Abstract
Elucidation of the genetic pathways that control red blood cell development has been a central goal of erythropoiesis research over the past decade. Notably, data from several recent studies have provided new insights into the regulation of erythroid gene transcription. Transcription profiling demonstrates that erythopoiesis is mainly controlled by a small group of lineage-restricted transcription factors (Gata1, Tal1, and Klf1). Binding site mapping using ChIP-Seq indicates that most DNA bound Gata1 and Tal1 proteins are contained within higher order complexes (Ldb1-complexes) that include the nuclear adapters Ldb1 and Lmo2. Ldb1-complexes regulate Klf1 and Ldb1-complex binding sites frequently co-localize with Klf1 at erythroid genes and cis-regulatory elements indicating strong functional synergy between Gata1, Tal1, and Klf1. Together with new data demonstrating that Ldb1 can mediate long-range promoter/enhancer interactions, these findings provide a foundation for the first comprehensive models for the global regulation of erythroid gene transcription.
Keywords: Erythropoiesis, Gata1, Tal1, Klf1, Ldb1-complexes, Transcriptional regulation, ChIP-Seq
Gata1, Tal1, and Klf1: the core erythroid transcription factors
Mammalian erythropoiesis is a dynamic, stepwise process that begins in multipotent hematopoietic progenitors (hematopoietic stem cells, HSCs) and ends with the generation of mature enucleated red blood cells. Erythrocyte development requires the coordinated expression and activity of several transcription factors that regulate terminal differentiation and induction of erythroid–specific genes. Among these, Gata1, Tal1 (Scl), and Klf1 (EKLF) have been shown to be exceptional, earning the designation erythroid ‘master-regulators’ [see Box1]. Gata1, Tal1, and Klf1 are each required for both primitive and definitive erythropoiesis. Indeed, absence of any one of these proteins in mice results in severe anemia and death by mid-gestation [1-6]. In addition, gene expression profiling has shown that Gata1, Tal1, and Klf1 are each required for β-globin (Hbb) and α-globin (Hba) gene expression as well as for the induction of a large number of other erythroid signature genes suggesting that Gata1, Tal1, and Klf1 function broadly and synergistically to regulate the erythroid transcriptional program [7-12].
Box 1. The core erythroid transcription factors.
Gata1
Gata1 (Gata binding protein 1) was the first erythroid “master regulator” identified and remains the best-studied hematopoietic transcription factor [59]. Gata1 is the founding member of the Gata family of transcription factors that each contain two zinc fingers and bind to a canonical GATA DNA motif. Gata1 can directly associate with several co-factors such as the zinc finger protein Friend of Gata1 (Fog1) through its N-terminal zinc finger while making contact with DNA via the C-terminal zinc finger [60]. Gata1 also harbors an N-terminal transcriptional activation domain and has been shown to interact with multiple transcription factors, co-activators, and co-repressors and is subject to a complex array of post-translational modifications [61, 62]. Deletion of Gata1 in male mice (Gata1 is located on the X chromosome) results in death by E11.5 (embryonic day 11.5) from severe anemia, demonstrating that Gata1 is essential for erythropoiesis [63]. Gata1 is expressed at low levels in common myeloid progenitors (CMP), but is highly up-regulated in megakaryocyte-erythroid progenitors (MEP) and erythroblasts. Gata1 activates a set of early erythroid genes, such as the erythropoietin receptor at the onset of erythropoiesis to sustain the proliferation and differentiation of immature erythroid cells. As these cells mature, Gata1 also induces later stage eyrthroid genes such as those encoding the hemoglobin and heme bio-synthesis proteins. Gata1 also plays a critical role in the terminal maturation of other hematopoietic lineages including megakaryocytes, eosinophils, and mast cells.
Tal1/Scl
T cell acute lymphocytic leukemia 1 protein (Tal1 and also known as Scl) was discovered through its involvement in translocation events that give rise to T-cell acute lymphoblastic leukemia. Tal1 belongs to the family of basic helix-loop-helix (bHLH) transcription factors characterized by a 50-residue HLH protein interaction domain, preceded by a 10-residue basic region that binds DNA [64]. Tal1 forms heterodimers with ubiquitous bHLH factors, such as the E2a-proteins E12 and E47 as a pre-requisite to DNA binding at E-box (CANNTG) motifs. Follow-up loss-of-function studies demonstrated a critical role for Tal1 in early hematopoietic specification as Tal1 null mice die at E9.5 from lack of yolk sac hematopoiesis [3, 4]. Despite the requirement for Tal1 in hematopoietic stem cell generation, it is not specifically required for hematopoietic stem cell survival, multipotency, or long-term repopulating in mice due to functional redundancy with the related b-HLH protein, Lyl1 [65]. However, there are severe defects in erythroid and megakaryocytic development in the absence of Tal1 [2, 66]. Mice expressing a DNA-binding deficient form of Tal1 survive beyond the E9.5 time point when Tal1 deficient embryos die. These mutant mice displayed signs of anemia consistent with the idea that DNA binding is dispensable for HSC generation but is important for proper erythroid maturation [7].
Klf1/EKLF
Erythroid Kruppel-like factor (EKLF; henceforth referred to as Klf1) is the founding member of the KLF family of proteins that comprises 17 different transcription factors that function in diverse tissues and play many critical biological roles [67, 68]. The KLF family is characterized by three similar C2H2-type zinc fingers at the C-terminus that form a DNA binding domain that binds the consensus sequence CC[A/C]C[A/G]CCC. Klf1 also contains an N-terminal transactivation domain consisting of an acidic-patch and proline rich region. Klf1 is remarkably erythroid lineage restricted, being marginally expressed in CMPs, upregulated in their MEP progeny, and reaching peak expression levels at the mature erythroblast stage [10, 69]. Expression of Klf1 in megakaryocyte-erythroid progenitors restricts megakaryocyte development and promotes erythropoiesis [70, 71]. Klf1 function is critical for the proper development of the erythroid lineage as supported by knockout experiments in mice. Klf1 null mice die at E14-15 of gestation due to lack of proper definitive erythropoiesis [5, 6]. Although the role of Klf1 in regulating β-globin gene expression has been the major focus of its characterization, recent studies have confirmed Klf1 as one of the primary players in establishing and maintaining global erythroid gene regulation. Combined approaches using expression profiling and ChIP-Seq in fetal liver erythroid cells have demonstrated that Klf1 targets hundreds of genes and acts primarily as a transcriptional activator [10, 11].
