Abstract
AIM: To investigate the expression and clinical relevance of inhibitor of differentiation (ID) proteins in biliary tract cancer.
METHODS: ID protein expression was analyzed in 129 samples from patients with advanced biliary tract cancer (BTC) (45 extrahepatic, 50 intrahepatic, and 34 gallbladder cancers), compared to normal controls and correlated with clinical an pathological parameters.
RESULTS: ID1-3 proteins are frequently overexpressed in all BTC subtypes analyzed. No correlation between increased ID protein expression and tumor grading, tumor subtype or treatment response was detected. Survival was influenced primary tumor localization (extrahepatic vs intrahepatic and gall bladder cancer, OS 1.5 years vs 0.9 years vs 0.7 years, P = 0.002), by stage at diagnosis (OS 2.7 years in stage I vs 0.6 years in stage IV, P < 0.001), resection status and response to systemic chemotherapy. In a multivariate model, ID protein expression did not correlate with clinical prognosis. Nevertheless, there was a trend of shorter OS in patients with loss of cytoplasmic ID4 protein expression (P = 0.076).
CONCLUSION: ID protein expression is frequently deregulated in BTC but does not influence clinical prognosis. Their usefulness as prognostic biomarkers in BTC is very limited.
Keywords: Biliary tract cancer, Cholangiocarcinoma, Inhibitor of differentiation, Prognostic factors
Core tip: Cholangiocarcinoma present as heterogeneous tumors with generally poor prognosis. Molecular changes that drive tumor development are poorly understood, and no valid prognostic markers other than stage and performance status have been identified. Here we analyzed the protein expression of the four inhibitor of differentiation (ID)-proteins by immunohistochemistry in 129 patients with advanced biliary tract cancer, which showed a deregulated ID protein expression in cancer cells and this protein expression partly correlated with the overall survival of patients. Therefore the ID-proteins maybe useful prognostic markers.
INTRODUCTION
Cholangiocarcinomas/biliary tract cancers (BTC) form a heterogeneous group of tumors consisting of intrahepatic mass forming type biliary tract cancer (IHC), perihilar Klatskin tumors, extrahepatic BTC (EHC), and gallbladder cancer (GBC)[1,2]. More than 50% of tumors are diagnosed at an advanced stage. Prognosis is dismal with a mean overall survival of 7 to 8 mo. Although expression of oncogenes such as K-ras, c-myc, c-neu, c-met, and bcl-2, or inactivation of tumor suppressor genes like p53 in BTC has been described[3,4], the pathogenesis and molecular biology of this tumor entity is poorly understood. A recent analysis of liver-fluke associated biliary tract cancer on eight tumors and matched normal tissue identified mutations in three signaling pathways, namely histone modification, G protein activation, and genomic instability[5]. Validation of 15 of the identified 187 mutated genes in another 46 cases of BTC confirmed the role of p53, K-ras, and SMAD-4, and identified additional mutations in genes not prior associated to BTC[5].
The inhibitor of DNA-binding (ID) proteins, ID1-4, are members of the larger family of basic Helix-Loop-Helix (bHLH) transcription factors, which share a basic domain necessary for DNA-binding[6]. ID proteins lack this DNA-binding domain and inhibit transcription of target genes such as p21CIP1/WAF1, p16INK4B, and pRb by forming DNA-binding incompetent heterodimers with other bHLH factors. Various cellular processes are regulated by individual ID-proteins: Inhibition of cellular differentiation by interference with differentiation-specific bHLH and non-bHLH transcription factors[7], extension of cellular life span[8-10], regulation of angiogenesis[11] and maintenance of embryonic and adult stem cells[12], and chromosomal instability[13,14]. ID expression is deregulated in many tumors including pancreatic cancer[6] a malignancy somehow related to BTC. In some cases ID-expression is associated with poor clinical prognosis[15]. Until now, no data on ID protein expression in biliary tract cancer is available, although methylation of the ID4 promoter has been reported in some instances[16].
To investigate the role of ID proteins in BTC we analyzed the expression of ID proteins 1-4 in tumor specimen from 129 patients with advanced BTC and in 9 normal controls by immunohistochemistry (IHC).
MATERIALS AND METHODS
Archival tumor samples and controls
The institutional database of the University Medical Center Freiburg, Germany, was retrospectively searched for patients presenting with advanced BTC between 1996 and 2007. Archival hematoxylin and eosin (HE) stained slides from routinely processed paraffin-embedded samples collected at time of the initial diagnosis were reviewed to verify the diagnosis and to choose representative blocks for further evaluation. Control tissue samples included normal liver with normal biliary structures obtained from 9 male autopsy cases that had died without any evidence of liver or cardiac disease. Samples from tonsils, lymph nodes and colon mucosa were used as positive controls. The study was in accordance with the ethical standards set by the institutional ethics committee and was conducted according to the declaration of Helsinki.
Staging
Staging was performed according to UICC/AJCC recommendations.
