Abstract
Aims
Myotonic dystrophy types 1 and 2 (DM1 and DM2) are multisystem disorders caused by similar repeat expansion mutations, with similar yet distinct clinical features. Aberrant splicing of multiple effector genes, as well as dysregulation of transcription and translation, have been suggested to underlie different aspects of the complex phenotypes in DM1 and DM2. Ca2+ plays a central role in both muscle contraction and control of gene expression, and recent expression profiling studies have indicated major perturbations of the Ca2+ signaling pathways in DM. Here we have further investigated the expression of genes and proteins involved in Ca2+ metabolism in DM patients, including Ca2+ channels and Ca2+ binding proteins.
Methods
We used patient muscle biopsies to analyze mRNA expression and splicing of genes by microarray expression profiling and RT-PCR. We studied protein expression by immunohistochemistry and immunoblotting.
Results
Most of the genes studied showed mRNA up-regulation in expression profiling. When analyzed by immunohistochemistry the Ca2+ release channel ryanodine receptor was reduced in DM1 and DM2, as was calsequestrin 2, a sarcoplasmic reticulum lumen Ca2+ storage protein. Abnormal splicing of ATP2A1 was more pronounced in DM2 than DM1.
Conclusions
We observed abnormal mRNA and protein expression in DM affecting several proteins involved in Ca2+ metabolism, with some differences between DM1 and DM2. Our protein expression studies are suggestive of a post-transcriptional defect(s) in the myotonic dystrophies.
Keywords: Myotonic dystrophy type 1 (DM1), myotonic dystrophy type 2 (DM2), skeletal muscle, calcium metabolism
Introduction
Myotonic dystrophy type 1 (DM1) and type 2 (DM2) are multisystem disorders, with core symptoms of progressive muscle weakness, myotonia, cataracts, heart conduction defects and endocrine dysfunction [1, 2]. DM1 and DM2 are caused by repeat expansion mutations residing in transcribed, non-coding regions of their respective genes. DM1 is caused by a (CTG)n in the 3´ UTR of DMPK [3–5] and DM2 by a (CCTG)n in intron 1 of ZNF9 [6, 7]. DMPK and ZNF9 are expressed in a wide variety of tissues, which is thought to underlie the multi-organ involvement in both diseases. In spite of larger expansions, DM2 is generally milder compared to DM1, and DM2 lacks a congenital form or clear anticipation [2, 8].
Since transcription of mutant repeats appears to be necessary and sufficient to cause disease, DM1 and DM2 are commonly considered toxic RNA gain-of-function diseases [9]. Accumulation of pathogenic (CUG)DM1/(CCUG)DM2 in ribonuclear foci leads to the sequestration of trans-acting nuclear proteins, including MBNL1 [10, 11], and the up-regulation of CUGBP1 levels by PKC-mediated hyperphosphorylation [12]. Dysregulation of these splice factors has been linked to altered pre-mRNA processing of various DM effector genes, favoring the expression of their embryonic mRNA and protein isoforms [13]. Aberrant splicing of multiple skeletal muscle genes has been reported in both DM1 and DM2 [10, 14–23]. More recently, additional pathomechanisms disrupting cellular homeostasis have been implicated, including dysregulation of transcription, translation and protein degradation [13, 24–27]. In addition, haploinsufficiency of DMPK in DM1 and ZNF9 in DM2 has been identified [4, 26, 28–32].
Although similar in many respects, there are clear phenotypic differences between the two diseases: compared to DM1 the clinical presentation of DM2 is more variable in both severity and range of clinical symptoms (see Table 1 in [17] for detailed description of clinical features in DM1 and DM2) [2, 33, 34]. Proximal muscles and type 2 fibres are more severely affected in DM2, whereas in DM1 distal muscles and type 1 fibres are more involved [1, 35, 36]. In DM2 myalgic pains can be the most disabling symptom [2, 37], although little is known about their underlying molecular pathophysiology. Recently we showed divergent aberrant splicing of several myofibrillar proteins, including the fast skeletal muscle troponin T, fTnT (TNNT3) and Z-disc alternatively spliced PDZ-motif containing protein, ZASP (LDB3). These differences may in part account for differential skeletal muscle weakness and fibre type involvement in DM1 and DM2 [17].
Table 1.