Widespread binding of Gata1, Tal1, and Klf1 at erythroid genes and enhancers
Results from recent experiments in which chromatin immunoprecipitation coupled with massively parallel sequencing (ChIP-Seq) was used to map Gata1, Tal1, or Klf1 binding sites genome-wide in primary murine erythroblasts or in murine erythroid cell lines have provided important insights into the regulatory functions of these proteins in controlling the expression of erythroid genes [7-9, 12-16]. Each of these studies included transcriptional profiling by microarray or RNA-sequencing so that ChIP-Seq binding profiles could be correlated with gene expression. Although these experiments were performed by independent groups and, in many cases, with different erythroid cell populations, several general conclusions can nevertheless be drawn from comparative analysis of the data. First, Gata1, Tal1, and Klf1 each bind at or near, and are required for the induction of, a large cohort of erythroid genes including Hba and Hbb. Second, Gata1, Tal1, and Klf1 ChIP-Seq peaks strongly correlate with the presence of their respective consensus DNA binding motifs [see Box 1]. Third, although Gata1, Tal1, and Klf1 frequently bind near transcription start sites of erythroid genes, most binding sites are either within introns of target genes or at intergenic regions often far removed from any known gene (notably, some of these distal sites are within well characterized erythroid cis-regulatory elements including the Hbb locus control region (LCR) and the Hba MARE regulatory domain). Fourth, considerable Gata1, Tal1, and Klf1 binding site convergence can be inferred by the presence of consensus DNA binding motifs for one or both of the other two factors at Gata1, Tal1, or Klf1 ChIP-Seq sites. For example, Tal1 binding E-box (CANNTG) motifs are frequently detected near Gata1 ChIP-Seq binding sites, especially those that are at or near genes that are induced by Gata1 [13, 14]. Likewise, after E-boxes, GATA motifs were the most prevalent consensus sequences identified within Tal1 ChIP-Seq peaks [7].
Analysis of merged ChIP-Seq runs from independent studies confirmed frequent co-occupancy of binding sites by Gata1, Tal1, and Klf1. For example, comparison of Gata1 and Klf1 ChIP-seq results revealed striking binding site overlap with approximately 48% of all Klf1 ChIP-Seq peaks located within one kilobase (kb) of Gata1 binding sites [8]. In another study, the authors noted strong positive correlation of DNA occupancy by Gata1 and Tal1 over a 66 megabase region of chromosome 7 that includes a large number of key erythroid genes [9]. Finally, merged analysis of independently generated Gata1, Tal1, and Klf1 ChIP-Seq runs identified >300 genes that were co-occupied by all three factors [16].
In general, co-binding by more than one of the three transcription factors is associated with gene induction (positive regulation) rather than repression. Notably, several groups found that most Gata1 induced genes were co-occupied by both Gata1 and Tal1 [9, 12, 17, 18]. Similarly, co-binding by Gata1 and Klf1 was found to be strongly associated with gene activation [8, 10]. Collectively, these observations suggested that Gata1, Tal1, and Klf1 function together to positively regulate erythroid gene expression and establish erythroid lineage identity.
Ldb1-complexes are major instruments of Gata1 and Tal1 regulated erythroid gene activation
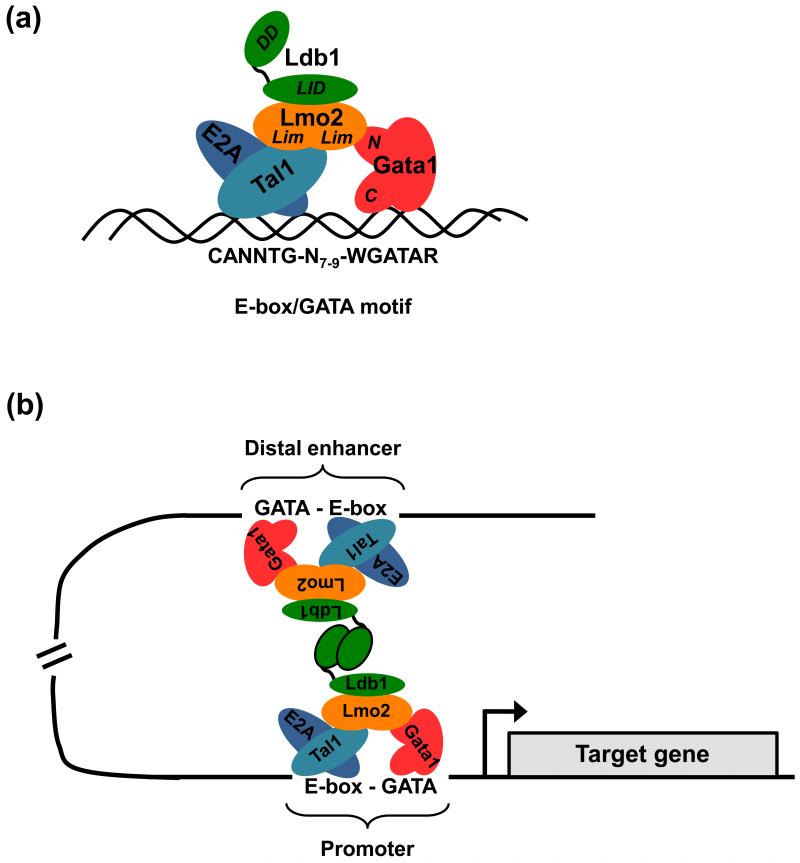
An intriguing observation from several of the aforementioned ChIP-Seq studies was the frequent detection of a paired E-box/GATA DNA motif within DNA fragments bound by either Gata1 [13], Tal1 [7], or Klf1 [8]. This paired motif, which consists of a preferentially ordered and spaced partial or complete E-box and a consensus Gata1 binding sequence [(CANN)TG-N7-9-WGATAR], matches the consensus binding site for a multimeric erythroid protein complex (herein designated Ldb1-complex) that contains Gata1 and Tal1 in addition to the nuclear adapter proteins LIM domain binding protein 1 (Ldb1) and LIM domain only 2 (Lmo2) [Fig. 1A] [19].
Figure 1. The structure and function of erythroid Ldb1-complexes.