Immunohistochemistry
Fresh serial sections were cut at 2 μm from the original diagnostic paraffin-embedded tissue blocks, mounted on Superfrost Plus slides, dried overnight at 37 °C, deparaffinized in xylene and rehydrated through a graded series of alcohol solutions. Antigen retrieval was performed in Target Retrieval Solution (ID1, ID2, ID4: pH6; ID3: pH8; Dako, Glostrup, Denmark) at 95 °C for 60 min. Immunostaining was performed using a semi-automatically autostainer (Dako). The sections were incubated for 60 min at room temperature with primary polyclonal rabbit antibodies against relevant ID proteins (Santa Cruz Biotechnology, Santa Cruz, CA, United States) at different dilutions (ID1, C-20: sc-488 at 1:150; ID2: C-20: sc-489 at 1:1000; ID3, C-20: sc-490 at 1:100; ID4: H-70: sc-13047 at 1:100). After washing with PBS, the samples were incubated with biotinylated goat anti-polyvalent antibodies for 15 min and subsequently by streptavidin alkaline phosphatase for 15 min according to the labeled streptavidin-biotin (LSAB) method. The ID protein binding sections was visualized by K 5005, Fast-Red-Chromogen (Dako) detection for 10 min. Nuclei were counterstained with Mayer´s hemalaun solution. Vascular smooth muscle cells served as internal positive controls for the ID proteins. As a negative control, the primary antibody was omitted, with all other experimental conditions kept constant. The staining specificity was further confirmed by blocking the antibody binding to the antigen with the corresponding blocking peptides for ID1, ID2 and ID3 (Santa Cruz). No blocking peptide was commercially available for ID4.
Scoring of the immunohistochemical staining
Two blinded observers independently evaluated all cases. For cases with discordant scoring results, a consensus was obtained by reevaluating the slides. When the tumor samples showed a heterogeneous staining pattern, the “hot spots” showing the highest staining intensity were selected for further quantification. Cytoplasmic and nuclear immunolabeling were evaluated. Cytoplasmic expression was scored according to the staining intensity (0, negative; 1, low; 2, moderate to 3, strong). Nuclear staining was scored according to the percentage of positive nuclei. At least 100 nuclei were assessed in the hot spots at high magnification (× 400). The percentage of positive cells was rated as follows: 0: 0%-10%, 1: 11%-50%, 2: 51%-80%, and 3: 81%-100% of tumor cell nuclei positive.
Statistical analysis
Statistical analysis was performed using SPSS 15.0 statistical software (SPSS, Inc, Chicago, Illinois). Both the analysis of the association of ID1-4 expression levels in BTC with the overall survival (OS) and the correlation of tumor subtypes with OS were performed by the chi-square test with P < 0.05 considered as statistically significant. Overall survival was defined as the interval from date of diagnosis (histopathology) until death from any cause. Additionally to the analysis with the whole study cohort (n = 129), the patients were divided into the two following subgroups: Patients treated by chemotherapy (n = 64) and patients not treated by chemotherapy (n = 56). Survival estimates were calculated using the Kaplan-Meier method. To test for independent relevance of the candidate prognostic factors a multivariate Cox proportional hazards regression model was fit for each of the ID-proteins. All baseline characteristics plus cytoplasmic and nuclear expression of the respective ID-protein were included in the model. Tumor stage and grading were dichotomized (1/2 vs 3/4).
RESULTS
We identified 129 cases of advanced BTC (unresectable at time of presentation in our institution) that had been treated at our institution between June 1998 and June 2005. Thirty-five percent (n = 45) were of extrahepatic origin (EHC), 39% (n = 50) were of intrahepatic origin (IHC), and 26% (n = 34) were gallbladder cancers (GBC). Median age at diagnosis was 62.2 years (range 32-84 years) with even gender distribution (48% female, 52% male). The majority of patients had developed metastatic disease; most tumors were G2 tumors (63%). Patients were treated with best supportive care (n = 56) and various chemotherapeutic regimens (n = 64). Nine patients had become secondary resectable (R0). Chemotherapy regimens were 5-FU or gemcitabine based, often in combination with cisplatin or oxaliplatin. Response to treatment was monitored by CT, MRI or ultrasound according to the standard WHO criteria (WHO, 1979). Eleven patients (17%) showed a partial remission, 27 (42%) had stable disease, and 26 patients (41%) had progressive disease despite chemotherapy. Follow-up data were available from all 129 patients with a median follow-up of 16.5 mo. The median overall survival for all patients was 18.3 mo (Table 1).
Table 1.
Patient characteristics, survival and best response to treatment n (%)
| Age (yr) | Median (62.2) | Range (32-84) | ||
| Sex | Male | Female | ||
| 67 (51.9) | 62 (48.1) | |||
| BTC subtype | EHC | IHC | GBC | Total |
| 45 (34.9) | 50 (38.8) | 34 (26.4) | 129 | |
| Stage | I | II | III | IV |
| 10 (7.8) | 20 (15.5) | 40 (31.0) | 65 (50.4) | |
| Grade | G1 | G2 | G3 | |
| 7 (5.4) | 81 (62.8) | 41 (31.8) | ||
| Treatment | BSC | Cx | Sx | Total |
| 56 (43.4) | 64 (49.6) | 9 (7.0) | 129 | |
| Response to Cx | PR | SD | PD | |
| 11 (17.2) | 27 (42.2) | 26 (40.6) | ||
| OS | BSC | Cx | Sx | All |
| (mo) | 15.1 | 17.7 | 41.8 | 18.3 |
BSC: Best supportive care; Sx: Surgery; Cx: Chemotherapy; PR: Partial remission; SD: Stable disease; PD: Progressive disease; EHC: Extrahepatic BTC; IHC: Intrahepatic BTC; GBC: Gallbladder cancer.
