Introduction
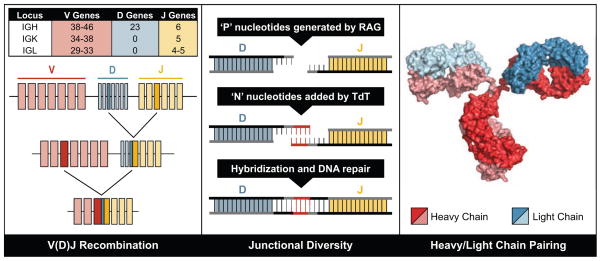
The ability to generate a specific and long-lived antibody response is a key element of acquired immunity and is a necessary component for the prevention or resolution of disease caused by most viruses [1]. Specificity of antibody responses for particular pathogens is achieved by the development of a diverse repertoire of recombined antibody variable genes that encode antibodies that can recognize an enormous number of potential epitopes. Diversity in the antigen combining site of the B cell receptor repertoire (and thus also in the corresponding secreted antibody repertoire) is mediated by three principal mechanisms that are illustrated in Figure 1: (1) random pairing of heavy and light chains to form the antigen-binding site in the immunoglobulin molecule; (2) combinatorial diversity generated by V(D)J recombination, which together with heavy and light chain pairing results in approximately 2.3 × 106 different possible combinations; (3) junctional diversity generated by P- and N-nucleotide addition or deletion at recombination sites during V(D)J processing by isoforms of the enzyme terminal deoxynucleotidyl transferase (TdT), which theoretically results in 1011 different antibody specificities [2]. Somatic hypermutation, a fourth mechanism of diversification, introduces point mutations into the rearranged immunoglobulin variable domain after B cell activation. Additional functional diversity in secreted antibodies is conferred by differences between isotypes after class switching, since the Fc region of immunoglobulins determines the valency of the antibody combining sites and many functions such as complement fixation, and interaction with various Fc receptors or the polyimmunoglubilin receptor. Following diversification of the repertoire, longevity of particular B cells is mediated by complex regulatory functions.
Figure 1.
Diversity in the antigen-combining site of the B cell receptor repertoire (and thus also in the corresponding secreted antibody repertoire) is mediated by three principal molecular mechanisms, illustrated in the three panels, left, middle, and right.
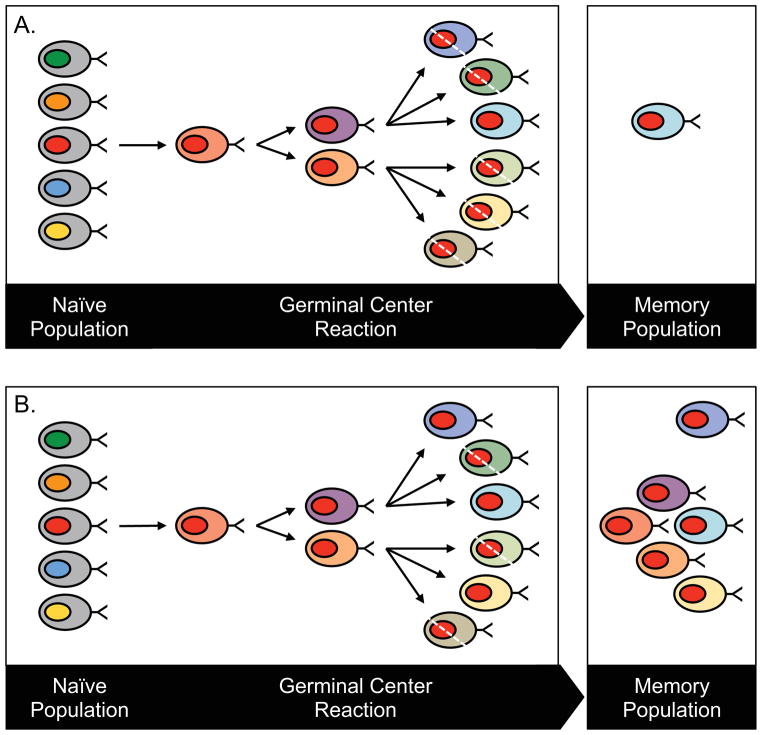
In years past, immunologists understood diversification of B cell populations specific to particular foreign antigens to involved a burst of diversification within a clone of B cells in the activated germinal center, followed by a selection for survival of the highest affinity clone and drastic loss of related somatic variants with lower affinity. Although this “single winner” model did correctly describe the typical panel of B cell clones isolated from experimental studies using isolation of hybridomas and monoclonal antibodies (mAbs), the technical approach to isolation of mAbs likely biased such studies toward the isolation of only the most avidly binding antibodies. Emerging techniques using high-throughput DNA and RNA sequence analysis are increasingly revealing that this paradigm is not correct, and instead human B cell repertoires maintain very large populations of somatic variants within clones [3]; see Figure 2. It may seem metabolically wasteful and counter-intuitive that the immune system would allow hundreds or thousands of related clones to persist in circulation when many of those variants possess many fewer somatic mutations than the most mature clones, and thus by inference likely have lower affinity of binding for the inciting epitope. There may be method in this madness, however, if persisting diversity in the B cell