(a) Model of the core erythropoietic Ldb1-complex. The zinc finger DNA binding protein Gata1 and a heterodimer of the basic helix-loop-helix (b-HLH) proteins Tal1 and E2a bind to a paired E-box—WGATAR motif (W is A/T; R is A/G) with a restricted spacing of 7-9 bp. The dual LIM (Lin11, Isl-1 & Mec-3) domain protein Lmo2 bridges and associates with both Gata1 and the bHLH factors while the LIM interacting protein Ldb1 associates with Lmo2. Gata1 and Tal1 are described in more detail in Box 1. Ldb1 is a transcription co-factor widely expressed throughout embryonic and adult tissues. Ldb1 has no known enzymatic or nucleic acid-binding function, but rather it seems to act as an interface for specific protein interactions [29]. Ldb1 achieves this through its two predominant functional domains: an N-terminal self-association (dimerization) domain and a C-terminal region that interacts with the LIM domain that is common to a large family of proteins that have important roles in tissue development. Lmo2 is a small protein composed of two LIM domains and is expressed in a variety of tissues including hematopoietic precursors as well as many but not all hematopoietic lineages. In vitro binding studies have shown that association of Lmo2 with Tal1 increases the affinity of Tal1 to bind E2a and in turn, more stably bind to the E-box sequence [75]. Lmo2 also binds to the N-terminal zinc finger of Gata1, thereby forming a bridge between Tal1 and Gata1 [45]. Furthermore, it has been demonstrated that Gata1 can recruit additional proteins, such as Fog1 (Zfpm1), to the complex, through its N-terminal zinc finger [45]. A major function of the Ldb1-complex is to provide a stable scaffold through which the hematopoietic transcription factors Tal1 and Gata1 act together to regulate erythroid gene transcription. (b) An illustration of Ldb1-mediated juxtaposition of two Ldb1-complexes bound to DNA at sites far apart from each other. Ldb1 can dimerize through its self-association domain facilitating DNA looping and juxtaposition of two Ldb1-complexes. This property of Ldb1 suggests a model whereby enhancers can communicate with distal promoters (in cis or possibly in trans) via Ldb1-complex mediated association. The Ldb1 self-association domain can also form trimeric structures as well as dimers and these types of higher order structures are likely relevant in instances where multiple Ldb1-complexes are assembled near a gene (e.g. the Hbb locus) [30].
That such higher order complexes may be important for erythroid gene expression is supported by data derived from Ldb1 and Lmo2 gene ablation studies in mice. Homozygous germline deletion of Ldb1 in mice results in a pleiotropic phenotype with arrested development and death at embryonic day 9.5-10 [20]. Among the multiple abnormalities observed was a defect in the expansion of the yolk sac including the absence of blood islands, indicating a role for Ldb1 in hematopoietic development. Further analysis demonstrated that Ldb1 is necessary for primitive erythropoiesis as Ldb1 null yolk sac cells are incapable of generating erythroid colonies in in vitro culture assays [21]. Using mice harboring conditional deletion alleles of Ldb1 and expressing either an embryonic endothelial/hematopoietic lineage-specific Cre (Tie2-Cre) or an inducible Mx1-Cre, it was shown that Ldb1 is also required for both fetal and adult definitive erythropoiesis [21]. Deletion of Ldb1 resulted in a significant reduction of megakaryocyte-erythroid progenitors, megakaryocytes and erythroblasts. Lmo2 null mice also display hematopoietic defects that phenocopy Ldb1 deletion in that both primitive and definitive erythropoiesis are virtually absent [22, 23].
To determine if co-binding of Gata1 and Tal1 is mediated by Ldb1-complexes on a genome-wide scale, two groups performed ChIP-Seq studies to map Ldb1, Gata1, and Tal1 binding sites in erythroid cells [24, 25]. Remarkably, both studies reported strong overlap of Gata1, Tal1, and Ldb1 peaks. Co-occupancy was especially high at sites within or near known erythroid genes; for example, 84% of Ldb1, 79% of Gata1, and 84% of Tal1 binding sites at erythroid “fingerprint” genes were occupied by the other two factors [25]. Genes involved in all aspects of erythropoiesis including transcriptional regulation (Klf1, E2f2, Zfpm1, Sox6), heme/hemoglobin biosynthesis (Alad, Alas2, Cpox, Ppox Fech, Hbb), cytoskeletal organization (Add2, Ank1, Epb4.1, Epb4.2, Tmod), and ion or solute transport (Slc4a1, Slc25a37, Aqp1, Aqp9) as well as virtually all known erythroid enhancer elements contained Ldb1 complex-binding sites [24, 25]. Ldb1-complex bound genes were mainly induced during terminal erythroid differentiation, and Ldb1 knockdown confirmed that activation of these genes is Ldb1-dependent [25]. Further, since nearly all of the genes bound by Ldb1-complexes were previously shown to require Gata1 and/or Tal1 for their induction [7, 9], these results advocated for a model in which Ldb1-complexes represent key structures by which Gata1 and Tal1 positively regulate erythroid gene transcription.
Ldb1-complexes mediate long-range promoter/enhancer interactions
A consistent finding from studies using ChIP-Seq to map the binding profiles of essential erythropoietic transcription factors is that these proteins frequently bind at sites located far from potential target genes. Indeed, as mentioned above, most Gata1, Tal1, Klf1, and Ldb1-complex binding sites identified by ChIP-Seq are intronic or within intergenic regions [7, 8, 13, 14, 16, 24, 25]. Even at these distal locations, core erythroid transcription factors tend to co-occupy the same sites and are accompanied by epigenetic enhancer marks suggesting that these regions function as regulatory elements.
The Hbb LCR establishes contact with promoters of the actively transcribed Hbb genes through chromatin looping [26]. Gata1, Tal1, and Klf1 are bound to the Hbb LCR and are each required for loop formation as well for β-globin transcription [27, 28]. However, these factors alone do not provide a satisfactory model to account for looping and distal interactions. On the other hand, Ldb1 contains a self-association domain that is capable of facilitating the formation of stable long-range promoter/enhancer interactions through Ldb1-mediated oligomerization [29, 30] [Fig. 1B]. Experiments employing ChIP, Chromosome Conformation Capture (3C), and shRNA-mediated knockdown of Ldb1 in murine erythroleukemia (MEL) cells have shown that Ldb1 is physically present at the Hbb LCR and Hbb promoters and that Ldb1 is required for loop formation and for transcriptional activation of Hbb genes [31]. A reciprocal experiment demonstrated that enforced Ldb1 dimerization was sufficient for LCR-promoter looping and Hbb transcription even in the absence of Gata1 [32]. Ldb1 is also required for migration of the Hbb locus to regions of active transcription in the nucleus [33]. In addition to the Hbb promoters and LCR, Ldb1-complexes bind to distal regulatory elements upstream of the Myb gene [34]. 3C experiments demonstrated that these regulatory elements are brought into proximity with the Myb promoter in an Ldb1-dependent manner [34]. Novel long-range cis-interactions between the Hbb promoter and Ldb1-complexes bound to the promoters of other erythorid genes including Uros and Tspan32 were also recently identified [24]. Thus, in addition to providing Gata1 and Tal1, key transcription factors necessary for erythropoiesis, Ldb1-complexes provide a mechanism for promoter/enhancer interactions through Ldb1 self-association.