To identify prognostic subgroups standard clinical prognostic variables were correlated with clinical outcome. As expected from clinical experience, site of the primary tumor clearly influenced prognosis. Patients with EHC had a significantly longer OS compared to patients with IHC and GBC (Median OS 1.5 years vs 0.9 years vs 0.7 years, P = 0.002) (Figure 1). Overall survival from time of diagnosis was also influenced by stage at diagnosis (Stage I: 2.7 years vs stage II: 2.0 years vs stage III 0.9 years vs stage IV 0.6 years, P < 0.001). Resection status also influenced survival, which was longer in patients with completely resected (R0) tumors compared to incompletely resected (R1/R2) and not surgically treated tumors (OS 2.0 years vs 1.3 years vs 0.6 years, P < 0.001). Likewise, OS was better in patients whose tumors responded to chemotherapy (PR 2.0 years vs SD 1.3 years vs PD 0.6 years, P = 0.003). To exclude an influence of curative resection (n = 9) on the results, a sensitivity analysis excluding these cases was performed that confirmed the results (P = 0.004). Because of the small sample size no additional subgroup analyses were performed.
Figure 1.
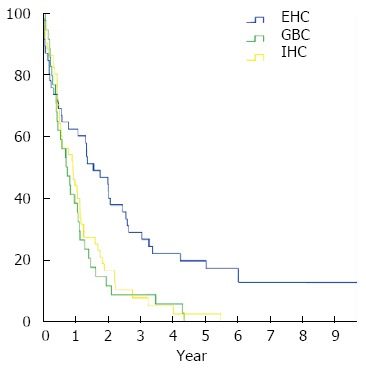
Survival of patients with biliary tract cancer by primary tumor localization. Kaplan-Meier estimates of overall survival in patients with extrahepatic biliary tract cancer (BTC) (n = 45, blue line), intrahepatic BTC (n = 50, yellow line), and gall bladder cancer (n = 34, green line).
ID proteins are frequently expressed in BTC
To determine the role of the ID proteins in BTC, we analyzed ID protein expression and subcellular localization of ID proteins in archival samples from all 129 patients collected at initial diagnosis. Normal bile ducts expressed only low amounts of ID proteins with the exception of ID4, where strong nuclear expression of ID4 was detected in all cases analyzed. In contrast to normal controls, high levels of ID1, ID2 and ID3 were detected in the majority of BTC cases (Figure 2). Table 2 summarizes the results.
Figure 2.
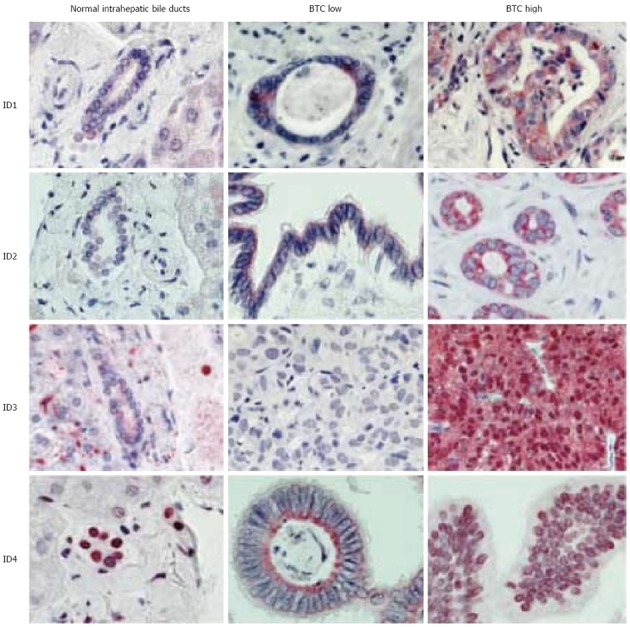
Inhibitor of differentiation protein expression in normal intrahepatic bile ducts and biliary tract cancer. Representative photographs. Depicted are representative stains of normal intrahepatic bile ducts, low and high expressing biliary tract cancer cases for each inhibitor of differentiation (ID) protein.
Table 2.
Inhibitor of differentiation protein expression and tumor characteristics n (%)
| Total (n) | IHC | EHC | GBC | Stage I | Stage II | Stage III | Stage IV | G1 | G2 | G33 | |
| ID1 neg | 17 | 6 (5) | 6 (5) | 5 (4) | 1 (1) | 2 (2) | 4 (3) | 10 (8) | 1 (1) | 9 (7) | 7 (5) |
| ID1 pos | 112 | 44 (34) | 39 (30) | 29 (22) | 9 (7) | 18 (14) | 30 (23) | 55 (43) | 6 (5) | 72 (56) | 34 (26) |
| ID2 neg | 29 | 13 (10) | 9 (7) | 7 (5) | 1 (1) | 3 (2) | 9 (7) | 16 (12) | 2 (2) | 19 (15) | 8 (6) |
| ID2 pos | 100 | 37 (29) | 36 (28) | 27 (21) | 9 (7) | 17 (13) | 25 (19) | 49 (38) | 5 (4) | 62 (48) | 33 (26) |
| ID3 neg | 25 | 9 (7) | 9 (7) | 7 (5) | 1 (1) | 7 (5) | 3 (2) | 14 (11) | 2 (2) | 15 (12) | 8 (6) |
| ID3 pos | 104 | 41 (32) | 36 (28) | 27 (21) | 9 (7) | 13 (10) | 31 (24) | 51 (40) | 5 (4) | 66 (51) | 33 (36) |
| ID4 neg | 26 | 11 (9) | 9 (7) | 6 (5) | 2 (2) | 7 (6) | 9 (7) | 8 (7) | 1 (1) | 15 (12) | 10 (8) |
| ID4 pos | 96 | 35 (29) | 35 (29) | 26 (21) | 8 (7) | 13 (11) | 23 (19) | 52 (43) | 6 (5) | 61 (50) | 29 (24) |
EHC: Extrahepatic biliary tract cancer (BTC); IHC: Intrahepatic BTC; GBC: Gallbladder cancer; G: Grade.
