 7SK snRNP (Figure 2B). Remarkably, we found that Cdk9 dephosphorylation by PPM1G (but not D496A) at Thr186 releases P-TEFb from the 7SK snRNP, as shown by the presence of the inhibitory 7SK snRNP components (devoid of P-TEFb) in the supernatant of a P-TEFb purification. Collectively, our data strongly supports a functional link between Cdk9 T-loop dephosphorylation by PPM1G and the release of P-TEFb from the 7SK snRNP in vitro.
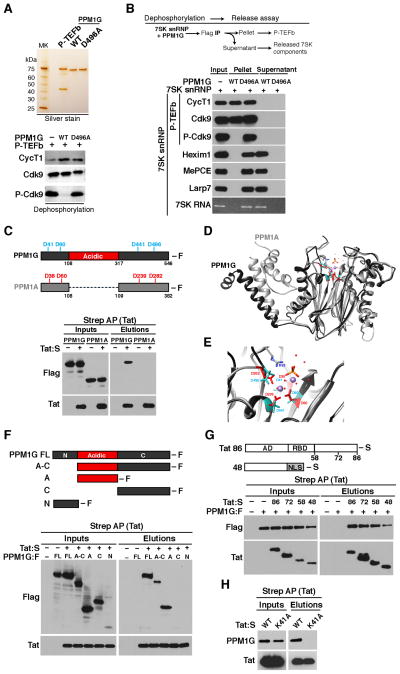
Figure 2. Tat selectively recruits the PPM1G phosphatase to disassemble the 7SK snRNP and release P-TEFb through Cdk9 T-loop dephosphorylation.
(A) Top, P-TEFb and wild-type PPM1G or catalytically inactive mutant (D496A) were purified and visualized by silver stain. Bottom, P-TEFb was incubated with PPM1G or D496A under dephosphorylation conditions and Cdk9 T-loop phosphorylation at Thr186 (P-Cdk9) was monitored by Western blot.
(B) Enzymatic release assay of P-TEFb from the 7SK snRNP. 7SK-bound P-TEFb (CycT1:S–Cdk9:F) complexes were purified from mammalian cells and incubated with PPM1G or D496A under dephosphorylation conditions. Subsequent purification of P-TEFb using Flag beads was done to monitor released (supernatant) and retained (pellet) components by Western blot.
(C) Tat selectively binds PPM1G but not PPM1A. Top, PPM1G possesses an acidic-region intervening the N- and C-terminal phosphatase domain. The position of the four Asp (D) residues required for metal coordination and catalysis are shown above the schemes. Bottom, Strep-tagged Tat and Flag-tagged PPM1G or PPM1A were co-transfected into 293T cells, affinity purified using Strep beads and analyzed by Western blot.
(D) Superimposition of the PPM1A structure (Das et al., 1996) and structure prediction of PPM1G lacking its acidic-region.
(E) Close-up view of the PPM1A-PPM1G superimposition showing the catalytic core in the PPM1A structure (red) and structure prediction of PPM1G (light blue). The two Mn+ ions (violet) and the phosphate group (orange) are shown. In PPM1A, Arg33 (R33) side-chains form H-bonds with the phosphate ion (orange).
(F) Tat binds to the PPM1G acidic domain. Strep-tagged Tat and Flag-tagged PPM1G (full-length (FL) or domains) were co-transfected into 293T cells, affinity purified using Strep beads and analyzed by Western blot.
(G) Tat domain mapping. Strep-tagged Tat full-length (Tat86) or domains (AD: activation domain; RBD: RNA-binding domain; NLS: SV40 T-Ag nuclear localization signal) and Flag-tagged PPM1G were co-transfected into 293T cells, affinity purified using Strep beads and analyzed by Western blot.
(H) Strep-tagged wild-type (WT) Tat or a non-functional K41A mutant and Flag-tagged PPM1G were-co-transfected into 293T cells, affinity purified using Strep beads and analyzed by Western blot. See also Figure S2.
Because PPM1G and P-TEFb do not stably associate in vitro or in cells (data not shown), we reasoned that Tat mediates the recruitment of PPM1G to the 7SK snRNP to dephosphorylate the T-loop of Cdk9 thereby releasing P-TEFb for gene activation. To test this hypothesis, we co-transfected Tat along with PPM1G or PPM1A (another family member shown to dephosphorylate P-TEFb in vitro (Wang et al., 2008)), and observed that Tat selectively associates with PPM1G but not PPM1A (Figure 2C). Through studies with bacterially expressed Tat and PPM1G, as well as tandem affinity purification of the Tat-PPM1G complex from mammalian cells, we confirmed that Tat and PPM1G physically interact (Figures S2A–B).
The structure of PPM1G is currently unknown, but PPM1A folds with a catalytic domain composed of a central beta-sandwich that binds two Mn2+ ions surrounded by α-helices (Das et al., 1996) (Figure 2D). The phosphatase domains of PPM1A, and the structure prediction of PPM1G, contain the four Asp residues required for metal coordination and catalysis (Figures 2D–E), but PPM1G distinguishes itself from the other PPM-family members with an internal acidic-domain (Figure 2C). Co-transfection of Tat along with several PPM1G domains demonstrated that the acidic-domain in PPM1G is necessary and sufficient for Tat binding (Figure 2F), explaining Tat’s specificity for PPM1G over PPM1A.
To further define how Tat binds the enzyme, we co-transfected PPM1G with several Tat fragments and determined that the activation domain (residues 1–48) is necessary and sufficient for PPM1G recognition (Figure 2G). In agreement, mutations in the activation domain that eliminate Tat transactivation (such as K41A) (D’Orso et al., 2012) abolish the Tat-PPM1G interaction (Figure 2H).
Recruitment of PPM1G by Tat activates the transition into elongation by disassembling the HIV promoter-bound 7SK snRNP complex
The finding that PPM1G associates with the Tat activation domain suggests that Tat may recruit the enzyme to the 7SK snRNP complex during HIV transcription. To determine if PPM1G is required for Tat activation, we utilized a genetic interaction approach. HeLa cells were transfected with short interfering RNAs (siRNAs) to knockdown PPM1G, Cdk9 (used as a positive control), or a non-target control siRNA, and we measured Tat-mediated transcriptional activity on an HIV LTR luciferase reporter (Figure 3A). Interestingly, knockdown of PPM1G and Cdk9 reduced the Tat activation step (~4–5-fold) without affecting Tat levels (Figures 3A and S3A). Because PPM1G appears to be implicated in transcription, we examined if the other components of the Tat-7SK RNP complex identified by mass spectrometry (Figure 1) also play a similar role. Interestingly, knockdown of Sart3, NPM1 and hnRNP-F affect the Tat activation step in a reporter assay, thus implicating these factors, like PPM1G, in HIV transcription (Figure S3B).