Ldb1-complexes positively regulate Klf1
As noted above, the phenotype of Klf1−/− mice [5, 6], together with the results of gene expression profiling studies, have identified a critical role for Klf1 in erythropoiesis and in the induction of many erythroid genes [10, 11, 35-37]. Interestingly, ChIP-Seq experiments revealed that Klf1 is a target of Ldb1-complexes [25]. In addition, knockdown of Ldb1 markedly attenuates induction of Klf1 in MEL cells indicating that Klf1 is directly regulated by Ldb1-complexes [25]. This result is consistent with the previous finding that Gata1 regulates the expression of Klf1 and that Klf1 transcription is dependent upon a paired E-box/GATA motif within the Klf1 promoter [38, 39]. Interestingly, known cis-regulatory elements for the Gata1, Tal1, and Lmo2 genes are also occupied by Ldb1-complexes in erythroid progenitors suggesting a positive auto-regulatory role for Ldb1-complexes in the expression of these subunits [24, 25].
Ldb1-complexes cooperate with Klf1 to activate transcription of erythroid genes through distinct regulatory mechanisms
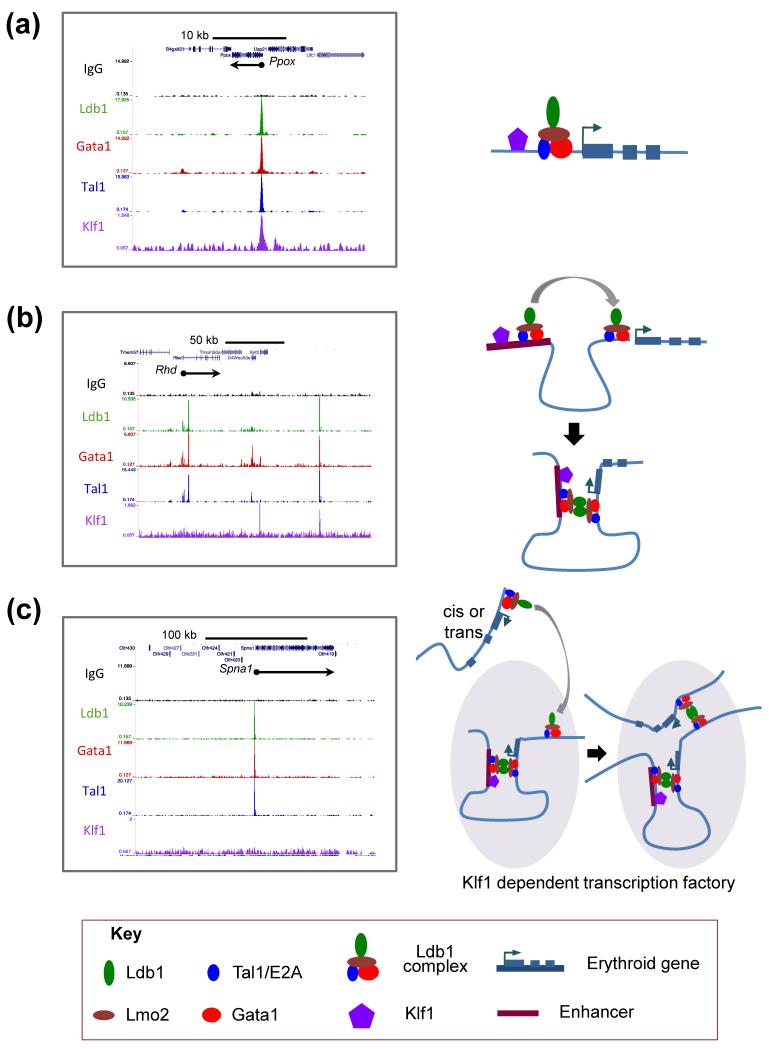
Comparative analysis of transcriptional profiling data from three studies [8, 25, 40] identified a cohort of 62 erythroid genes that are co-regulated by Ldb1-complexes and by Klf1 (i.e. genes that are significantly down-regulated in both Klf1−/− fetal liver erythroblasts and in differentiated MEL cells where Ldb1 expression is reduced by shRNA) [Table 1]. Strikingly, Ldb1-complex binding sites were detected at or within 10 kb of 85% (53/62) of these co-regulated genes in primary murine erythroid cells [25] indicating that Ldb1-complexes function primarily to directly regulate erythroid gene transcription. In most cases (e.g. Hba, Hbb, Gypc, Alad, Add2, Ermap, Urod, and Ppox) ChIP-Seq data revealed that both Klf1 and Ldb1-complexes bind to promoter-proximal sites, a configuration most consistent with a classical “feed-forward” mechanism of transcriptional co-regulation [41] [Fig. 2A]. However, a second group of co-regulated genes (e.g. Ank1, Slc25a37, Ctsb, Rhd, and Gypa) were bound by Ldb1-complexes but not Klf1 near their TSS; yet, in each case, prominent co-localized Ldb1-complex and Klf1 binding was detected within the intron(s) of the same gene or at intergenic sites located 10-100 kb away at known or suspected cis-regulatory elements [Fig. 2B]. Similar to what has been shown at the Hbb and Myb loci [31, 32, 34], Ldb1-mediated dimerization likely facilitates promoter/enhancer interactions necessary to bring Klf1 into proximity with the promoter enabling transcriptional activation of these genes [Fig. 2B]. A third group of co-regulated genes were bound by Ldb1-complexes, but no Klf1 binding was detected at or within 100 kb of the gene [Fig. 2C]. This configuration could reflect that Klf1 dependence is indirect (i.e. transcription is controlled by a Klf1-dependent factor but not directly by Klf1). Notwithstanding, recent data suggest that a more complex mechanism is responsible for the regulation of many of these genes including Arrb1, Fech, Uros, Kcnn4, Spna1, Spnb1, Kel, Tmcc2, and Cpox. In definitive erythroid cells, each of the aforementioned genes is recruited to active sites of transcription (designated transcriptional interactomes, active chromatin hubs, or transcription factories) near the Hba Hbb loci [40]. Previous work has shown that Ldb1-complexes [18, 24, 31] and Klf1 [8, 42, 43] bind to the Hbb promoters, and the α- and β-globin regulatory elements. Moreover, both Ldb1 and Klf1 are required to establish long-range promoter/LCR interactions at these loci and are also required for α- and β-globin gene transcription [28, 31]. We speculate, based on these findings, that Klf1-dependent genes such as Spna1 are recruited by Ldb1-complexes to sites of active transcription at the Hba and Hbb loci [Fig. 2C]. A corollary of this model (as depicted in Fig. 2C) is that although Klf1 is necessary for initiating transcription at active chromatin hubs, it may not be directly required for the transcription of genes that are subsequently recruited to these sites. It is also likely that sites of Ldb1-complex/Klf1 co-binding other than those at the Hba and Hbb loci function as erythroid transcriptional interactomes. Finally, recent data indicate that Ldb1 proteins may preferentially form trimers or higher order oligomers [30] raising the possibility that individual Ldb1 complex-bound enhancer modules could be recruited to more than one target gene or that multiple Ldb1-complex bound regulatory elements can be recruited to genes with a single Ldb1-complex binding site. Accordingly, these data suggest that Ldb1-complexes may function in several distinct ways to orchestrate transcriptional activation on a global scale during terminal erythropoiesis.