Detailed analysis of expression levels and subcellular localization of the ID proteins showed clear differences between normal bile ducts and BTC. While expression of ID1, ID2, and ID3 was undetectable or low in normal tissue, similarly high cytoplasmic and nuclear ID1 expression was detected in 63.6%, and 61.3% of BTC cases (Figure 3). Likewise, high ID2 expression was detected, which was mainly cytoplasmic (77.5% cytoplasmic vs 4.7% nuclear). ID3 was highly expressed in the cytoplasm (40.3%) and even more pronounced in the nucleus (68.3%). In contrast, all normal controls analyzed expressed high levels of nuclear ID4, but only low levels of cytoplasmic ID4. In BTC, this predominant nuclear ID4 staining was reduced to 77.8%.
Figure 3.
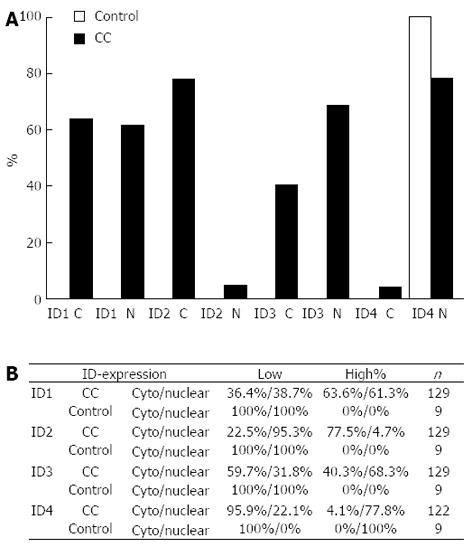
Proportion of inhibitor of differentiation protein expression in biliary tract cancer and normal intrahepatic bile ducts. A: Percentage of biliary tract cancer (BTC) with high expression of the respective inhibitor of differentiation (ID) protein in BTC (black bars) and normal intrahepatic bile ducts (control; grey bar); B: Summary of ID protein expression. Overall ID protein expression ID protein expression was scored in low (0+1) and high (2+3) and assessed for nuclear and cytoplasmic (cyto) staining pattern. Cytoplasmic staining intensity was scored 0 (negative) to 3 (strong), nuclear expression was scored based on the percentage of positive nuclei (0: 0%-10%; 1: 11%-50%; 2: 51%-80%; and 3: 81%-100%). For ID4, only 122 samples could be analyzed. C: Cytoplasmic expression; N: Nuclear expression.
To investigate the clinical relevance of the above findings, ID protein expression was correlated with overall survival (OS), tumor grade, tumor stage, prior chemotherapy, and response to chemotherapy. Correlation of ID expression with OS was calculated for the whole study cohort (n = 129; n = 122 for ID4) and for the subgroups of patients treated with chemotherapy (n = 64) and patients not treated by chemotherapy (n = 56). While neither cytoplasmic nor nuclear ID1 expression was correlated with OS, a clear trend for shorter OS was observed for nuclear negative (n = 27) vs nuclear positive (n = 102) cases (0.9 years vs 1.2 years, P = 0.058) (Figure 4A). Further subgroup analyses identified a strong correlation of ID1 expression and overall survival in chemotherapy naïve patients (n = 56). Here, patients without nuclear (n = 10) ID1 expression had an OS of 2.1 years compared to 0.5 years in patients with nuclear (n = 46) ID1 expression (P = 0.001) (Figure 4B). ID2 did not have a prognostic value for the overall study population (P = 0.79 for cytoplasmic expression, P = 0.28 for nuclear expression). Nevertheless, in the subgroup of patients who had received chemotherapy (n = 64) patients without nuclear ID2 expression (n = 51) had a significantly better prognosis than patients with nuclear ID2 expression (n = 13), with OS of 1.3 years and 0.5 years, respectively (P = 0.001) (Figure 4C). As for ID1 and ID2, ID3 expression in the overall study population did not correlate with OS (P = 0.28 for cytoplasmic, and P = 0.44 for nuclear ID3 expression). The subgroup of patients who had received chemotherapy (n = 64) patients without cytoplasmatic ID3 expression (n = 36) had a slightly better prognosis than patients with cytoplasmatic ID3 expression (n = 28), with OS of 1.0 years and 1.1 years, respectively (P = 0.037) (Figure 4D). Using the same subgroup analyses as for the other IDs, no relevant subgroup was identified. Quite strikingly, while nuclear ID4 expression did not correlate with OS (P = 0.27), cytoplasmic ID4 expression seemed to correlate with prognosis (Figure 4E). Overall survival in patients with cytoplasmic ID4 expression (n = 60) was 1.2 years compared with 0.6 years in patients without cytoplasmic ID4 expression (n = 62, P = 0.001) in univariate analysis. This was reproduced in the subgroup of patients who had not received prior chemotherapy (n = 56). Here, OS was 1.1 years vs 0.5 years in patients with cytoplasmic (n = 23) vs no cytoplasmic (n = 33) ID4 expression (P = 0.025) (Figure 4F). Analyses of ID1-4 expression and other clinical-pathological variables failed to show any significant correlation (data not shown). Specifically, ID expression levels were similar in EHC, IHC, and GBC.
Figure 4.