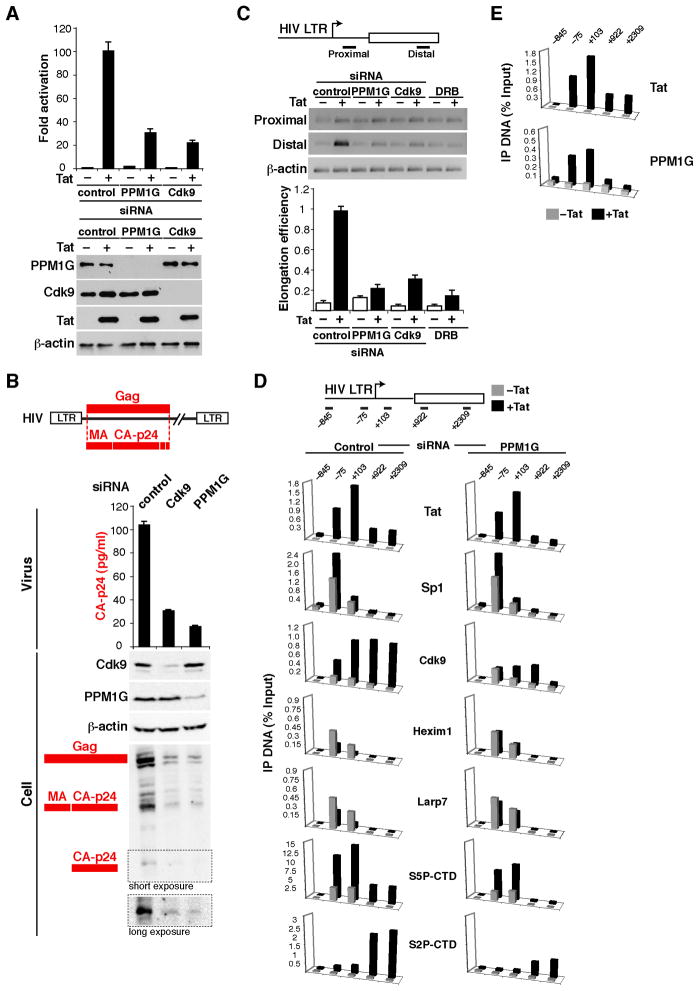
Figure 3. Recruitment of PPM1G by Tat mediates disassembly of the promoter-bound 7SK snRNP and activates the transition into elongation.
(A) Knockdown of PPM1G antagonizes Tat-mediated HIV transcription activation. Upper panel, HeLa cells were co-transfected with an HIV LTR-Firefly luciferase and Flag-tagged Tat plasmid after siRNA knockdown. HIV luciferase activity was normalized to CMV Renilla luciferase (Mean ± SEM are shown, n=3). Lower panel, HeLa samples were used for Western blots with the indicated antibodies; β-actin was used as an internal loading control.
(B) Production of viral particles from cells. 293T cells were sequentially transfected with three different siRNAs and a full-length HIV proviral DNA (NL4-3). Only Gag is shown in the scheme. Viral particles released to the supernatant were quantified by p24 ELISA (Mean ± SEM are shown, n=4) and the levels of intracellular Cdk9, PPM1G, β-actin and HIV Gag were detected by Western blot.
(C) Transcription elongation assay with the schematic of RT-PCR products. The gel shows the products resulting from transfection of a HeLa LTR-FFL cell line with three different siRNAs or pre-treated with DRB in the presence of an empty (−) or Tat plasmid (+). The graph indicates the calculated elongation efficiencies expressed in arbitrary units (AU) standardized to β-actin (Mean ± SEM are shown, n=3).
(D) ChIP assay to analyze the distribution of Tat and co-factors at the HIV locus in a HeLa LTR- FFL cell line transfected with a control or PPM1G siRNA along with a mock (gray bars) or Flag-tagged Tat (black bars) plasmid. The position of the amplicons used in ChIP-qPCR is shown with the schematic of the HIV locus. Values represent the average of three independent experiments. The SEM is less than 10% and not shown for simplicity.
(E) ChIP assay to analyze the distribution of Tat and PPM1G at the HIV locus in a HeLa LTR-FFL cell line transfected with a mock (grey bars) or Tat (black bars) plasmid. Values represent the average of three independent experiments. The SEM is less than 10% and not shown for simplicity. See also Figure S3.
To put our findings in the context of HIV, we assayed the amount of infectious particles produced from 293T cells transfected with a provirus DNA after knockdown of PPM1G or Cdk9. Similar to Cdk9 knockdown, which affects proviral transcription (Ott et al., 2011), loss of PPM1G resulted in a sharp decrease in the production of viral particles, as revealed by a reduction in the levels of the p24 Capsid (CA) protein (Figure 3B). The processing of the Gag precursor into the smaller products CA and Matrix (MA) is required for viral assembly and egress from the infected cell. To evaluate whether the reduction of extracellular p24 is due to improper Gag processing, we monitored intracellular Gag levels by Western blot and observed that the ratio of processed to unprocessed Gag is not affected by the loss of PPM1G (Figure 3B). Collectively, these results support a transcriptional, but not a post-translational role, for PPM1G in the viral life cycle.
Because PPM1G is involved in HIV transcription and Tat functions to relieve the block at the elongation step, we asked if PPM1G promotes the initiation-to-elongation switch at the viral promoter. To test this, we used a quantitative elongation assay to monitor levels of promoter-proximal and -distal transcripts, which approximate the extent of initiation and elongation, respectively (Figure 3C). Even in the presence of Tat, PPM1G knockdown markedly decreased (~5-fold) the production of promoter-distal transcripts, thus negatively affecting the transcription elongation step. This elongation blockage by PPM1G knockdown phenocopies the effect of Cdk9 knockdown or kinase inactivation with the Pol II transcription elongation inhibitor DRB (Kim et al., 2002; Marshall and Price, 1992). These results functionally link the release of P-TEFb from the 7SK snRNP by Tat and PPM1G with activation of the elongation step.
We then asked whether decreased elongation due to PPM1G knockdown correlated with reduced P-TEFb recruitment and Pol II phosphorylation at the HIV promoter during Tat activation. To assess this, we performed a ChIP assay on the HeLa-LTR cell line by depleting PPM1G in the absence or presence of a functional Tat (Figure 3D). Assembly of HIV transcription pre-initiation complexes at the promoter (demonstrated by Sp1 occupancy and levels of the initiating Ser-5 CTD phosphorylated Pol II (S5P-CTD)), were minimally affected by PPM1G knockdown. However, PPM1G knockdown markedly reduced the Tat-mediated increase in Pol II S2P-CTD levels and P-TEFb occupancy in downstream regions (lowered by a factor of ~2.5–9-fold), consistent with a role of PPM1G in elongation. Strikingly, this finding correlates well with the inability of Tat to disassemble the 7SK snRNP (Hexim1 and Larp7) at the promoter. We noticed that levels of the 7SK snRNP are reduced non-stoichiometrically with Tat probably because not all the cells express Tat after transfection.
To test if Tat mediates recruitment of PPM1G to the HIV promoter we performed a ChIP assay on the HeLa-LTR cell line in the absence or presence of Tat (Figure 3E). While in the absence of Tat there is no detectable PPM1G, Tat recruits PPM1G to the promoter (~4-fold over no Tat). Together, Tat mediates the enzymatic disassembly of the 7SK snRNP from the viral promoter in vivo to release P-TEFb for transcription elongation.
Tat binds released, dephosphorylated P-TEFb and stimulates phosphorylation of the Cdk9 T-loop to activate the transition into elongation
The release of P-TEFb from the promoter-assembled 7SK snRNP, upon dephosphorylation of the Cdk9 T-loop, indicates that Tat might capture dephosphorylated P-TEFb prior to the assembly of Tat:TAR:P-TEFb elongation complexes (Figure 1A). To test whether the phosphorylation state of the Cdk9 T-loop plays any role in Tat binding, we purified P-TEFb and obtained homogeneous T-loop phosphorylated and unphosphorylated kinase preparations that were incubated with Tat in an in vitro binding assay. We observed that Tat binds both P-TEFb forms in vitro (Figure 4A), demonstrating that Tat can capture dephosphorylated P-TEFb upon its release from the promoter-bound 7SK snRNP.