repertoire allows the subject to respond to antigenic variation in the target, such as antigenic drift in acute infections like influenza or persistent escape by point mutations during chronic infections with viruses like HIV-1 or hepatitis C. Dealing with the enormous sequence and structural plasticity of the protective antigens of these viruses (such as influenza hemagglutinin, HIV-1 gp140, or hepatitis envelope protein) likely requires an equivalent breadth of diversity of antigen combining sites in the responding B cell population. Therefore, recent observations that human B cell repertoires engage pathogens with large clonal families of highly related combining sites, which we have termed “antibody swarms”, makes sense from a strategic standpoint for the immune system. Studying the diverse antibody response to antigen as a swarming population instead of as a one-to-one, specific interaction informs our understanding of disease and immunity in a new way. In recent years, key studies have leveraged new technological advances in gene sequencing and microfluidics to provide evidence regarding the mechanisms of repertoire diversification, the size of the antibody repertoire and methods of repertoire regulation shared by different individuals. These studies are the foundation upon which further applications will be developed.
Figure 2.
[A] Classical models of somatic hypermutation conceive of rapid generation of variants in the activated germinal center followed by a severe down-selection of number of variants, resulting in selection of only the clones with the most avidly binding B cell receptors for survival. [B] Newer repertoire studies using large-scale sequence analysis reveal that human B cell repertoires retain large number of variants with diverse numbers of point mutations within clones, even in the peripheral blood.
Sequencing the antibody variable gene repertoire
Many next-generation sequencing techniques are available today; specifications for three of the most commonly used current techniques are detailed in Table 1. No doubt the capabilities and proprietary formats of these types of technologies will continue to evolve rapidly. These methods can be used to determine the sequence of recombined antibody variable genes amplified from primary cell or tissue samples, generating large sequence databases. It is possible to sequence recombined genes isolated from genomic DNA by PCR, or from transcribed genes using cDNA made from mRNA by reverse transcription and amplified by PCR. The resulting amplicon sequences are determined by high-throughput amplicon DNA sequencing technologies, and then analyzed with some type of specialized antibody variable gene sequence analysis software platform. Several web-based software approaches to antibody gene analysis are available, such as IMGT V-QUEST, SoDA and JOINSOLVER, which identify the inferred V, D and J gene segments used during recombination and resolve P- and N-nucleotides, providing robust data for further study [4–6].
Table 1.
Characteristics of three of the most commonly used current next-generation sequencing techniques
| Roche 454 GS FLX Titanium* | Illumina MiSeq** | Illumina HiSeq 2500‡ | |
|---|---|---|---|
| Read Length | Up to 600 bp | Up to 500 bp (250 × 2) | Up to 200 bp (100 × 2) |
| Output | 450 Mb | 7.5–8.5 Gb | 540–600 Gb |
| Reads Run | 700,000 | 15 Million | 3 Billion |
| Quality | Consensus accuracy of 99.995% | > 75% bases above Q30 | > 80% bases above Q30 |
“GS FLX+ System.” 454 Life Sciences, a Roche Company. Web. Accessed 05 Aug 2013.