Table 1. Ldb1-complex and Klf1 co-activated genes.
| Gene Ontology Term | Ldb1-complex/Klf1 Co-activated Genes |
|---|---|
| Membrane/ Cytoskeleton/ Blood group |
Cd24a, Cd47, Cd59a, Ermap, Gypa, Gypc, Kcnn4, Kel, Mgll, Rhd, Slc4a1, Slc2a4, Slc22a4, Tmcc2, Vamp5, Fam210b, Sppl2b |
| Cytoskeleton | Add2, Ank1, Epb4.1, Epb4.2, Spna1, Spnb1 |
| Heme Synthesis & Transport/ Mitochondrial |
Abcb10, Abcg2, Alad, Alas2, Blvrb, Bzrap1, Cat, Cpox, Fech, Hagh, Hmbs, Ppox, Urod, Vdac1 |
| Hb/Iron Procurement |
Fxn, Hba, Hbb, Ppox, Slc25a37/mitoferrin, Slc11a2/Dmt1, Steap3, Tfrc |
| Apoptosis/ Survival/Cell cycle |
Cdkn2c/p 18INK4c, Ctsb, Dlgap5, Pim1, Prdx2, Ptp4a3, Rad23a, Rgcc |
| Cytoplasmic | Arrb1, Dck, Pcx, Stx2, Ube2c, Ubap1 |
| Nuclear/ Transcription |
Cdyl, E2f2, Mafk |
Genes included in the table are those significantly down-regulated in Ldb1 shRNA-mediated knock down Murine Erythroleukemia cells and in Klf1−/−fetal liver cells. Color code: Black, genes bound by Ldb1-complex(es) (gene body ± 10 Kb) and by Klf1; Blue, genes bound only by Klf1; Red, genes bound only by Ldb1-complex(es) (gene body ± 10 Kb); Green, genes not bound by Ldb1-complexes or Klf1. Gene list was compiled from published articles [8, 25, 40, 58].
Figure 2. Hypothetical models for the cooperation of Ldb1-complexes and Klf1 in the regulation of erythroid gene transcription.
Genome-wide mapping of Ldb1-complex and Klf1 binding by ChIP-Seq suggests several possible mechanisms by which Klf1 functions in concert with the Ldb1-complex to activate erythroid genes known to be dependent on both Klf1 and Ldb1 for their expression. Images on the left represent raw ChIP-Seq read data for Ldb1, Gata1, Tal1, and Klf1 [8, 25] transformed into a density plot for each factor and loaded into the UCSC genome browser as a custom track. Models corresponding to the binding profiles are depicted on the right. (a) Left, example of an erythroid gene (Ppox) where an Ldb1-complex and Klf1 bind in close proximity to each other and to the transcription start site of a gene to directly activate transcription. Right, model depicting the direct regulation of Ppox transcription by the Ldb1-complex and Klf1. (b) Left, example of an erythroid gene (Rhd) where an Ldb1-complex but not Klf1 binds at the promoter and where both an Ldb1-complex and Klf1 bind to a distal enhancer. Right, model depicting the recruitment of Klf1 to the Rhd promoter through dimerization of the Ldb1 self-association domain. (c) Left, example of an erythroid gene (Spna1) where Ldb1-complex binding is detected at the gene but Klf1 binding is not detected within 100 kilobases of the gene. Right, model depicts Ldb1-complex mediated recruitment of Spna1 to a transcriptional hub near the Hbb gene. In this model, the transcriptional hub serves as a nexus where Klf1-dependent genes are brought into locations of direct Klf1 recruitment through the self-interactions of Ldb1-complexes.
Role of Ldb1-complexes in gene repression
Although Ldb1-complex binding strongly correlates with gene activation, in a few cases binding of Ldb1-complexes has been associated with gene repression. For example, Ldb1-complexes bind to the Lyl1 and Egr1 promoters and knockdown of Ldb1 in MEL cells causes both genes to be significantly up-regulated [25]. An emerging concept is that whereas “core” Ldb1/Lmo2/Tal1/Gata1 Ldb1-complexes function mainly as transcriptional activators, repressive potential can be conferred by recruitment of additional factors by subunits of the Ldb1-complex. Gata1 can associate with the zinc finger protein Friend of Gata1 (Fog1/Zfpm1), which is capable of recruiting the nucleosome remodeling and deacetylase (NuRD) complex and the co-repressor CtBP providing a mechanism for Gata1-dependant gene repression [44]. It was recently shown that Gata1 can simultaneously interact with Lmo2 and Fog1 demonstrating that Fog1, and presumably other associated factors, can be recruited to Ldb1 complexes through Gata1 [45]. Ldb1 complexes can also potentially acquire repressive activity through Tal1-mediated recruitment of Cbfa2t3 (Eto2), which can bind histone deacetylases [46-48]. Interestingly, ChIP-Seq data have shown that Eto2 and the related protein Mtgr1 bind to Ldb1-complexes in undifferentiated MEL cells but disassociate upon terminal erythroid differentiation when most erythroid genes are strongly induced [24].
The polycomb repressive complex 2 (PRC2) has also recently been implicated in Gata1-mediated erythroid gene repression [9, 14]. Using an estrogen receptor-inducible system to induce Gata1 nuclear localization, it was observed that a subset of the Gata1-repressed genes show enrichment of repressive H3K27me3 histone modification and loss of co-binding of Tal1 with Gata1 [9, 14]. Gata1 can physically associate with Suz12 and Ezh2, two core subunits of PRC2, and Suz12, in turn, can recruit the transcriptional repressor Gfi-1b [49]. Interestingly, Gata1 binding sites within several Gata1-repressed genes, including c-Kit, Gata2, and c-Myb, were co-occupied by Gfi-1b [14]. The loss of Tal1 (and presumably also Lmo2 and Ldb1) at these sites may facilitate or be a consequence of the formation of repressive Gata1/Gfi-1b/PRC2 complexes.