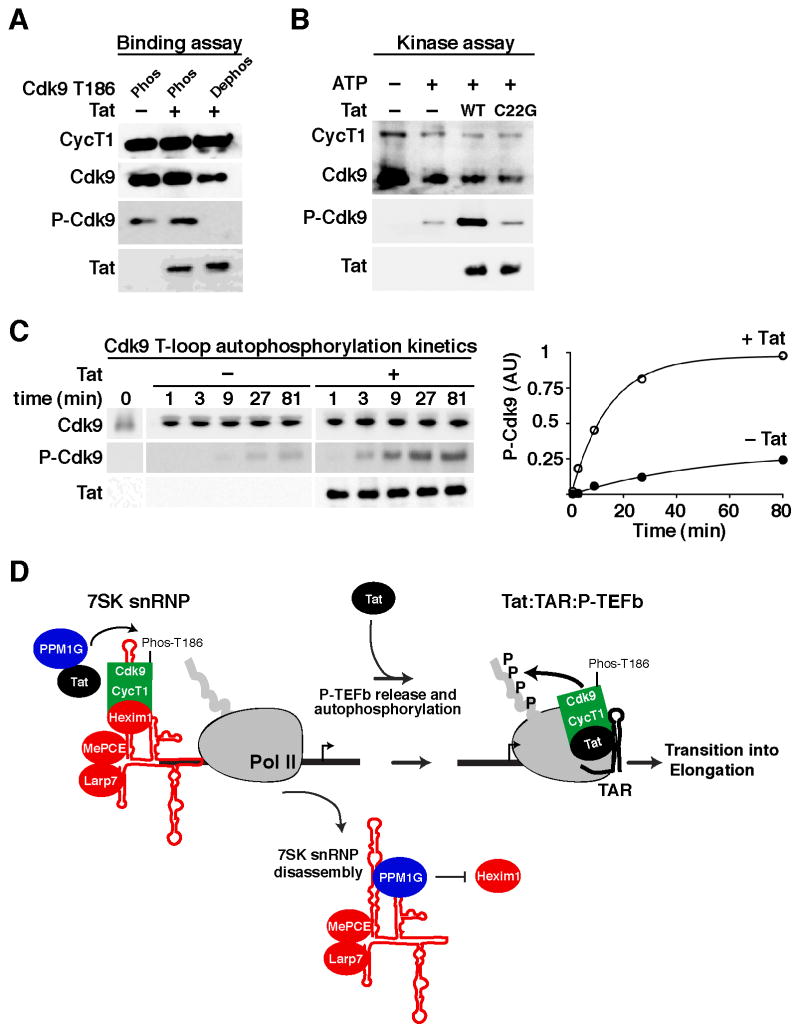
Figure 4. Tat binds dephosphorylated P-TEFb and stimulates Cdk9 T-loop phosphorylation to activate the transition into elongation.
(A) P-TEFb containing a phosphorylated (Phos) or dephosphorylated (Dephos) Cdk9 T-loop (T186) was incubated with Tat. Cdk9 was subsequently purified using Flag beads to monitor for Tat binding by Western blot.
(B) Dephosphorylated P-TEFb was incubated with or without ATP, or with ATP along with wild-type (WT) Tat or a non-functional C22G mutant in kinase conditions and analyzed by Western blot.
(C) Kinetics of Cdk9 T-loop autophosphorylation. Left panel, dephosphorylated P-TEFb was incubated with or without Tat in kinase conditions. Fractions were removed and assayed for Cdk9 T-loop phosphorylation (P-Cdk9). Right panel, levels of P-Cdk9 standardized to total Cdk9 were expressed in arbitrary units (AU).
(D) Model of Tat-mediated enzymatic release of P-TEFb from the 7SK snRNP. Tat recruits PPM1G to the HIV promoter-bound 7SK snRNP. PPM1G dephosphorylates the T-loop of Cdk9 (Phos-T186) to disassemble the 7SK snRNP and release P-TEFb. Released P-TEFb is captured by Tat, which stimulates Cdk9 T-loop autophosphorylation to form transcription elongation competent complexes on TAR. See also Figure S4.
Because Cdk9 T-loop dephosphorylation renders an inactive kinase, we hypothesized that, by binding dephosphorylated P-TEFb, Tat can stimulate autophosphorylation of the Cdk9 T-loop. To test this possibility, we incubated dephosphorylated P-TEFb in kinase buffer with ATP: (i) alone, (ii) with Tat or (iii) with a non-functional C22G mutant, and measured levels of Cdk9 autophosphorylation at Thr186. Interestingly, Tat, but not the C22G mutant, stimulates (~5-fold) Cdk9 autophosphorylation in vitro in conditions where the kinase activity is in the linear range (Figures 4B–C).
Collectively, we propose an enzymatic model of P-TEFb transfer from the promoter-assembled 7SK snRNP to TAR during HIV transcription activation (Figure 4D). In a first step, Tat recruits PPM1G to the promoter to dephosphorylate the T-loop of Cdk9 and release P-TEFb from the 7SK snRNP. In a second step, Tat binds dephosphorylated P-TEFb to stimulate autophosphorylation of the Cdk9 T-loop and transfers the locally released kinase to TAR to assemble transcription elongation complexes. This enzymatic mechanism of 7SK snRNP disassembly takes into consideration the low levels of Tat during infection and is favored over the previously proposed competition model where excess Tat physically captures P-TEFb from the 7SK snRNP (Barboric et al., 2007; D’Orso and Frankel, 2010; Krueger et al., 2010; Sedore et al., 2007). In fact, we found that the competitive release of P-TEFb by Tat occurs primarily due to protein re-assortment in vitro, but not in cells, since Tat and P-TEFb ectopically expressed in separate plates can efficiently associate leading to Hexim1 displacement (Figure S4).