“MiSeq Benchtop Sequencer Specifications.” Illumina: sequencing and array-based solutions for genetic research. Web. Accessed 05 Aug 2013.
“HiSeq 2500/1500 Specifications.” Illumina: sequencing and array-based solutions for genetic research. Web. Accessed 05 Aug 2013.
Antibody heavy and light chain pairing is an important aspect of the diversification of the antibody repertoire, and it has been shown that antibody heavy chains are capable of pairing with many light chains [7]. Therefore, identifying the correct heavy and light chain pairing partners during repertoire sequencing will be of critical importance to future efforts to understand repertoire diversity. Currently, technical limitations prevent large-scale sequence analysis of naturally paired heavy and light chain genes. There are two principal approaches that are being pursued currently to accomplish the task of pairing heavy and light chain genes on a massive scale. The first approach aims to pair the heavy and light chain sequences from separately sequenced repertoires using informatic approximations, while the other approach aims to link the sequences during variable gene amplification by PCR, followed by sequence analysis of both chains in one amplicon.
Indexed sequencing protocols can be readily applied to barcode both the heavy and light chain sequences from a single sample, after which the heavy and light chain sequences can be paired. One study paired heavy and light chain variable gene sequences according to their relative frequencies within the repertoire, with a majority (21/27, or 78%) of the pairings tested generating antigen-specific antibodies [8]. A second study found that heavy and light chain pairs could be identified often using an evolution-based analysis, wherein coevolution of the heavy and light chains resulted in correlations between both the frequency and topology of the corresponding phylogenetic tree branches [7]. In either case, although these techniques may allow isolation of antigenic binding antibodies, they do not assuredly retain endogenous the original heavy and light chain gene pairing information.
Recently, techniques were developed to retain the endogenous pairing information by linking the heavy and light chains during gene amplification [9,10]. In one study, single B cells were lysed in isolation using a high-density microwell plate, after which mRNA transcripts were captured on magnetic beads for emulsion PCR amplification with linking primers [9]. This process annealed the heavy and light chain complementarity determining region 3 (CDRH3 and CDRL3, respectively) sequences together into one amplicon for next-generation sequencing. A similar technique was employed by another study, which used advances in microfluidics to successfully accomplish on-chip single-cell RT-qPCR [10]. While published results are limited to 300 single-cell RT-qPCR measurements per run, the success of this protocol suggests that the chip could be scaled up to more than 1,000 measurements per chip. While these techniques likely highlight the future of antibody repertoire studies, the current read lengths of next generation sequencing limits the application to only CDRH3:CDRL3 paired sequences. Longer read lengths will be required to identify full-length antibody variable gene sequences that can be used to synthesize cDNA encoding the native sequence of the original antibody including all six CDRs.
Mechanisms of repertoire diversification
Next-generation sequence analysis of the antibody variable gene repertoire broadens our understanding of the critical V(D)J recombination events that are central to antibody repertoire diversification. While the mechanisms of recombination activating gene (RAG)-mediated V(D)J recombination are relatively well understood, rare events that occur during V(D)J recombination have been difficult to study using individual antibodies isolated by hybridoma or single B cell sorting techniques because of the limited scale of such techniques. In contrast, rare genetic events representing additional methods of repertoire diversification are observed readily in large antibody gene repertoire sequence databases generated by next-generation sequencing.
For example, V(DD)J recombination events that appear to violate the 12/23 rule of recombination, occurring when the 12-bp recombination signal sequences (RSS) flanking the D gene segment incorrectly pair to allow fusion of two D gene segments. V(DD)J recombination has been observed in both in vitro and in vivo systems, but accurate calculations of the frequencies of these events were difficult to establish in the past. Furthermore, it was unclear if the perceived V(DD)J recombinations were instead artifacts of random N-additions that simply mimicked natural D gene genomic sequences [11]. In one study, human peripheral blood antibody repertoires collected using Roche 454 technology were analyzed using stringent criteria that revealed that V(DD)J recombination events occur in approximately 1 in 800 circulating B human cells [11]. A second study of human peripheral blood antibody repertoires generated with Illumina HiSeq technology found that tandem D gene sequences occur in human pro-B cells more frequently than would be expected by random chance [12]. Additionally, these V(DD)J recombination events appear to be selected against during B cell development, occurring at much lower frequencies in the population of productive antibody sequences. Analysis of the larger data set produced by Illumina HiSeq found V(DD)J recombination events in approximately 1 in 25,000 B cells.