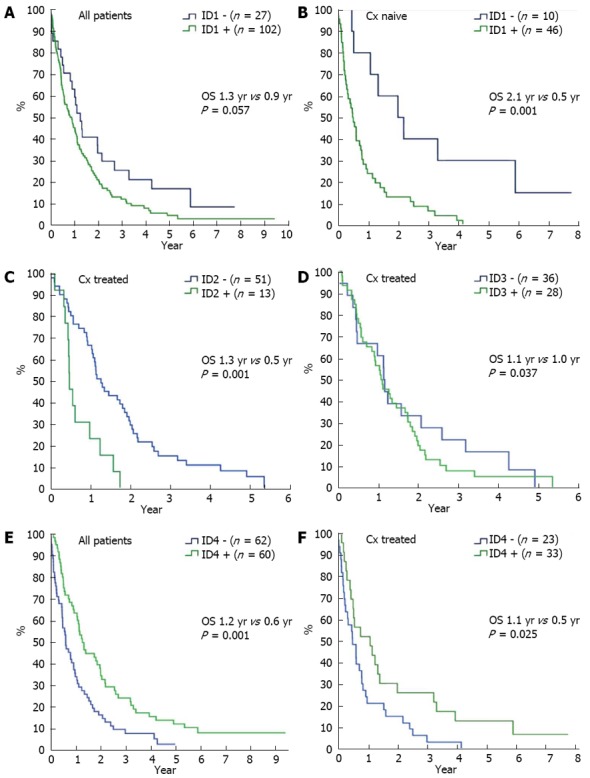
Kaplan-Meier survival estimates for overall survival from time of diagnosis for patients with biliary tract cancer expressing ID1-ID4 with or without systemic chemotherapy. A: Survival in patients (n = 129) with (green line) and without nuclear ID1 expression (blue line); B: Survival in patients who have not received systemic chemotherapy (n = 56) with (green line) or without nuclear ID1 expression (blue line); C: Survival in patients who have received systemic chemotherapy (n = 64) with (green line) or without nuclear ID2 expression (blue line); D: Survival in patients who have not received systemic chemotherapy (n = 64) with (green line) or without cytoplasmic ID3 expression (blue line); E: Survival in patients (n = 122) with (green line) and without cytoplasmic ID4 expression (blue line); F: Survival in patients who have not received systemic chemotherapy (n = 56) with (green line) or without cytoplasmic ID4 expression (blue line). Cx: Chemotherapy.
To test the relevance of the above findings a multivariate Cox proportional hazards regression model was used. Multivariate testing confirmed the importance of tumor localization, surgical treatment, and response to chemotherapy as factors influencing survival (Table 3). Patients with extrahepatic BTC have the best prognosis (HR = 0.32, 95%CI: 0.18-0.6, P < 0.0005), as have patients who had been treated surgically (HR = 0.3, 95%CI: 0.17-0.55, P < 0.0001). Patients responding to chemotherapy with a partial remission (PR) or disease stabilization (SD) likewise seem to have a better clinical prognosis (HR = 0.43, 95%CI: 0.20-0.91, P = 0.267; and HR 0.4, 95%CI: 0.22-0.71, P = 0.002, respectively). The effects of the ID proteins observed in univariate testing did not reach statistical significance in this multivariate model. Only tumors with loss of cytoplasmatic ID4 expression showed a trend for shorter survival (HR = 0.68, 95%CI: 0.45-1.04, P = 0.0736).
Table 3.
Multivariate analysis of clinical characteristics and inhibitor of differentiation protein expression
|
ID1 |
ID2 |
ID3 |
ID4 |
|||||||||
| HR | 95%CI | P value | HR | 95%CI | P value | HR | 95%CI | P value | HR | 95%CI | P value | |
| Gender (male) | 1.17 | 0.76-1.80 | 0.4717 | 1.07 | 0.71-1.64 | 0.7363 | 1.03 | 0.68-1.56 | 0.8877 | 1.29 | 0.82-2.02 | 0.267 |
| IHC | 0.57 | 0.33-0.97 | 0.0399 | 0.66 | 0.39-1.11 | 0.1193 | 0.72 | 0.42-1.23 | 0.2312 | 0.75 | 0.44-1.28 | 0.295 |
| EHC | 0.32 | 0.18-0.60 | 0.0003 | 0.36 | 0.20-0.64 | 0.0005 | 0.36 | 0.20-0.63 | 0.0003 | 0.32 | 0.18-0.57 | 0.0001 |
| Stage III-IV | 1.51 | 0.80-2.83 | 0.2037 | 1.5 | 0.80-2.80 | 0.2051 | 1.72 | 0.91-3.25 | 0.0928 | 1.24 | 0.65-2.35 | 0.5129 |
| Grade 3 | 1.57 | 0.99-2.50 | 0.0532 | 1.58 | 1.01-2.47 | 0.0471 | 1.43 | 0.90-2.27 | 0.1326 | 1.53 | 0.96-2.45 | 0.0732 |
| Surgery | 0.3 | 0.17-0.55 | < 0.0001 | 0.35 | 0.20-0.61 | 0.0003 | 0.33 | 0.18-0.61 | 0.0004 | 0.31 | 0.17-0.55 | < 0.0001 |
| Age | 0.99 | 0.97-1.01 | 0.2327 | 0.99 | 0.97-1.01 | 0.3835 | 0.99 | 0.97-1.01 | 0.3118 | 0,99 | 0.97-1.02 | 0.6452 |
| Response to Cx PD | 0.81 | 0.44-1.47 | 0.4836 | 0.83 | 0.45-1.53 | 0.5442 | 0.65 | 0.35-1.20 | 0.1669 | 0.7 | 0.37-1.31 | 0.261 |
| Response to Cx SD | 0.4 | 0.22-0.71 | 0.002 | 0.4 | 0.22-0.72 | 0.0021 | 0.33 | 0.19-0.60 | 0.0002 | 0.37 | 0.20-0.67 | 0.0012 |
| Response to Cx PR | 0.43 | 0.20-0.91 | 0.0267 | 0.48 | 0.23-0.98 | 0.0426 | 0.46 | 0.22-0.94 | 0.0329 | 0.5 | 0.25-1.01 | 0.0531 |
| Nuclear low | 0.93 | 0.46-1.88 | 0.8447 | 1.41 | 0.80-2.50 | 0.2333 | 1.32 | 0.62-2.79 | 0.4721 | 0.94 | 0.38-2.33 | 0.8979 |
| Nuclear high | 1.2 | 0.65-2.20 | 0.5563 | 1.51 | 0.19-11.84 | 0.696 | 0.97 | 0.47-2.00 | 0.9275 | 0.63 | 0.27-1.47 | 0.2813 |
| Cytoplasmic low | 0.81 | 0.34-1.91 | 0.6276 | 0.59 | 0.07-4.80 | 0.6195 | 1.0 | 0.60-1.68 | 0.9948 | 0.68 | 0.45-1.04 | 0.0736 |
| Cytoplasmic high | 0.81 | 0.34-1.90 | 0.6261 | 0.64 | 0.08-5.04 | 0.6703 | 0.71 | 0.33-1.54 | 0.3866 | 0.31 | 0.03-2.85 | 0.3028 |
n = 129, number of events = 119, r2 = 0.386 for ID1-3, r2 = 0.392 for ID4; EHC: Extrahepatic biliary tract cancer (BTC); IHC: Intrahepatic BTC; Cx: Chemotherapy; PR: Partial remission; SD: Stable disease; PD: Progressive disease; ID: Inhibitor of differentiation.