PPM1G is a transcriptional co-activator of NF-κB in the inflammatory pathway
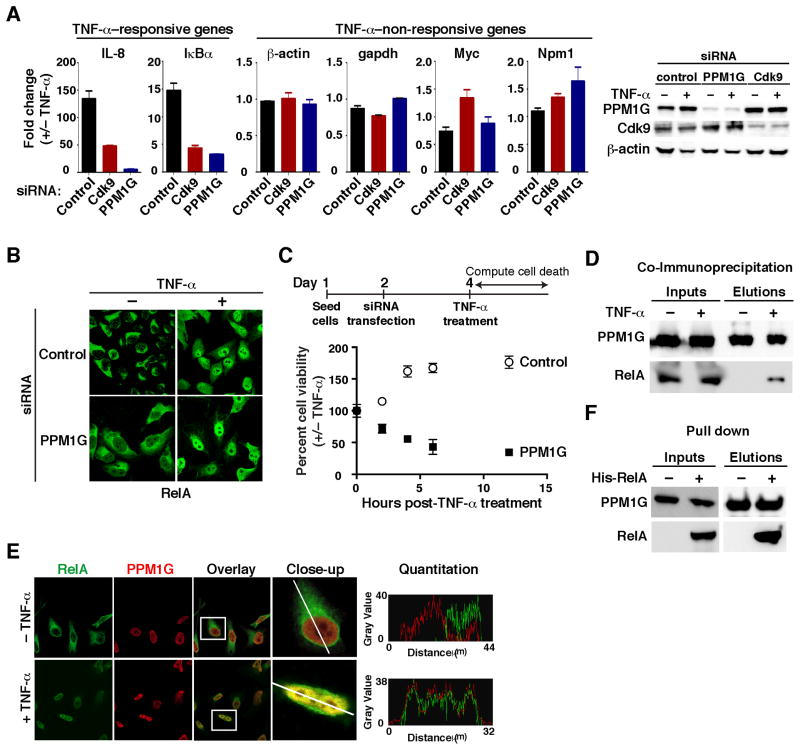
Since P-TEFb is a general elongation factor, we asked whether PPM1G is required in the context of transcriptional programs activated during cellular responses. We examined the inflammatory pathway where the pro-inflammatory cytokine tumor necrosis factor-alpha (TNF-α) triggers signaling cascades that converge on the activation of the transcription factor NF-κB (Ghosh and Karin, 2002). Upon TNF-α stimulation, NF-κB (RelA) rapidly translocates from the cytoplasm into the nucleus to activate a set of inflammatory-responsive genes in a P-TEFb–dependent manner (Barboric et al., 2001; Nissen and Yamamoto, 2000). Interestingly, PPM1G knockdown sharply downregulates (~7–15-fold) the induction of two RelA target genes (IL-8 and IκBα) upon TNF-α treatment, without affecting several non-responsivegenes (Figure 5A). Although PPM1G knockdown virtually abolishes the activation of inflammatory-responsive genes, it does not alter RelA translocation to the nucleus upon TNF-α treatment (Figure 5B), implying that PPM1G functions as a nuclear regulator of NF-κB activity. To rule out possible RNAi off-target effects, we used two siRNAs to target a non-coding region in PPM1G and observed that subsequent transfection of a siRNA-resistant PPM1G construct restored the stimulus-dependent activation of inflammatory-responsive genes (Figure S5). These results suggest that PPM1G is an essential nuclear co-factor of NF-κB for the normal activation of the inflammatory transcriptional program.
Figure 5. PPM1G is a nuclear transcriptional co-activator of NF-κB in the inflammatory pathway.
(A) HeLa cells were transfected with the indicated siRNAs and treated with or without TNF-α. Left panel, the expression of TNF-α–responsive and –non-responsive genes (normalized to Rpl19) was measured by RT-qPCR and plotted as fold change +/−TNF-α (Mean ± SEM are shown, n=3). Right panel, Western blots showing the validation of the knockdown.
(B) Knockdown of PPM1G in HeLa cells does not alter RelA translocation to the nucleus upon TNF-α treatment.
(C) PPM1G knockdown sensitizes cells to TNF-α–induced apoptosis. HeLa cells were transfected with the indicated siRNAs. At 48 hrs post-transfection, cells were treated with or without TNF-α and incubated for the indicated time points. The plot shows percentage of viable cells and is expressed as fold difference +/−TNF-α (Mean ± SEM are shown, n=3).
(D) PPM1G and RelA interact in cells. 293T cells were transfected with a Strep-tagged PPM1G plasmid and cells were treated with or without TNF-α, crosslinked with formaldehyde and lysed under denaturing conditions. Strep AP was performed to analyze the interaction between PPM1G and RelA.
(E) RelA and PPM1G co-localize in a stimulus-dependent manner in the cell nuclei. HeLa cells were treated with or without TNF-α and confocal images are shown. A single cross-sectional image was acquired (line) and emission intensities were quantified to analyze the relative co-localization along the path indicated (histograms).
(F) RelA and PPM1G directly bind. Bacterially synthesized RelA was incubated with Strep-tagged PPM1G and the final elution was analyzed by Western blot. See also Figure S5.
Upon stimulation with pro-inflammatory cytokines, NF-κB induces the transcription of a number of anti-apoptotic genes (Barkett and Gilmore, 1999; Ghosh and Karin, 2002; Sidoti-de Fraisse et al., 1998). To investigate the biological significance of PPM1G in these processes, we analyzed its involvement in the apoptotic response regulated by NF-κB (Figure 5C). To this end, we transfected HeLa cells with control and PPM1G siRNAs and, two days later, the cells were treated with or without TNF-α. We assayed for cell death at different time intervals post-treatment and then computed a cumulative cell death index by plotting the effect of TNF-α on cell viability. Treatment with TNF-α was not toxic to cells transfected with control siRNA; however, PPM1G knockdown drastically sensitized cells to TNF-α–induced apoptosis (Figure 5C). This suggests that NF-κB requires PPM1G, in addition to P-TEFb (Barboric et al., 2001), to stimulate transcription elongation of pro-survival genes to prevent apoptosis induced by pro-inflammatory cytokines.
Given the fact that Tat physically recruits PPM1G to stimulate HIV transcription and that PPM1G is required for activation of inflammatory-responsive genes, we asked whether RelA binds PPM1G. To test this, we crosslinked cells after TNF-α treatment to eliminate any possible in vitro protein reassortment during the affinity purification step. Interestingly, we found that transfected PPM1G stably associates with endogenous RelA in a stimulus-dependent manner (Figure 5D). Supporting this result, endogenous PPM1G and RelA proteins colocalize in the nucleus upon TNF-α treatment (Figure 5E) and bacterially synthesized RelA directly binds PPM1G in vitro (Figure 5F). Taken together, these results functionally link PPM1G and NF-κB during activation of the inflammatory transcriptional program.
Recruitment of PPM1G by NF-κB to the promoter-assembled 7SK snRNP mediates the activation of inflammatory-responsive genes
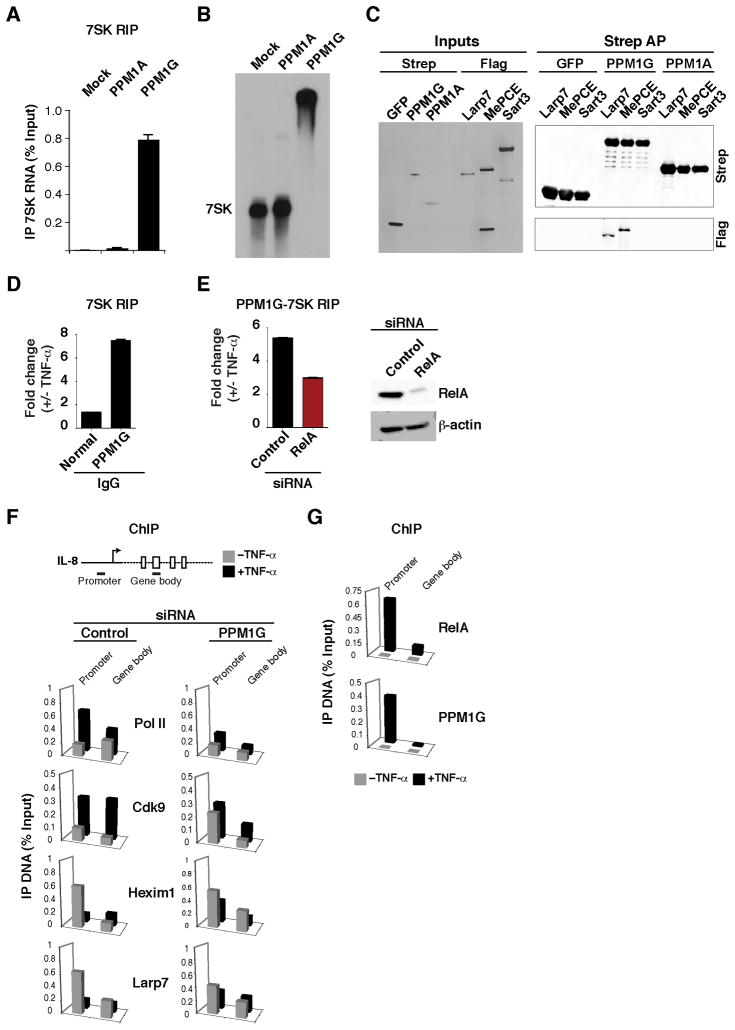
The idea that PPM1G co-purifies with the 7SK snRNP prompted us to characterize the interaction of the enzyme with the RNA and core snRNP components. To examine the interaction with 7SK RNA, we performed a RIP assay from cells transfected with Strep-tagged PPM1G and PPM1A (Figure 6A). Notably, we found that PPM1G, but not PPM1A, associates with 7SK in cells, but not with U6 snRNA (data not shown), demonstrating binding specificity. To test whether PPM1G directly associates with 7SK RNA, we purified recombinant PPM1G and PPM1A proteins from bacteria and incubated them with a radiolabeled 7SK RNA. Interestingly, PPM1G, but not PPM1A, directly binds 7SK RNA in in vitro gel shift assays (Figure 6B). In addition to binding 7SK RNA, we observed that PPM1G, but not PPM1A, directly interacts (in the absence of 7SK RNA) with MePCE and Larp7 core snRNP components in vitro (Figure 6C), demonstrating that PPM1G can make multiple contacts with the 7SK snRNP complex. In support of the physical interaction data, we observed that knockdown of PPM1G, but not PPM1A, affects transcription of inflammatory-responsive genes by NF-κB (Figure 5A and data not shown).