Function of Ldb1 complexes in other hematopoietic lineages
Several recent studies have identified important roles for Ldb1-complexes in non-erythroid hematopoietic lineages. Notably, DNA binding complexes that include Ldb1, Tal1, and Gata2 in lieu of Gata1 have been shown to regulate a transcriptional program required for HSC maintenance [50]. Whereas Gata1 is essential for erythropoiesis, Gata2, which is highly expressed in hematopoietic progenitors, performs an equally critical role in the generation and maintenance of HSCs [51]. Commitment of HSCs to the erythroid lineage is associated with an event known as the ‘Gata-switch’ that involves the induction of Gata1 and the Gata1-mediated repression of Gata2 [see Box2] [52]. Substitution of Gata1 for Gata2 within Ldb1-complexes results in induction of Klf1 and global erythroid gene activation. Thus, the modular design of Ldb1-complexes enables acquisition of distinct gene regulatory activities in HSCs and erythroblasts through the stage-specific assembly of different Ldb1-complexes that incorporate either Gata2 or Gata1. Ldb1-complexes that contain Gata2 are likely required for hematopoietic specification in the embryo and are also necessary for proper development of hemangioblasts, the common progenitors of hematopoietic and endothelial cells [53]. Megakaryocytes and mast cells have been shown to require Gata1, Gata2, Tal1, and Ldb1 for their normal development indicating that Ldb1-complexes perform key functions in these lineages [21, 54-57]. In agreement with this idea, Gata1, Tal1, and Ldb1 co-occupancy has been observed at several key megakaryocyte genes, including Mpl, αIIb, GpIa, Mc-Cpa, FcεR1-b, Pf4 [18], and Itga2b [25].
Box 2. The ‘Gata Switch’ Model.
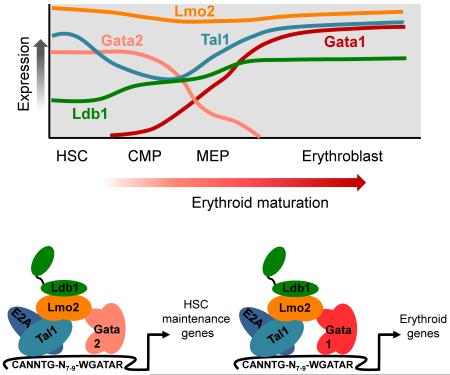
Gata2 plays a critical role in the emergence and maintenance of hematopoietic stem cells (HSCs) as well as in the specification of early erythroid progenitors [72]. The zinc fingers of Gata2 and Gata1 share a high degree of similarity and they both bind a consensus GATA DNA motif (see Box 1). Gata2 can partially restore primitive erythropoiesis in the absence of Gata1 [73]. However, the N and C termini of Gata1 and Gata2 are quite divergent suggesting that Gata1 and Gata2 interact with a unique set of co-factors. Gata1 and Gata2 are involved in a key regulatory loop during erythropoiesis designated the ‘Gata-switch’ [74]. Gata2 directly activates its own gene in HSCs but in the early stages of erythropoiesis Gata1 is induced [Figure I, top]. As a consequence, Gata1 replaces Gata2 at both genes, resulting in repression of Gata2 transcription and increased Gata1 transcription. Studies using ChIP-Seq to interrogate the binding profiles of Gata2 and Gata1 in erythroid precursor cells have found that they share many chromatin sites and also reveal that this exchange in Gata factor binding is widespread [12, 13]. Thus, an elegant “switch” of Gata factors triggers erythroid differentiation. In HSCs, Gata2 functions in large part within the context of Ldb1-complexes to control expression of genes responsible for HSC maintenance [50] whereas Ldb1-complexes that contain Gata1 regulate the expression of a large cohort of erythroid signature genes [24, 25] [Figure I, bottom]. Consequently, the Ldb1-complex serves as a core structure through which the “Gata switch” operates to regulate erythroid lineage commitment and differentiation.
Figure I. ‘Gata switch’ model
Top, graph representing the expression of Ldb1-complex components as hematopoietic progenitor cells differentiate toward the erythroid lineage. Notably, Gata2 expression is high and Gata1 expression is low in the earliest progenitor cells, but as cells become committed to the erythroid lineage, Gata1 expression is induced while Gata2 is repressed. Relative expression levels were deduced from microarray data available at www.BioGps.org and RNA-Seq data available on the UCSC Genome Browser.
Bottom, effect of the Gata switch on the subunit composition of Ldb1-complexes. In HSCs, Ldb1-complexes that contain Gata2 regulate expression of HSC maintenance genes. As a result of the Gata switch during erythropoiesis, Gata1 is incorporated into the Ldb1-complex to activate expression of erythroid genes.
Concluding remarks
In large part attributable to the advent of new technologies enabling genome-wide DNA binding site profiling, the past few years have witnessed dramatic advances in our understanding of the genetic regulatory mechanisms controlling erythropoiesis. One of the most intriguing findings is that two major erythroid transcription factors, Gata1 and Tal1, often function cooperatively within higher order Ldb1-nucleated protein complexes to activate erythroid genes. Whether, or to what extent, the regulatory properties of Gata1 and/or Tal1 are affected by their inclusion within Ldb1-complexes remains to be determined. An important role of Ldb1-complexes is to facilitate, via Ldb1-mediated oligomerization, long-distance interactions including the juxtaposition of erythroid promoters and enhancer elements and the recruitment of erythroid genes to transcriptional interactomes near the Hba and Hbb genes. Whether Ldb1 oligomerization can facilitate trans as well as cis chromosomal interactions is unclear and it is also currently unknown if a single complex can recruit multiple regulatory elements through Ldb1 oligomerization. Other important questions for future investigation are whether Ldb1-compelxes are required for the formation of erythroid transcriptional interactomes and if Ldb1-complex mediated associations are static or are instead dynamic and transient. Although the core subunits of erythroid Ldb1-complexes, which include Ldb1, Lmo2, Gata1, Tal1, and E2a, have been identified, clear evidence exists for Ldb1-complex modularity that can potentially modify or fundamentally alter the regulatory properties of Ldb1-complexes. Identifying the precise subunit composition and binding sites of Ldb1-complexes at different stages of hematopoiesis and correlating these data with gene expression profiling represents an important goal of ongoing and future studies.
Highlights.
Ldb1-complexes are primary instruments of Gata1 and Tal1 mediated erythroid gene activation.