DISCUSSION
Our analysis of 129 cases of biliary tract cancer confirmed the importance of primary tumor localization, tumor stage, resection status and response the treatment as strong predictors for patients’ prognosis. This difference in survival was independent of other confounding factors such as stage, resection, or response to chemotherapy. We believe that our findings are valid, as also patients who had not been curatively resected had a significantly longer OS (P = 0.004). Hence, information of primary BTC localization should be prospectively evaluated and used for stratification in clinical trials in advanced BTC to verify its prognostic relevance. It is hard to speculate on factors influencing this different prognosis. One report could hint to a role of the multidrug resistance proteins (MRP) showing lower levels of MRP3 in EHC compared to gallbladder carcinomas[17]. Alternatively, differential gene expression as of matrix metalloproteinases and growth factors might contribute to the different biological behavior of EHC[3,18].
While data on ID protein expression in BTC was scarce, more comprehensive data is available for ID expression in hepatocellular carcinoma (HCC). In HCC, ID1 is frequently expressed and higher ID1 expression was reported to correlate with decreased p16INK4A expression[19]. ID protein expression decreased with loss of differentiation, suggesting a role of ID proteins in early carcinogenesis[20]. Contrasting these results are data that imply a role of ID1 in progression, metastasis, and tumor vessel formation[19,21-23]. Immunohistochemical analysis of liver tissue from 112 patients with liver cirrhosis showed elevated ID1 expression in 38% (n = 42) of patients. These patients were at higher risk of developing HCC[23]. In an analysis of 80 matched pair biopsies of HCC and normal liver, and cirrhotic and chronic hepatitis samples no ID1-protein expression was observed in normal liver, whereas moderate to strong ID1 expression was detected in 50% of HCCs. In 60 matched pairs of primary tumor and metastasis, expression in the metastases was higher (90%) than in the primary tumors (42%), correlating with increased VEGF expression[22]. In a nude mouse model ID1 induced VEGF by stabilizing HIF alpha, and antisense-inhibition of ID1 resulted in decreased tumor growth due to decreased VEGF expression and decreased tumor vascularization[22].
Analysis of ID protein expression in our cohort of cholangiocarcinomas showed expression of all four ID proteins. ID4 was the only ID protein expressed in normal bile ducts. Neither overall ID protein expression levels nor subcellular localization correlated with clinical prognosis. While loss of ID4 protein expression was frequently detected and patients with ID4 negative tumors had a trend for shorter OS this needs to be confirmed in a larger sample. The role of ID4 on tumor development is still not fully understood. Recent studies revealed that the ID4 gene can be silenced through promoter hypermethylation in various tumors, including BTC[16,24-30], suggesting a tumor suppressive role. On the other hand ID4 was identified as an upstream regulator of BRCA1 in breast and ovarian cancer[31]. Also supporting an oncogenic role of ID4 are reports describing activating translocations of ID4 in some patients with bladder cancer and a subset of patients with acute lymphoblastic leukemia[32-34]. Our data suggest a tumor suppressive role of ID4 in BTC, as patients without or with low cytoplasmic levels of ID4 had shorter overall survival. While hypothesis generating, this must be confirmed and validated in a larger patient population. In summary, we have shown that ID protein expression is deregulated in biliary tract cancer but that their use as prognostic biomarkers is very limited.
ACKNOWLEDGMENTS
The authors thank Blum H, and Mertelsmann R for continuous support.
COMMENTS
Background
Cholangiocarcinoma present as heterogeneous tumors with generally poor prognosis. Molecular changes that drive tumor development are poorly understood, and no valid prognostic markers other than stage and performance status have been identified.
Innovations and breakthroughs
To investigate the role of inhibitor of differentiation (ID) proteins in biliary tract cancer (BTC) we analyzed the expression of ID proteins 1-4 in tumor specimen from 129 patients with advanced BTC and in 9 normal controls by immunohistochemistry.
Applications
Authors have shown that ID protein expression is deregulated in biliary tract cancer but that their use as prognostic biomarkers is very limited.
Peer review
The authors described the expressions of ID1-4 proteins in specimens of cholangiocarcinoma and compared to the normal control. An interesting paper looking for new histological parameters in cholangiocarcinoma.