Summary of mRNA expression measured by microarray expression profiling
| Gene | Probe sets n |
Probe sets dysregulated n |
Fold-change DM vs. N |
Fold-change DM1 vs. DM2 |
|---|---|---|---|---|
| RYR1 | 1 | 0 | NS | NS |
| CACNA1S | 1 | 0 | NS | NS |
| ATP2A1 | 3 | 2 | (1.47–1.85)a 1.66b | NS |
| ATP2A2 | 4 | 2 | (1.47–2.04) 1.76 | (1.57–1.68) 1.63 |
| CASQ1 | 1 | 1 | 1.81 | NS |
| CASQ2 | 1 | 1 | 2 | NS |
| TRDN | 3 | 3 | (2.09–2.86) 2.55 | 1.79 |
| JPH1 | 2 | 2 | (1.67–1.88) 1.78 | 1.93 |
| PPP3CA | 9 | 3 | (1.63–6.01) 3.15 | NS |
| NFATC3 | 4 | 0 | NS | NS |
Range;
Average;
NS, not significant for fold-change values <1.2.
Timely and effective Ca2+ storage, release and re-uptake in excitation-contraction coupling are essential for proper muscle functioning [38]. Key proteins regulating the Ca2+ flux include the voltage-sensitive Ca2+ release complex constituents ryanodine receptors (RYR) and dihydropyridine receptor (DHPR), the sarcoplasmic/endoplasmic reticulum (SR) Ca2+ re-uptake ATPase (SERCA) and the Ca2+ storage protein calsequestrin (CASQ) in the SR lumen [38]. Because free Ca2+ is cytotoxic, its tight regulation is essential. In addition to muscle contraction, Ca2+ is an important secondary messenger in cell signaling, regulating a variety of cellular functions from development to cell growth and differentiation [39, 40]. Several studies have consistently identified Ca2+ signaling as the most affected pathway in DM in human patients and mouse models [17, 41, 42]. Many myogenic transcription factors are Ca2+-sensitive, and we recently reported aberrant mRNA expression and splicing of the MEF2 family of transcription factors, SIX1 and NFATC4 in DM1 and DM2 [17, 42]. Aberrant splicing of RYR1 and ATP2A1 (SERCA1), which may contribute to muscle wasting through the disruption Ca2+ homeostasis, have been observed in skeletal muscle of DM1 patients and the HSALR DM1 mice [20, 43]. Here we show similar aberrant splicing in skeletal muscle of DM2 patients, with differences in relative proportions of ATP2A1 splice variants between DM1 and DM2. In addition, we show dysregulation of several factors involved in Ca2+ homeostasis, both at mRNA expression and protein levels including RYR1, DHPR (CACNA1S), CASQ2, TRDN, JPH1, calcineurin (PPP3CA) and NFATC3.
Materials and Methods
Patients
Enrollment of patients was approved by the respective local institutional review boards. Muscle biopsies were obtained after informed consent from the patients. DM1 and DM2 patients are summarized in Supplemental Table 1. Altogether, DM1 (n = 21), DM2 (n = 34), other neuromuscular disease (NMD) controls (n = 7), and healthy controls (n = 20) were included. In DM1 and control groups, both distal and proximal muscles were included. Only proximal muscles of DM2 patients were available, because distal muscles are rarely affected in DM2. Each group included both males and females. DM1 and DM2 patients with matched age, gender and stage of disease were used. Different sample sets were used for protein analyses and mRNA analyses using expression profiling and splice variant analysis.