These preliminary studies offer a glimpse into the depth of information made available by antibody repertoire analysis. While rare, there are unusual recombination events that contribute to repertoire diversification with unusual structural elements, such as the formation of long CDR3 loops, which are important in the recognition and neutralization of viruses such as HIV. Repertoire sequencing has delineated particular areas of structural plasticity in immunoglobulins that accommodate insertions and deletions [13], however the studies reveal that most long CDRH3 loops are formed at the time of recombination through use of long D and J segments and extended N addition regions, not by insertions [14].
Predicting the repertoire size
Repertoire diversification leads to the generation of a large population of unique antibody sequences. It is theorized that the repertoire may contain up to 1011 sequences, however laboratory studies suggest the circulating population B cells contains far fewer sequences.
One study applied the “birthday paradox” from probability theory, which concerns the probability of two people in a population of n random individuals sharing a birthday, the paradox being that it takes far fewer individuals than would otherwise be assumed to generate a 99% probability that two share a birthday. The study estimated that there are minimally 2 × 106 unique rearrangements in the peripheral blood compartment [15]. This algorithm, however, does not estimate the upper boundary of unique sequences due to the possibility of very low copy number sequences that are not observed using current sequencing techniques.
Roche 454 method sequencing can be used to generate large antibody sequence databases from human PBMC samples. Using these data, the total number of productive CDRH3 sequences in each of two healthy human subjects was calculated following a simple algorithm [16]. The number of unique sequences added to the repertoire per 1,000 additional sequences was counted and found to decrease regularly, following a pattern of logarithmic decay. The point where no additional unique sequences would be observed was calculated, and that value was expanded to encompass the total blood volume of a human adult. The upper bound of the circulating human CDRH3 repertoire was estimated to be between 3 and 9 million unique sequences [16]. As stated previously, the four principal recognized mechanism underlying antibody diversification result in a theorized population of 1011 possible antibody sequences, far more than this technique predicted in the circulating repertoire.
Global repertoire regulation across individuals
Regulatory mechanisms exist that account for the inconsistency between the theorized number of possible recombined antibody sequences and the actual number of unique sequences observed to be circulating in the blood. For example, it is known that self-reactive antibodies are removed from the population by negative selection of B cells during early B cell development. Antibody repertoire studies have shown recently that these mechanisms of regulation seem common among many individuals, suggesting that global regulatory mechanisms may be more sophisticated than previously theorized.
One study quantitated the presence of the same CDRH3 amino acid sequence in two different individuals, also referred to as the overlap of sequences between two repertoires [16]. Synthetic CDRH3 repertoires then were generated computationally using knowledge-based rules developed from actual human antibody repertoires. The number of sequences the two synthetic data sets shared was related directly to the mutation rate used to develop those data sets. From these data, the researchers were able to determine that the overlap between two different CDRH3 repertoires occurs significantly more frequently than would be expected by chance, supporting the possibility of a global mechanism for antibody repertoire regulation [16]. It is now possible to conceive of several types of repertoires (see Figure 3): 1) Private repertoires, derived from the clones of one donor, 2) Shared (or public) repertoires, representing antibody sequences found in more than one donor, and 3) Global repertoires, representing the collection of all antibody sequences in a population of subjects.
Figure 3.

Types of repertoires: Private, from one donor, 2) Shared, sequences found in two or more donors, and 3) Global, the sequences in a population of subjects.
Cell surface markers can be used to sort naïve and memory B cell subsets prior to high throughput sequencing. In one study, such sequence data then was analyzed in the context of V(D)J recombinations to find that the hypothesized global mechanism of regulation results in increased oligoclonality in memory repertoire subsets when compared to the naïve B cell subset repertoire [17]. Furthermore, phylogenetic clustering revealed that subset repertoires cluster exclusively in an inter-donor dependent manner among four donors, revealing that the similarities between inter-donor repertoire subsets were significantly greater than the similarities between intra-donor repertoire subsets.