Figure 6. PPM1G binds the 7SK snRNP and participates in the NF-κB–mediated disassembly of the promoter-assembled snRNP at inflammatory-responsive genes.
(A) HeLa cells were transfected with an empty vector (Mock) or Strep-tagged PPM1A or PPM1G plasmids and levels of 7SK RNA co-immunoprecipitating were quantified by RT-qPCR (Mean ± SEM are shown, n=3).
(B) PPM1G binds 7SK RNA. Gel shift assay between in vitro synthesized 7SK RNA and His-tagged, bacterial PPM1A or PPM1G. Mock denotes incubation of 7SK RNA in binding buffer with a purification from bacterial cells transformed with an empty vector.
(C) PPM1G binds 7SK snRNP core components. Left panel shows a silver stained gel of input proteins purified from 293T in the presence of RNase A. The Flag-tagged proteins were incubated with GFP, PPM1G or PPM1A bound to Strep beads. Right panel shows Western blots of the Strep AP.
(D) Stimulus-dependent recruitment of PPM1G to the 7SK RNA. HeLa cells were treated with or without TNF-α and endogenous PPM1G was immunoprecipitated using a specific serum and levels of PPM1G-bound 7SK RNA were quantified by RT-qPCR. Normal IgG was used as negative control (Mean ± SEM are shown, n=4).
(E) PPM1G-7SK RIP. Left panel, HeLa cells were transfected with control or RelA siRNAs and treated with or without TNF-α and a RIP assay was performed to quantify the amount of endogenous PPM1G-bound 7SK RNA by RT-qPCR (Mean ± SEM are shown, n=3). Right panel Western blots to validate the knockdown.
(F) ChIP assay to analyze the distribution of Pol II and P-TEFb-7SK snRNP subunits at the IL-8 locus. HeLa cells were transfected with a control or PPM1G siRNA and treated with (black bars) or without (grey bars) TNF-α. Values represent the average of three independent experiments. The SEM is less than 5% and not shown for simplicity.
(G) ChIP assay to analyze the distribution of RelA and PPM1G at the IL-8 locus in HeLa cells treated with (black bars) or without (grey bars) TNF-α. Values represent the average of four independent experiments. The SEM is less than 5% and not shown for simplicity. See also Figure S6.
All aspects of transcription and its regulation involve dynamic events where regulatory components are recruited to their target genes (Hager et al., 2009). Thus, although PPM1G has RNA-binding activity per se, we examined the possibility of a stimulus-dependent PPM1G-7SK RNA interaction upon activation of the inflammatory transcriptional program. Using a RIP assay, we observed that treatment of HeLa cells with TNF-α induces the recruitment of endogenous PPM1G to 7SK RNA (Figure 6D).
To test whether there is a functional link between RelA and the recruitment of PPM1G to the 7SK RNA, we used RNAi to knockdown RelA and compared the levels of PPM1G-7SK RNA association with and without TNF-α using a RIP assay (Figure 6E). Intriguingly, knockdown of RelA reduced (by a factor of ~2-fold) the PPM1G-7SK RNA association in the presence of TNF-α, which correlates with the decreased expression of RelA target genes (data not shown). Thus, RelA, in addition to binding its target genes, plays a key role in the stimulus-dependent recruitment of PPM1G to the 7SK RNA to activate transcription.
To further define the role of PPM1G during activation of the inflammatory transcriptional program, we knocked down PPM1G from cells treated with or without TNF-α and examined Pol II and co-factor recruitment by ChIP at the IL-8 locus (Figure 6F). PPM1G knockdown reduces (~2–4-fold) the TNF-α–mediated increase of Pol II and P-TEFb occupancy in the gene body, which correlates with reduced gene expression (Figure 5A). Interestingly, similar to the HIV promoter, we observed that the inhibitory 7SK snRNP components (Larp7 and Hexim1) are recruited to the promoter and TNF-α treatment promotes their eviction. This TNF-α–mediated ejection of the 7SK snRNP relies on PPM1G since knockdown of the enzyme resulted in retention of both Hexim1 and Larp7 at the IL-8 promoter.
To test the model that PPM1G is required to directly activate transcriptional pause release through eviction of the 7SK snRNP at the IL-8 locus, we examined PPM1G recruitment by ChIP (Figure 6G). Interestingly, we observed that PPM1G is recruited to the IL-8 promoter, but not to the gene body, upon TNF-α stimulation, mirroring the occupancy profile of RelA. This result demonstrates that the recruitment of PPM1G by RelA to the IL-8 promoter is inducible and argues a role for PPM1G in activating the transition into elongation.
Because P-TEFb is a general elongation factor, we examined the requirement of PPM1G for several Pol II transcribed genes. As has been previously reported, Cdk9 knockdown antagonized the activation of heat shock-responsive genes (Lis et al., 2000); however, knockdown of PPM1G had no significant impact on activation of Hsp70 and Hsp90 upon heat shock stress (Figure S6), thus demonstrating that PPM1G is a regulator of selective cellular transcriptional programs.
DISCUSSION
The compartmentalization of factors enables rapid coordination of molecular reactions (Francastel et al., 2000; Hager et al., 2009). Transcription is one example of molecular crowding where a paused Pol II, negative elongation factors and co-activators are collectively assembled at gene promoters to facilitate rapid gene expression. P-TEFb is a pleiotropic co-activator of gene expression initially thought to be recruited to promoters upon gene activation. Recent studies, however, revealed a close association between P-TEFb as part of the inhibitory 7SK snRNP at gene promoters and this association is positively correlated with the degree to which Pol II pauses to prevent spurious transcription elongation (Barboric and Lenasi, 2010; D’Orso and Frankel, 2010; Ji et al., 2013). Thus, the 7SK snRNP complex is not simply a reservoir of inactive P-TEFb that roams the nucleus, but rather keeps a primed P-TEFb kinase at promoters of inducible genes.