Ldb1 oligomerization facilitates long-range promoter/enhancer interactions.
Ldb1-complexes and Klf1 co-occupy and co-regulate many erythroid genes.
Acknowledgments
This work was supported by the Intramural Research Programs of Eunice Kennedy Shriver NICHD [Project number1ZIAHD001803-19 (to P.E.L.)]. The authors thank Karl Pfeifer for critical review of the manuscript.
Abbreviations
- HSC
hematopoietic stem cell
- CMP
common myeloid progenitor
- MEP
megakaryocyte-erythroid progenitor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pevny L, et al. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 2.Schlaeger TM, et al. Tie2Cre-mediated gene ablation defines the stem-cell leukemia gene (SCL/tal1)-dependent window during hematopoietic stem-cell development. Blood. 2005;105:3871–3874. doi: 10.1182/blood-2004-11-4467. [DOI] [PubMed] [Google Scholar]
- 3.Robb L, et al. Absence of yolk sac hematopoiesis from mice with a targeted disruption of the scl gene. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:7075–7079. doi: 10.1073/pnas.92.15.7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porcher C, et al. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996;86:47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- 5.Nuez B, et al. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375:316–318. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 6.Perkins AC, et al. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 7.Kassouf MT, et al. Genome-wide identification of TAL1’s functional targets: insights into its mechanisms of action in primary erythroid cells. Genome research. 2010;20:1064–1083. doi: 10.1101/gr.104935.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tallack MR, et al. A global role for KLF1 in erythropoiesis revealed by ChIP-seq in primary erythroid cells. Genome research. 2010;20:1052–1063. doi: 10.1101/gr.106575.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng Y, et al. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome research. 2009;19:2172–2184. doi: 10.1101/gr.098921.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tallack MR, et al. Novel roles for KLF1 in erythropoiesis revealed by mRNA-seq. Genome research. 2012;22:2385–2398. doi: 10.1101/gr.135707.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hodge D, et al. A global role for EKLF in definitive and primitive erythropoiesis. Blood. 2006;107:3359–3370. doi: 10.1182/blood-2005-07-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu W, et al. Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. Genome Res. 2011;21:1659–1671. doi: 10.1101/gr.125088.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujiwara T, et al. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Molecular cell. 2009;36:667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu M, et al. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Molecular cell. 2009;36:682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papadopoulos GL, et al. GATA-1 genome-wide occupancy associates with distinct epigenetic profiles in mouse fetal liver erythropoiesis. Nucleic acids research. 2013;41:4938–4948. doi: 10.1093/nar/gkt167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wontakal SN, et al. A core erythroid transcriptional network is repressed by a master regulator of myelo-lymphoid differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3832–3837. doi: 10.1073/pnas.1121019109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wozniak RJ, et al. Molecular hallmarks of endogenous chromatin complexes containing master regulators of hematopoiesis. Molecular and cellular biology. 2008;28:6681–6694. doi: 10.1128/MCB.01061-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tripic T, et al. SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood. 2009;113:2191–2201. doi: 10.1182/blood-2008-07-169417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wadman IA, et al. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. The EMBO journal. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukhopadhyay M, et al. Functional ablation of the mouse Ldb1 gene results in severe patterning defects during gastrulation. Development. 2003;130:495–505. doi: 10.1242/dev.00225. [DOI] [PubMed] [Google Scholar]
- 21.Li L, et al. A requirement for Lim domain binding protein 1 in erythropoiesis. The Journal of experimental medicine. 2010;207:2543–2550. doi: 10.1084/jem.20100504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamada Y, et al. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:3890–3895. doi: 10.1073/pnas.95.7.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warren AJ, et al. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell. 1994;78:45–57. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- 24.Soler E, et al. The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes & development. 2010;24:277–289. doi: 10.1101/gad.551810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, et al. Ldb1-nucleated transcription complexes function as primary mediators of global erythroid gene activation. Blood. 2013;121:4575–4585. doi: 10.1182/blood-2013-01-479451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tolhuis B, et al. Looping and interaction between hypersensitive sites in the active beta-globin locus. Molecular cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 27.Drissen R, et al. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes & development. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vakoc CR, et al. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Molecular cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 29.Matthews JM, Visvader JE. LIM-domain-binding protein 1: a multifunctional cofactor that interacts with diverse proteins. EMBO reports. 2003;4:1132–1137. doi: 10.1038/sj.embor.7400030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cross AJ, et al. LIM domain binding proteins 1 and 2 have different oligomeric states. Journal of molecular biology. 2010;399:133–144. doi: 10.1016/j.jmb.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Song SH, et al. A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Molecular cell. 2007;28:810–822. doi: 10.1016/j.molcel.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng W, et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149:1233–1244. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song SH, et al. Multiple functions of Ldb1 required for beta-globin activation during erythroid differentiation. Blood. 2010;116:2356–2364. doi: 10.1182/blood-2010-03-272252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stadhouders R, et al. Dynamic long-range chromatin interactions control Myb proto-oncogene transcription during erythroid development. The EMBO journal. 2012;31:986–999. doi: 10.1038/emboj.2011.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drissen R, et al. The erythroid phenotype of EKLF-null mice: defects in hemoglobin metabolism and membrane stability. Molecular and cellular biology. 2005;25:5205–5214. doi: 10.1128/MCB.25.12.5205-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nilson DG, et al. Major erythrocyte membrane protein genes in EKLF-deficient mice. Experimental hematology. 2006;34:705–712. doi: 10.1016/j.exphem.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 37.Pilon AM, et al. Genome-wide ChIP-Seq reveals a dramatic shift in the binding of the transcription factor erythroid Kruppel-like factor during erythrocyte differentiation. Blood. 2011;118:e139–148. doi: 10.1182/blood-2011-05-355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anderson KP, et al. Multiple proteins binding to a GATA-E box-GATA motif regulate the erythroid Kruppel-like factor (EKLF) gene. The Journal of biological chemistry. 1998;273:14347–14354. doi: 10.1074/jbc.273.23.14347. [DOI] [PubMed] [Google Scholar]