Conclusions
Recent and ongoing development of high-throughput amplicon sequence analysis techniques is providing a new and detailed view of the complexity and composition of human B cell repertoires. These technologies will continue to evolve, with the most likely next leap the acquisition of the ability to link heavy and light chain sequences at high throughput with high facility. Proteomics sequencing of the expressed antibody repertoire in serum is on the horizon [18]. Robust computational methods for modeling the structure and function of antibodies, such as Rosetta, are starting to provide important insights [19]. Early views of the human B cell repertoire suggest that the size of antigenic-specific clones that persist after exposure to foreign pathogens is much larger than previously thought. In contrast, and perhaps paradoxically, the size of the total antibody repertoire in circulating B cells may be orders of magnitude smaller than predicted. Future studies will need to address how large epitope-specific “swarms” of somatic variants can be maintained in a repertoire of relatively small and fixed size without compromising responses to future exposures. The high level of concordance of the structure and size of repertoires between individuals suggests that there are strong regulatory programs that we understand only in part.
Highlights.
New technologies now allow determination of millions/billions of antibody sequences
An unexpectedly high number of somatic variants for each B cell clone is sustained
The size of total expressed antibody repertoires is much smaller than anticipated
Shared features of repertoires between individuals suggests high level regulation
Emerging proteomics and computational methods complement DNA sequencing
Acknowledgments
This work was supported by Defense Threat Reduction Agency (U.S. Department of Defense) Award Number HDTRA1-13-1-0034 and NIAID/NIH grant R01 AI106002. JAF is supported by the Vanderbilt Virology Training Program (T32 AI 89554, NIAID/NIH). JEC is the Ann Scott Carell chair at Vanderbilt University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
(Papers of special interest (*) or outstanding interest (**).
- 1.Lefranc M-P, Lefranc G. The Immunoglobulin FactsBook. Academic Press; 2001. [Google Scholar]
- 2.Crotty S, Ahmed R. Immunological memory in humans. Sem Immunol. 2004;16:197–203. doi: 10.1016/j.smim.2004.02.008. [DOI] [PubMed] [Google Scholar]
- *3.Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Briney BS, Smith SA, Basler CF, Crowe JE., Jr Epitope-specific human influenza antibody repertoires diversify by B cell intraclonal sequence divergence and interclonal convergence. J Immunol. 2011;187:3704–11. doi: 10.4049/jimmunol.1101823. This is one of the first papers to reveal the persistence of highly related somatic variants of antibodies within clones responding to a virus, and also shows that genetically related but independent clones converge on common sequences during epitope-specific responses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lefranc MP, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, Wu Y, Gemrot E, Brochet X, Lane J, et al. IMGT, the international ImMunoGeneTics information system. Nucl Acids Res. 2009;37:D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munshaw S, Kepler TB. SoDA2: A hidden Markov model approach for identification of immunoglobulin rearrangements. Bioinformatics. 2010;26:867–72. doi: 10.1093/bioinformatics/btq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Souto-Carneiro MM, Longo NS, Russ DE, Sun H, Lipsky PE. Characterization of the human immunoglobulin heavy chain antigen binding complementarity determining region 3 using a newly-developed software algorithm, JOINSOLVER. JImmunol. 2004;172:6790–6802. doi: 10.4049/jimmunol.172.11.6790. [DOI] [PubMed] [Google Scholar]
- *7.Zhu J, Ofek G, Yang Y, Zhang B, Louder MK, Lu G, McKee K, Pancera M, Skinner J, Zhang Z, et al. Mining the antibodyome for HIV-1–neutralizing antibodies with next-generation sequencing and phylogenetic pairing of heavy/light chains. PNAS. 2013;110:6470–6475. doi: 10.1073/pnas.1219320110. Using phylogenetic predictions, the authors explored a high-throughput sequence database to identify new virus-neutralizing antibodies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reddy ST, Ge X, Miklos AE, Hughes RA, Kang SH, Hoi KH, Chrysostomou C, Hunicke-Smith SP, Iverson BL, Tucker PW, et al. Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat Biotech. 2010;28:957–961. doi: 10.1038/nbt.1673. [DOI] [PubMed] [Google Scholar]