Our findings elucidate a functional link between the recruitment of PPM1G by transcription factors and the release of P-TEFb from the promoter-assembled 7SK snRNP during gene activation. PPM1G dephosphorylates the Cdk9 T-loop to enzymatically disassemble the snRNP during activation of the HIV and cellular transcriptional programs by Tat and NF-κB, respectively. Other cellular phosphatases have been previously described to dephosphorylate P-TEFb in vitro and to dissociate Hexim1 from P-TEFb in response to signal-dependent activation (Ammosova et al., 2005; Chen et al., 2008; Wang et al., 2008); however, the mechanisms of transcription activation in vivo are poorly understood. Here we demonstrate that Tat and NF-κB recruit PPM1G to the promoter-assembled 7SK snRNP to release P-TEFb from the snRNP providing a functional link between Cdk9 dephosphorylation and activation of specific elongation programs.
At the HIV promoter, the Tat-TAR interaction plays a key role in the disassembly of the 7SK snRNP in vivo, and thus the nascent RNA might coordinate the recruitment of the Tat-PPM1G complex to the HIV promoter to release P-TEFb from the 7SK snRNP. This could explain why TAR deletion impairs the eviction of the 7SK snRNP even in the presence of Tat (D’Orso and Frankel, 2010). Our results are consistent with a model whereby upon disassembly of the 7SK snRNP, Tat captures released P-TEFb in an unphosphorylated state and enhances the rate of Cdk9 T-loop autophosphorylation to induce a permissive kinase state for TAR binding. Several previous observations are consistent with this model of activation: (i) Tat contacts the Cdk9 T-loop in the Tat–P-TEFb structure (Tahirov et al., 2010), and (ii) phosphorylation of the T-loop is critical for the folding and autophosphorylation of its CTD, a prerequisite for the assembly of Tat:TAR:P-TEFb (Baumli et al., 2012; Garber et al., 2000). Thus, our results suggest that the local transfer of P-TEFb from 7SK to TAR at the viral promoter follows a stepwise Cdk9 dephosphorylation and rephosphorylation process.
Similar to Tat, we show that NF-κB utilizes P-TEFb (Barboric et al., 2001; Hargreaves et al., 2009; Luecke and Yamamoto, 2005) and PPM1G during activation of the inflammatory transcriptional program. The inducible recruitment of PPM1G by NF-κB to promoters of inflammatory-responsive genes disassembles the 7SK snRNP and releases P-TEFb for transcription elongation. We provide evidence that knockdown of PPM1G impairs disassembly of the snRNP, thereby decreasing the transcription of inflammatory-responsive genes and sensitizing cells to cytokine-induced apoptosis. Interestingly, the inducible disassembly of the 7SK snRNP at the IL-8 gene mimics the Tat-mediated snRNP eviction at the HIV promoter during the switch to elongation (D’Orso and Frankel, 2010). Although NF-κB operates in a TAR-independent manner, binding of NF-κB to its responsive elements, and cooperation with RNA-processing factors that bind the nascent RNA (Ji et al., 2013) might play an analogous role to Tat. At difference to the IL-8 gene, which requires P-TEFb to be induced during TNF-α signaling, the IκBα gene appears to be activated in a P-TEFb-independent manner in lung- and kidney-derived cell lines (Amir-Zilberstein et al., 2007; Luecke and Yamamoto, 2005). Because our assay was performed in HeLa, it is unclear whether the discrepancy in the requirement of P-TEFb is due to cell-type specific differences.
Interestingly, PPM1G assembles into the 7SK snRNP in a reversible- and stimulus-dependent manner through direct contacts with both the RNA and core snRNP components. Because PPM1G binds the snRNP tightly, in the absence of P-TEFb and Hexim1, the enzyme might also block the reassociation of P-TEFb–Hexim1 onto the 7SK RNA, thereby maintaining a local pool of free P-TEFb (Barboric et al., 2007) to sustain multiple rounds of elongation. When Tat levels or the inflammatory stimuli are decreased, PPM1G could dissociate from the 7SK snRNP, thereby allowing for the recruitment of P-TEFb and Hexim1 into the snRNP to reset the system to the basal, repressed state. Thus, we propose that the reversible nature of the PPM1G-7SK snRNP interaction facilitates both the initiation and termination of the elongation programs.
In summary, we report that the primary function of the promoter-assembled 7SK snRNP is to keep a primed P-TEFb kinase. We thus argue that the 7SK snRNP, along with paused Pol II, should be viewed as a mechanism for rapid and inducible gene expression through transcriptional pause release. The regulation of transcription elongation by the P-TEFb kinase has been extensively studied over the past decade (Zhou et al., 2012), with much of our current understanding owed to the HIV system (Ott et al., 2011). Once again, using Tat we discovered an unprecedented role for PPM1G as a transcriptional regulator of P-TEFb during cellular responses, a pathway that HIV exploits for infection of the host cell.
EXPERIMENTAL PROCEDURES
Cell culture
HeLa, HeLa-LTR (D’Orso and Frankel, 2010), and 293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) at 37°C with 5% CO2.
Plasmids
The plasmids used in this study are listed in Table S2. Site directed mutants were made using 100 ng of the indicated plasmid, 5′-phosphorylated oligonucleotides, and Phusion High-Fidelity PCR master mix (NEB).
Transcription reporter assays
For the reporter assays, HeLa cells were transfected using Polyjet (SignaGen) with 25 ng of a firefly luciferase (FFL) reporter plasmid, 1 ng of a cytomegalovirus (CMV)-Renilla (RL) luciferase plasmid and 1 ng of a Tat-expressing plasmid as described (D’Orso et al., 2012). Reporter activities are presented as fold activation relative to reporter alone and normalized to RL. For the RNAi assays, 24 hrs post-siRNA transfection, cells were retransfected with a Tat plasmid and luciferase levels were measured using the Dual-Luciferase Reporter Assay (Promega) on a FLUOStar Optima 96-well plate reader (BMG Labtech).
RNAi assays
HeLa cells were transfected with the indicated siRNAs (Table S3) at a final concentration of 100 μM using Lipofectamine 2000 (Invitrogen). Knockdown efficiency was evaluated by RT-qPCR (Table S4) and Western blot (Table S5).
RNA extraction and reverse transcription quantitative (RT)-PCR
Total RNA was isolated using TRIzol (Invitrogen) and RNeasy plus spin columns (Qiagen). First-strand cDNA was synthesized using M-MuLV Reverse Transcriptase (NEB) and quantitative PCR was performed with a SybrGreen master mix on an ABI7500 instrument. Ct values and fold change were calculated as described (Schmittgen and Livak, 2008).
Elongation assay
Total RNA was extracted from the HeLa-LTR cell line transfected with the indicated siRNAs. Transcripts were analyzed by RT-PCR as described (D’Orso et al., 2012).
ChIP assays
ChIP assays were performed as previously described (D’Orso and Frankel, 2010). The HeLa-LTR cell line was sequentially transfected with siRNAs (day 1) and a Tat:Flag plasmid (day 2) and proteins lysates were prepared (day 3). For ChIP at the IL-8 locus, HeLa cells were treated with or without TNF-α.