Footnotes
P- Reviewers: Chow WK, Esposito I, Lin ZY, Ramia JM, Wakai T S- Editor: Wen LL L- Editor: A E- Editor: Zhang DN
References
- 1.Malhi H, Gores GJ. Cholangiocarcinoma: modern advances in understanding a deadly old disease. J Hepatol. 2006;45:856–867. doi: 10.1016/j.jhep.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Groen PC, Gores GJ, LaRusso NF, Gunderson LL, Nagorney DM. Biliary tract cancers. N Engl J Med. 1999;341:1368–1378. doi: 10.1056/NEJM199910283411807. [DOI] [PubMed] [Google Scholar]
- 3.Harder J, Waiz O, Otto F, Geissler M, Olschewski M, Weinhold B, Blum HE, Schmitt-Graeff A, Opitz OG. EGFR and HER2 expression in advanced biliary tract cancer. World J Gastroenterol. 2009;15:4511–4517. doi: 10.3748/wjg.15.4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nault JC, Zucman-Rossi J. Genetics of hepatobiliary carcinogenesis. Semin Liver Dis. 2011;31:173–187. doi: 10.1055/s-0031-1276646. [DOI] [PubMed] [Google Scholar]
- 5.Ong CK, Subimerb C, Pairojkul C, Wongkham S, Cutcutache I, Yu W, McPherson JR, Allen GE, Ng CC, Wong BH, Myint SS, Rajasegaran V, Heng HL, Gan A, Zang ZJ, Wu Y, Wu J, Lee MH, Huang D, Ong P, Chan-on W, Cao Y, Qian CN, Lim KH, Ooi A, Dykema K, Furge K, Kukongviriyapan V, Sripa B, Wongkham C, Yongvanit P, Futreal PA, Bhudhisawasdi V, Rozen S, Tan P, Teh BT. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat Genet. 2012;44:690–693. doi: 10.1038/ng.2273. [DOI] [PubMed] [Google Scholar]
- 6.Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer. 2005;5:603–614. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- 7.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 8.Alani RM, Hasskarl J, Grace M, Hernandez MC, Israel MA, Münger K. Immortalization of primary human keratinocytes by the helix-loop-helix protein, Id-1. Proc Natl Acad Sci USA. 1999;96:9637–9641. doi: 10.1073/pnas.96.17.9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang J, Gordon GM, Nickoloff BJ, Foreman KE. The helix-loop-helix protein id-1 delays onset of replicative senescence in human endothelial cells. Lab Invest. 2002;82:1073–1079. doi: 10.1097/01.lab.0000022223.65962.3a. [DOI] [PubMed] [Google Scholar]
- 10.Nickoloff BJ, Chaturvedi V, Bacon P, Qin JZ, Denning MF, Diaz MO. Id-1 delays senescence but does not immortalize keratinocytes. J Biol Chem. 2000;275:27501–27504. doi: 10.1074/jbc.C000311200. [DOI] [PubMed] [Google Scholar]
- 11.Lyden D, Hattori K, Dias S, Costa C, Blaikie P, Butros L, Chadburn A, Heissig B, Marks W, Witte L, et al. Impaired recruitment of bone-marrow-derived endothelial and hematopoietic precursor cells blocks tumor angiogenesis and growth. Nat Med. 2001;7:1194–1201. doi: 10.1038/nm1101-1194. [DOI] [PubMed] [Google Scholar]
- 12.Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 13.Hasskarl J, Duensing S, Manuel E, Münger K. The helix-loop-helix protein ID1 localizes to centrosomes and rapidly induces abnormal centrosome numbers. Oncogene. 2004;23:1930–1938. doi: 10.1038/sj.onc.1207310. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Di K, Zhang X, Han HY, Wong YC, Leung SC, Ling MT. Id-1 promotes chromosomal instability through modification of APC/C activity during mitosis in response to microtubule disruption. Oncogene. 2008;27:4456–4466. doi: 10.1038/onc.2008.87. [DOI] [PubMed] [Google Scholar]
- 15.Hasskarl J, Münger K. Id proteins--tumor markers or oncogenes? Cancer Biol Ther. 2002;1:91–96. doi: 10.4161/cbt.50. [DOI] [PubMed] [Google Scholar]
- 16.Uhm KO, Lee ES, Lee YM, Kim HS, Park YN, Park SH. Aberrant promoter CpG islands methylation of tumor suppressor genes in cholangiocarcinoma. Oncol Res. 2008;17:151–157. doi: 10.3727/096504008785114110. [DOI] [PubMed] [Google Scholar]
- 17.Rau S, Autschbach F, Riedel HD, Konig J, Kulaksiz H, Stiehl A, Riemann JF, Rost D. Expression of the multidrug resistance proteins MRP2 and MRP3 in human cholangiocellular carcinomas. Eur J Clin Invest. 2008;38:134–142. doi: 10.1111/j.1365-2362.2007.01916.x. [DOI] [PubMed] [Google Scholar]
- 18.Kirimlioğlu H, Türkmen I, Başsüllü N, Dirican A, Karadağ N, Kirimlioğlu V. The expression of matrix metalloproteinases in intrahepatic cholangiocarcinoma, hilar (Klatskin tumor), middle and distal extrahepatic cholangiocarcinoma, gallbladder cancer, and ampullary carcinoma: role of matrix metalloproteinases in tumor progression and prognosis. Turk J Gastroenterol. 2009;20:41–47. [PubMed] [Google Scholar]