- 39.Crossley M, et al. Regulation of the erythroid Kruppel-like factor (EKLF) gene promoter by the erythroid transcription factor GATA-1. The Journal of biological chemistry. 1994;269:15440–15444. [PubMed] [Google Scholar]
- 40.Schoenfelder S, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nature genetics. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shyu YC, et al. Chromatin-binding in vivo of the erythroid kruppel-like factor, EKLF, in the murine globin loci. Cell research. 2006;16:347–355. doi: 10.1038/sj.cr.7310045. [DOI] [PubMed] [Google Scholar]
- 43.Vernimmen D, et al. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. The EMBO journal. 2007;26:2041–2051. doi: 10.1038/sj.emboj.7601654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hong W, et al. FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. The EMBO journal. 2005;24:2367–2378. doi: 10.1038/sj.emboj.7600703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilkinson-White L, et al. Structural basis of simultaneous recruitment of the transcriptional regulators LMO2 and FOG1/ZFPM1 by the transcription factor GATA1. Proc Natl Acad Sci U S A. 2011;108:14443–14448. doi: 10.1073/pnas.1105898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuh AH, et al. ETO-2 associates with SCL in erythroid cells and megakaryocytes and provides repressor functions in erythropoiesis. Molecular and cellular biology. 2005;25:10235–10250. doi: 10.1128/MCB.25.23.10235-10250.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meier N, et al. Novel binding partners of Ldb1 are required for haematopoietic development. Development. 2006;133:4913–4923. doi: 10.1242/dev.02656. [DOI] [PubMed] [Google Scholar]
- 48.Goardon N, et al. ETO2 coordinates cellular proliferation and differentiation during erythropoiesis. The EMBO journal. 2006;25:357–366. doi: 10.1038/sj.emboj.7600934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saleque S, et al. The zinc-finger proto-oncogene Gfi-1b is essential for development of the erythroid and megakaryocytic lineages. Genes & development. 2002;16:301–306. doi: 10.1101/gad.959102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li L, et al. Nuclear adaptor Ldb1 regulates a transcriptional program essential for the maintenance of hematopoietic stem cells. Nat Immunol. 2011;12:129–136. doi: 10.1038/ni.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ling KW, et al. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. The Journal of experimental medicine. 2004;200:871–882. doi: 10.1084/jem.20031556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bresnick EH, et al. GATA switches as developmental drivers. The Journal of biological chemistry. 2010;285:31087–31093. doi: 10.1074/jbc.R110.159079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mylona A, et al. Genome-wide analysis shows that Ldb1 controls essential hematopoietic genes/pathways in mouse early development and reveals novel players in hematopoiesis. Blood. 2013;121:2902–2913. doi: 10.1182/blood-2012-11-467654. [DOI] [PubMed] [Google Scholar]
- 54.Mikkola HK, et al. Haematopoietic stem cells retain long-term repopulating activity and multipotency in the absence of stem-cell leukaemia SCL/tal-1 gene. Nature. 2003;421:547–551. doi: 10.1038/nature01345. [DOI] [PubMed] [Google Scholar]
- 55.Salmon JM, et al. Aberrant mast-cell differentiation in mice lacking the stem-cell leukemia gene. Blood. 2007;110:3573–3581. doi: 10.1182/blood-2006-10-053124. [DOI] [PubMed] [Google Scholar]
- 56.Cantor AB, et al. Antagonism of FOG-1 and GATA factors in fate choice for the mast cell lineage. The Journal of experimental medicine. 2008;205:611–624. doi: 10.1084/jem.20070544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Migliaccio AR, et al. GATA-1 as a regulator of mast cell differentiation revealed by the phenotype of the GATA-1low mouse mutant. The Journal of experimental medicine. 2003;197:281–296. doi: 10.1084/jem.20021149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tallack MR, Perkins AC. KLF1 directly coordinates almost all aspects of terminal erythroid differentiation. IUBMB life. 2010;62:886–890. doi: 10.1002/iub.404. [DOI] [PubMed] [Google Scholar]
- 59.Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21:3368–3376. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- 60.Trainor CD, et al. GATA zinc finger interactions modulate DNA binding and transactivation. The Journal of biological chemistry. 2000;275:28157–28166. doi: 10.1074/jbc.M000020200. [DOI] [PubMed] [Google Scholar]
- 61.Lee HY, et al. Controlling hematopoiesis through sumoylation-dependent regulation of a GATA factor. Molecular cell. 2009;36:984–995. doi: 10.1016/j.molcel.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lowry JA, Mackay JP. GATA-1: one protein, many partners. The international journal of biochemistry & cell biology. 2006;38:6–11. doi: 10.1016/j.biocel.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 63.Fujiwara Y, et al. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Green AR, Begley CG. SCL and related hemopoietic helix-loop-helix transcription factors. International journal of cell cloning. 1992;10:269–276. doi: 10.1002/stem.5530100504. [DOI] [PubMed] [Google Scholar]
- 65.Souroullas GP, et al. Adult hematopoietic stem and progenitor cells require either Lyl1 or Scl for survival. Cell stem cell. 2009;4:180–186. doi: 10.1016/j.stem.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hall MA, et al. The critical regulator of embryonic hematopoiesis, SCL, is vital in the adult for megakaryopoiesis, erythropoiesis, and lineage choice in CFU-S12. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:992–997. doi: 10.1073/pnas.0237324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Siatecka M, Bieker JJ. The multifunctional role of EKLF/KLF1 during erythropoiesis. Blood. 2011;118:2044–2054. doi: 10.1182/blood-2011-03-331371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McConnell BB, Yang VW. Mammalian Kruppel-like factors in health and diseases. Physiological reviews. 2010;90:1337–1381. doi: 10.1152/physrev.00058.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frontelo P, et al. Novel role for EKLF in megakaryocyte lineage commitment. Blood. 2007;110:3871–3880. doi: 10.1182/blood-2007-03-082065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tallack MR, Perkins AC. Megakaryocyte-erythroid lineage promiscuity in EKLF null mouse blood. Haematologica. 2010;95:144–147. doi: 10.3324/haematol.2009.010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bouilloux F, et al. EKLF restricts megakaryocytic differentiation at the benefit of erythrocytic differentiation. Blood. 2008;112:576–584. doi: 10.1182/blood-2007-07-098996. [DOI] [PubMed] [Google Scholar]
- 72.Vicente C, et al. The role of the GATA2 transcription factor in normal and malignant hematopoiesis. Critical reviews in oncology/hematology. 2012;82:1–17. doi: 10.1016/j.critrevonc.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 73.Fujiwara Y, et al. Functional overlap of GATA-1 and GATA-2 in primitive hematopoietic development. Blood. 2004;103:583–585. doi: 10.1182/blood-2003-08-2870. [DOI] [PubMed] [Google Scholar]
- 74.Grass JA, et al. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8811–8816. doi: 10.1073/pnas.1432147100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ryan DP, et al. Assembly of the oncogenic DNA-binding complex LMO2-Ldb1-TAL1-E12. Proteins. 2008;70:1461–1474. doi: 10.1002/prot.21638. [DOI] [PubMed] [Google Scholar]