- **9.DeKosky BJ, Ippolito GC, Deschner RP, Lavinder JJ, Wine Y, Rawlings BM, Varadarajan N, Giesecke C, Dörner T, Andrews SF, et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat Biotech. 2013;31:166–169. doi: 10.1038/nbt.2492. Identification of the natural pairings of heavy and light chain antibody genes will lead to the ability to both analyze and synthesize new antibodies at an unprecedented scale. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White AK, VanInsberghe M, Petriv OI, Hamidi M, Sikorski D, Marra MA, Piret J, Aparicio S, Hansen CL. High-throughput microfluidic single-cell RT-qPCR. Proc Nat Acad Sci USA. 2011;108:13999–14004. doi: 10.1073/pnas.1019446108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briney BS, Willis JR, Hicar MD, Thomas JW, 2nd, Crowe JE., Jr Frequency and genetic characterization of V(DD)J recombinants in the human peripheral blood antibody repertoire. Immunology. 2012;137:56–64. doi: 10.1111/j.1365-2567.2012.03605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larimore K, McCormick MW, Robins HS, Greenberg PD. Shaping of human germline IgH repertoires revealed by deep sequencing. J Immunol. 2012;189:3221–3230. doi: 10.4049/jimmunol.1201303. [DOI] [PubMed] [Google Scholar]
- 13.Briney BS, Willis JR, Crowe JE., Jr Location and length distribution of somatic hypermutation-associated DNA insertions and deletions reveals regions of antibody structural plasticity. Genes Immun. 2012;13:523–9. doi: 10.1038/gene.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Briney BS, Willis JR, Crowe JE., Jr Human peripheral blood antibodies with long HCDR3s are established primarily at original recombination using a limited subset of germline genes. PLoS One. 2012;7:e36750. doi: 10.1371/journal.pone.0036750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyd SD, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, Jones CD, Simen BB, Hanczaruk B, Nguyen KD, et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Science Translational Medicine. 2009;1:12ra23. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arnaout R, Lee W, Cahill P, Honan T, Sparrow T, Weiand M, Nusbaum C, Rajewsky K, Koralov SB. High-resolution description of antibody heavy-chain repertoires in humans. PLoS One. 2011;6:e22365. doi: 10.1371/journal.pone.0022365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Briney BS, Willis JR, McKinney BA, Crowe JE., Jr High -throughput antibody sequencing reveals genetic evidence of global regulation of the naïve and memory repertoires that extends across individuals. Genes Immun. 2012;13:469–473. doi: 10.1038/gene.2012.20. [DOI] [PubMed] [Google Scholar]
- 18**.Cheung WC, Beausoleil SA, Zhang X, Sato S, Schieferl SM, Wieler JS, Beaudet JG, Ramenani RK, Popova L, Comb MJ, et al. A proteomics approach for the identification and cloning of monoclonal antibodies from serum. Nat Biotechnol. 2012;30:447–52. doi: 10.1038/nbt.2167. Antibodies in serum, which are commonly measured in clinical studies, are usually secreted by long-lived plasma cells in the bone marrow, while repertoire studies using antibody genes are currently defined by sequencing mRNA or genomic DNA from peripheral blood B cells. This study suggests that it will be possible to sequence the amino acids in CDRs of antibody proteins in serum for comparative purposes. [DOI] [PubMed] [Google Scholar]
- 19**.Willis JR, Briney BS, DeLuca SL, Crowe JE, Jr, Meiler J. Human germline antibody gene segments encode polyspecific antibodies. PLoS Comput Biol. 2013;9:e1003045. doi: 10.1371/journal.pcbi.1003045. The scale of antibody sequences that can be obtained now exceeds our ability to test or validate the structure, function, or specificity of identified antibodies in the laboratory. This paper shows that Rosetta software suite can explore the structural diversity of the antibody repertoire accurately using massive computational experiments. The paper also shows that many of the framework residues encoded by germline gene sequences facilitate the polyspecific response that is essential to broad recognition of diverse epitopes by the naïve repertoire. [DOI] [PMC free article] [PubMed] [Google Scholar]