Tandem affinity purification and mass spectrometry
293T cells were transfected with Tat:Strep and Larp7:Flag plasmids. Tat complexes were sequentially purified using Strep-tactin resin (IBA Life Sciences) and Flag beads (Sigma). Gel slices and in-solution samples were analyzed by LC-MS/MS in the UTSW Proteomics Core.
Immunoprecipitations
Endogenous proteins were immunoprecipitated with the indicated antibodies conjugated to Protein G dynabeads (Invitrogen) from a HeLa whole cell extract as previously described (D’Orso et al., 2012). Isolation of co-purifying RNAs from immunoprecipitation material was performed as described (D’Orso et al., 2012).
In vitro phosphatase assay
In vitro dephosphorylation of P-TEFb (0.5 μg) was carried out in dephosphorylation buffer (20 mM Tris-HCl pH = 7.4, 150 mM NaCl, 5 mM Imidazole, 10 mM MnCl2, and 1 mM DTT) at 30°C for 2 hrs with 1 μg of the indicated phosphatase. Reactions were stopped by the addition of 2.5 mM EDTA and boiled at 90°C for 5 min in laemmli buffer. For the release assay, dephosphorylation reactions were diluted in half in buffer (20 mM Tris-HCl pH = 7.4, 150 mM NaCl, 0.05 % NP-40) and incubated with Flag beads overnight. The reaction products were electrophoresed on a SDS-PAGE gel and analyzed by Western blot.
P-TEFb dephosphorylation and Cdk9 T-loop phosphorylation assay
P-TEFb (CycT1:Strep + Cdk9:Flag) was purified using Strep-tactin resin from 293T cells. Cdk9 was dephosphorylated with PPM1G and P-TEFb was subsequently purified beads. Dephosphorylated P-TEFb was then incubated with bacterially synthesized Strep-tagged Tat or a C22G mutant in kinase buffer (10 mM Tris-HCl pH = 7.0, 150 mM KCl, 0.1 mg/ml BSA, 1.5 mM MgCl2, 0.5 mM ATP) at 30 C for the indicated time points. Kinase reactions were stopped with the addition of laemmli buffer and boiled for 10 min at 90 C. Quantification of relative phosphorylation intensities was done using Image J.
Protein expression and purification from bacterial cells
PPM1A, PPM1G and RelA were cloned into the pET30a vector (Novagen). BL21 DE3 cells were grown in LB medium and induced for protein expression with 1 mM at 30°C for 4 hrs. Cells were harvested by centrifugation and resuspended in lysis buffer (20 mM Tris-HCl pH = 7.5, 150 mM NaCl, 20 mM Imidazole, 0.2 mM EDTA, 1 mM PMSF, 2 mM MnCl2, 1 mM DTT, 5% glycerol and 1% NP-40). Lysis was carried out by sonication and clarified by centrifugation at 10,000g. The clarified supernatant was loaded onto a Ni-NTA affinity column (Qiagen), washed in buffer (20 mM Tris-HCl pH = 7.5, 250 mM NaCl, 20 mM Imidazole, 0.2 mM EDTA, 2 mM MnCl2, 1 mM DTT, 5% glycerol and 0.1% NP-40) and eluted with Imidazole. Recombinant HIV Tat was expressed as an N-terminal GST (D’Orso and Frankel, 2010), or C-terminal Strep, fusion.
In vitro transcription and RNA binding gel shift assays
7SK RNA was produced by in vitro run off transcription with T7 RNA polymerase (NEB). The transcription reaction mixture contained 0.35 μmol of [α-32P]-UTP 3000Ci/mmol (Perkin Elmer) along with non-radiolabelled nucleotides (500 μM ATP/CTP/GTP and 100 μM UTP). RNAs were purified on a 5% polyacrylamide/1X Tris-Glycine gel and eluted overnight at 4°C in elution buffer (0.6 M NaOAc pH = 6.0, 1 mM EDTA, and 0.01% SDS), ethanol precipitated, and resuspended in water. Protein and RNA were incubated for 20 min at 4°C in 10 μl binding buffer (10 mM HEPES-KOH pH = 7.5, 100 mM KCl, 1 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 10% glycerol, and 50 μg/ml yeast tRNA). Protein-RNA complexes were resolved on a 5% polyacrylamide/1X Tris-Glycine gel at 4°C.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde and permeabilized in 0.5% Triton X-100. Coverslips were blocked at 4°C for 1 hr in 1X PBS buffer containing 4% BSA and 5% normal goat serum. Primary antibodies used were: rabbit anti-RelA and mouse anti-PPM1G. Secondary antibodies were: goat anti-mouse Alexa-Fluor 546 and goat anti-rabbit Alexa-Fluor 488 (Invitrogen). Images were acquired on a Zeiss LSM 5 Pascal confocal microscope and analyzed on Image J.
Quantitation of TNF-α–induced apoptosis
HeLa cells were seeded in 6-well plates and transfected with the indicated siRNAs. At 48 hrs post-transfection, cells were treated with TNF-α and apoptosis was monitored over time through picture images (n=3). The graph was plotted by counting the cells within a defined area (3 images per time point (cell counts were averaged)). The percent of viable cells expressed as fold change +/− TNF-α was calculated.
De novo modeling and structure alignment
PPM1G structure prediction was done using the Robetta server (Kim et al., 2004). Structural similarities were identified after superimposition of the model into the crystal structures using MAMMOTH. Three PPM1G domains (N-terminal, acidic, and C-terminal) were modeled using four different structures. The N-terminal (residues 1–140) was modeled using the Ser/Thr phosphatase 2C (PDB 2i0O, chain A). Two regions in the acidic-domain (residues 141–231 and 232–324) were modeled using haemagglutinin esterase fusion protein (PDB 1FLC, chain B) and Contractile protein (PDB 1c1g, chain A) structures. The C-terminal (residues 325–546) was modeled using PPM1A structure (PDB 1A6Q, chain A). The four models were assembled into a full-length model packed using the Monte-Carlo algorithm with a backbone-dependent side-chain rotamer library. The predicted PPM1G structure (without the acidic domain) and PPM1A structure (PDB 1A6Q) (Das et al., 1996) were aligned and superimposed using the Matchmaker option within the UCSF Chimera molecular modeling package (Pettersen et al., 2004).
Supplementary Material
Highlights.
The 7SK snRNP complex keeps a primed P-TEFb kinase at promoters of inducible genes
PPM1G is a co-activator of viral (HIV Tat) and cellular (NF-κB) transcription factors
Transcription factors recruit PPM1G to the promoter-bound 7SK snRNP
PPM1G dephosphorylates the T-loop of the P-TEFb kinase to disassemble the 7SK snRNP
Acknowledgments
We thank N. Alto, M. Cobb, C.-M. Chiang, N. Conrad, E. Martinez and J. Espinosa for their comments. We are grateful to E. Burstein for the RelA plasmid and the AIDS Research and Reference Reagent Program for reagents. This work was supported by NIH R00AI083087 and The Welch Foundation I-1782 grants to I.D. and NIH training grant T32 GM008203 to R.P.M.