- 19.Lee TK, Man K, Ling MT, Wang XH, Wong YC, Lo CM, Poon RT, Ng IO, Fan ST. Over-expression of Id-1 induces cell proliferation in hepatocellular carcinoma through inactivation of p16INK4a/RB pathway. Carcinogenesis. 2003;24:1729–1736. doi: 10.1093/carcin/bgg145. [DOI] [PubMed] [Google Scholar]
- 20.Damdinsuren B, Nagano H, Kondo M, Yamamoto H, Hiraoka N, Yamamoto T, Marubashi S, Miyamoto A, Umeshita K, Dono K, et al. Expression of Id proteins in human hepatocellular carcinoma: relevance to tumor dedifferentiation. Int J Oncol. 2005;26:319–327. [PubMed] [Google Scholar]
- 21.Lee KT, Lee YW, Lee JK, Choi SH, Rhee JC, Paik SS, Kong G. Overexpression of Id-1 is significantly associated with tumour angiogenesis in human pancreas cancers. Br J Cancer. 2004;90:1198–1203. doi: 10.1038/sj.bjc.6601684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee TK, Poon RT, Yuen AP, Ling MT, Wang XH, Wong YC, Guan XY, Man K, Tang ZY, Fan ST. Regulation of angiogenesis by Id-1 through hypoxia-inducible factor-1alpha-mediated vascular endothelial growth factor up-regulation in hepatocellular carcinoma. Clin Cancer Res. 2006;12:6910–6919. doi: 10.1158/1078-0432.CCR-06-0489. [DOI] [PubMed] [Google Scholar]
- 23.Matsuda Y, Yamagiwa S, Takamura M, Honda Y, Ishimoto Y, Ichida T, Aoyagi Y. Overexpressed Id-1 is associated with a high risk of hepatocellular carcinoma development in patients with cirrhosis without transcriptional repression of p16. Cancer. 2005;104:1037–1044. doi: 10.1002/cncr.21259. [DOI] [PubMed] [Google Scholar]
- 24.Chan AS, Tsui WY, Chen X, Chu KM, Chan TL, Chan AS, Li R, So S, Yuen ST, Leung SY. Downregulation of ID4 by promoter hypermethylation in gastric adenocarcinoma. Oncogene. 2003;22:6946–6953. doi: 10.1038/sj.onc.1206799. [DOI] [PubMed] [Google Scholar]
- 25.Umetani N, Takeuchi H, Fujimoto A, Shinozaki M, Bilchik AJ, Hoon DS. Epigenetic inactivation of ID4 in colorectal carcinomas correlates with poor differentiation and unfavorable prognosis. Clin Cancer Res. 2004;10:7475–7483. doi: 10.1158/1078-0432.CCR-04-0689. [DOI] [PubMed] [Google Scholar]
- 26.Noetzel E, Veeck J, Niederacher D, Galm O, Horn F, Hartmann A, Knüchel R, Dahl E. Promoter methylation-associated loss of ID4 expression is a marker of tumour recurrence in human breast cancer. BMC Cancer. 2008;8:154. doi: 10.1186/1471-2407-8-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagiwara K, Nagai H, Li Y, Ohashi H, Hotta T, Saito H. Frequent DNA methylation but not mutation of the ID4 gene in malignant lymphoma. J Clin Exp Hematop. 2007;47:15–18. doi: 10.3960/jslrt.47.15. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Wang XQ, Xu XP, Lin GW. ID4 methylation predicts high risk of leukemic transformation in patients with myelodysplastic syndrome. Leuk Res. 2010;34:598–604. doi: 10.1016/j.leukres.2009.09.031. [DOI] [PubMed] [Google Scholar]
- 29.Chen SS, Claus R, Lucas DM, Yu L, Qian J, Ruppert AS, West DA, Williams KE, Johnson AJ, Sablitzky F, et al. Silencing of the inhibitor of DNA binding protein 4 (ID4) contributes to the pathogenesis of mouse and human CLL. Blood. 2011;117:862–871. doi: 10.1182/blood-2010-05-284638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu L, Liu C, Vandeusen J, Becknell B, Dai Z, Wu YZ, Raval A, Liu TH, Ding W, Mao C, et al. Global assessment of promoter methylation in a mouse model of cancer identifies ID4 as a putative tumor-suppressor gene in human leukemia. Nat Genet. 2005;37:265–274. doi: 10.1038/ng1521. [DOI] [PubMed] [Google Scholar]
- 31.Beger C, Pierce LN, Kruger M, Marcusson EG, Robbins JM, Welcsh P, Welch PJ, Welte K, King MC, Barber JR, et al. Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach. Proc Natl Acad Sci USA. 2001;98:130–135. doi: 10.1073/pnas.98.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bellido M, Aventín A, Lasa A, Estivill C, Carnicer MJ, Pons C, Matías-Guiu X, Bordes R, Baiget M, Sierra J, et al. Id4 is deregulated by a t(6; 14)(p22; q32) chromosomal translocation in a B-cell lineage acute lymphoblastic leukemia. Haematologica. 2003;88:994–1001. [PubMed] [Google Scholar]
- 33.Wu Q, Hoffmann MJ, Hartmann FH, Schulz WA. Amplification and overexpression of the ID4 gene at 6p22.3 in bladder cancer. Mol Cancer. 2005;4:16. doi: 10.1186/1476-4598-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russell LJ, Akasaka T, Majid A, Sugimoto KJ, Loraine Karran E, Nagel I, Harder L, Claviez A, Gesk S, Moorman AV, et al. t(6; 14)(p22; q32): a new recurrent IGH@ translocation involving ID4 in B-cell precursor acute lymphoblastic leukemia (BCP-ALL) Blood. 2008;111:387–391. doi: 10.1182/blood-2007-07-092015. [DOI] [PubMed] [Google Scholar]