Footnotes
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allemand E, Hastings ML, Murray MV, Myers MP, Krainer AR. Alternative splicing regulation by interaction of phosphatase PP2Cγ with nucleic acid-binding protein YB-1. Nat Struct Mol Biol. 2007;14:630–638. doi: 10.1038/nsmb1257. [DOI] [PubMed] [Google Scholar]
- Amir-Zilberstein L, Ainbinder E, Toube L, Yamaguchi Y, Handa H, Dikstein R. Differential regulation of NF-κB by elongation factors is determined by core promoter type. Mol Cell Biol. 2007;27:5246–5259. doi: 10.1128/MCB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammosova T, Washington K, Debebe Z, Brady J, Nekhai S. Dephosphorylation of CDK9 by protein phosphatase 2A and protein phosphatase-1 in Tat-activated HIV-1 transcription. Retrovirology. 2005;2:47. doi: 10.1186/1742-4690-2-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboric M, Lenasi T. Kick-sTARting HIV-1 transcription elongation by 7SK snRNP deporTATion. Nat Struct Mol Biol. 2010;17:928–930. doi: 10.1038/nsmb0810-928. [DOI] [PubMed] [Google Scholar]
- Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-κB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- Barboric M, Yik JH, Czudnochowski N, Yang Z, Chen R, Contreras X, Geyer M, Matija Peterlin B, Zhou Q. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res. 2007;35:2003–2012. doi: 10.1093/nar/gkm063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkett M, Gilmore TD. Control of apoptosis by Rel/ NF-κB transcription factors. Oncogene. 1999;18:6910–6924. doi: 10.1038/sj.onc.1203238. [DOI] [PubMed] [Google Scholar]
- Baumli S, Hole AJ, Wang LZ, Noble ME, Endicott JA. The CDK9 tail determines the reaction pathway of P-TEFb. Structure. 2012;20:1788–1795. doi: 10.1016/j.str.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Liu M, Li H, Xue Y, Ramey WN, He N, Ai N, Luo H, Zhu Y, Zhou N, et al. PP2B and PP1α cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev. 2008;22:1356–1368. doi: 10.1101/gad.1636008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Yang Z, Zhou Q. P-TEFb is tagged for inhibition through association with 7SK snRNA. J Biol Chem. 2004;279:4153–4160. doi: 10.1074/jbc.M310044200. [DOI] [PubMed] [Google Scholar]
- Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Orso I, Frankel AD. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat Struct Mol Biol. 2010;17:815–821. doi: 10.1038/nsmb.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Orso I, Jang GM, Pastuszak AW, Faust TB, Quezada E, Booth DS, Frankel AD. Transition Step during Assembly of HIV Tat:P-TEFb Transcription Complexes and Transfer to TAR RNA. Mol Cell Biol. 2012;32:4780–4793. doi: 10.1128/MCB.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das AK, Helps NR, Cohen PT, Barford D. Crystal structure of the protein serine/threonine phosphatase 2C at 2.0Å resolution. EMBO J. 1996;15:6798–6809. [PMC free article] [PubMed] [Google Scholar]
- Faust T, Frankel A, D’Orso I. Transcription control by long non-coding RNAs. Transcription. 2012;3:78–86. doi: 10.4161/trns.19349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francastel C, Schubeler D, Martin DI, Groudine M. Nuclear compartmentalization and gene activity. Nat Rev Mol Cell Biol. 2000;1:137–143. doi: 10.1038/35040083. [DOI] [PubMed] [Google Scholar]
- Garber ME, Mayall TP, Suess EM, Meisenhelder J, Thompson NE, Jones KA. CDK9 autophosphorylation regulates high-affinity binding of the HIV-1 Tat-P-TEFb complex to TAR RNA. Mol Cell Biol. 2000;20:6958–6969. doi: 10.1128/mcb.20.18.6958-6969.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Karin M. Missing pieces in the NF-κB puzzle. Cell. 2002;109:S81–96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager GL, McNally JG, Misteli T. Transcription dynamics. Mol Cell. 2009;35:741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Therien C, Bergeron D, Bourassa S, Greenblatt J, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Zhou Y, Pandit S, Huang J, Li H, Lin CY, Xiao R, Burge CB, Fu XD. SR Proteins Collaborate with 7SK and Promoter-Associated Nascent RNA to Release Paused Polymerase. Cell. 2013;153:855–868. doi: 10.1016/j.cell.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W526–531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Bourgeois CF, Isel C, Churcher MJ, Karn J. Phosphorylation of the RNA polymerase II carboxyl-terminal domain by CDK9 is directly responsible for human immunodeficiency virus type 1 Tat-activated transcriptional elongation. Mol Cell Biol. 2002;22:4622–4637. doi: 10.1128/MCB.22.13.4622-4637.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Mbonye U, Hokello J, Karn J. T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway. J Mol Biol. 2011;410:896–916. doi: 10.1016/j.jmb.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger BJ, Jeronimo C, Roy BB, Bouchard A, Barrandon C, Byers SA, Searcey CE, Cooper JJ, Bensaude O, Cohen EA, et al. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008;36:2219–2229. doi: 10.1093/nar/gkn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger BJ, Varzavand K, Cooper JJ, Price DH. The mechanism of release of P-TEFb and HEXIM1 from the 7SK snRNP by viral and cellular activators includes a conformational change in 7SK. PLoS One. 2010;5:e12335. doi: 10.1371/journal.pone.0012335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Price JP, Byers SA, Cheng D, Peng J, Price DH. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J Biol Chem. 2005;280:28819–28826. doi: 10.1074/jbc.M502712200. [DOI] [PubMed] [Google Scholar]
- Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- Luecke HF, Yamamoto KR. The glucocorticoid receptor blocks P-TEFb recruitment by NF-κB to effect promoter-specific transcriptional repression. Genes Dev. 2005;19:1116–1127. doi: 10.1101/gad.1297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancebo HS, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, et al. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Price DH. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol Cell Biol. 1992;12:2078–2090. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniz L, Egloff S, Ughy B, Jady BE, Kiss T. Controlling cellular P-TEFb activity by the HIV-1 transcriptional transactivator Tat. PLoS Pathog. 2010;6:e1001152. doi: 10.1371/journal.ppat.1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen RM, Yamamoto KR. The glucocorticoid receptor inhibits NF-κB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 2000;14:2314–2329. doi: 10.1101/gad.827900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M, Geyer M, Zhou Q. The control of HIV transcription: keeping RNA polymerase II on track. Cell Host Microbe. 2011;10:426–435. doi: 10.1016/j.chom.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Sedore SC, Byers SA, Biglione S, Price JP, Maury WJ, Price DH. Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucleic Acids Res. 2007;35:4347–4358. doi: 10.1093/nar/gkm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidoti-de Fraisse C, Rincheval V, Risler Y, Mignotte B, Vayssiere JL. TNF-α activates at least two apoptotic signaling cascades. Oncogene. 1998;17:1639–1651. doi: 10.1038/sj.onc.1202094. [DOI] [PubMed] [Google Scholar]
- Smith E, Shilatifard A. Transcriptional elongation checkpoint control in development and disease. Genes Dev. 2013;15:1079–1088. doi: 10.1101/gad.215137.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell. 2010;38:439–451. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirov TH, Babayeva ND, Varzavand K, Cooper JJ, Sedore SC, Price DH. Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature. 2010;465:747–751. doi: 10.1038/nature09131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dow EC, Liang YY, Ramakrishnan R, Liu H, Sung TL, Lin X, Rice AP. Phosphatase PPM1A regulates phosphorylation of Thr-186 in the Cdk9 T-loop. J Biol Chem. 2008;283:33578–33584. doi: 10.1074/jbc.M807495200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.