Microarray expression profiling
Preparation and labelling of RNA
The U133Plus2 GeneChip (Affymetrix, Santa Clara, CA) was used for RNA expression profiling. Total cellular RNA was extracted from muscle biopsies after mechanical homogenization using a shark-tooth pulveriser with TriZol (Invitrogen, Carlsbad, CA), according to the manufacturer's suggestions. The RNeasy kit (Qiagen, Valencia, CA) was used to further purify the RNA. The quality and integrity of the RNA was monitored using an Agilent BioAnalyzer and the RNA 6000 Nano LabChip (Agilent, Santa Clara, CA), and samples with a RIN (RNA integrity number) >7 were accepted for use. Five µg of total cellular RNA from each sample was used for cDNA synthesis according to the manufacturer’s protocol. The efficiency of cRNA synthesis was controlled by adding a mixture of in vitro transcribed cRNAs of cloned bacterial genes for lysA, pheB, thrB, and dap (American Type Culture Collection) as external controls. The Superscript II system (GIBCO/BRL) was employed for first-strand cDNA synthesis, which was performed at 42°C for 1 hr at a final concentration of 1 × first-strand synthesis buffer, 10 mM DTT, 500 µM dNTPs, 100 pmol of T7-(T)24 primer, and 200 units of reverse transcriptase. Second-strand cDNA synthesis was performed at 16°C for 2 hr at a final concentration of 1 × second-strand buffer, 250 µM dNTP, 65 U/ml DNA ligase, 250 units/ml DNA polymerase I, 13 U/ml RNase H. Reaction mixtures from the second-strand synthesis were purified with an Affymetrix cDNA purification column. Biotinylated UTP and CTP were used for in vitro transcription labelling, performed according to the manufacturer’s protocol (Enzo Diagnostics) for 16 hr at 37°C. cRNA purification column (RNeasy, Qiagen), was used to clean the amplified cRNA, and its quality was monitored with an Agilent BioAnalyzer. Labelled cRNAs were fragmented at 94°C for 35 min in 40 mM Tris-acetate, pH 8.1/100 mM KOAc/30mM Mg(OAc)2, and the fragmented cRNAs were hybridized to Affymetrix U133Plus2 GeneChips. The hybridization cocktail contained 10 µg of fragmented cRNA in 200 µl, with 50 pM control oligonucleotide B2, 0.1 mg/ml herring sperm DNA, 0.5 mg/ml acetylated BSA, 100 mM Mes, 20 mM EDTA, 0.01% Tween 20 (total Na+ = 1 M), and bacterial sense cRNA controls for bioB, bioC, bioD, and cre at 1.5, 5.0, 25, and 100 pM. After hybridization, the chips were scanned according to the manufacturer’s protocol. The gene expression data are available at www.ncbi.nlm.nih.gov/geo/ (GEO series numbers GSE7014) [17].
Data analysis
While we considered the expression profiles of the entire set of genes on the array, we decided to specifically focus on the subset of genes functionally associated with Ca2+ metabolism, in order to examine key genes associated with the two types of DM. Using the DChip software package (DNA-Chip analyzer) annotation tools (February 2006 build, http://biosun1.harvard.edu/complab/dchip/) and a series of queries to Gene Ontology along with information from the literature, we generated a list of 2,490 probe set IDs specific for 1,039 genes involved in Ca2+ metabolism. It is important to note, however, that the genes were not selected on the basis of a contrast between the DM groups, or on the basis of any particular direction to these contrasts.
A healthy control sample was used as the reference for normalization, which was performed with the Invariant Set Normalization method (PM-only model with DChip default settings). The expression values for each probe set were calculated with DChip model-based expression, and comparisons were made between sample groups using a fold-change (FC) cut-off FC ≥ 1.2, a lower bound (lb) limit lb = 90% (default), e-b, b-e difference thresholds of 100 (e experiment, b baseline), and a 50-permutations false discovery rate (FDR) calculation for each comparison. Clustering analyses on samples and differentially regulated genes were performed with the Euclidian Distance metric in DChip using default settings.
Pathway analysis of differentially regulated genes identified by expression profiling was performed with the Ingenuity Pathway Analysis software (IPA, Ingenuity Systems, Release Number: 5.5 – 2233, http://www.ingenuity.com). IPA analysis uses the right-tailed Fisher Exact Test to define the most relevant functional categories and canonical pathways (Ingenuity Pathway Analysis-Canonical Pathways, IPA-CP).
Splice variant analysis
cDNA Synthesis
Standard methods were used to synthesize cDNA from total muscle RNA. Briefly, 5 µg of RNA were digested with DNaseI (Ambion, Austin, TX, USA) according to the manufacturer’s protocol. SuperScript™ III First-strand cDNA Synthesis Kit (Invitrogen, Carlsbad, CA, USA) was used to generate cDNA of the DnaseI-treated RNA, half and half in separate reactions using random hexamer and oligo-(dT) priming according to the manufacturer. The resultant cDNAs were RNase H treated before use. A mixture of random hexamer and oligo-(dT) primed cDNA, in equal amounts, was diluted to 200 µl with molecular grade RNAse-free H2O. The possibility of genomic contamination was eliminated by testing the cDNAs for a known genomic marker by PCR.
RT-PCR
In order to measure the proportions of the splice variants present in the muscle samples, RT-PCR was performed, with fluorescently (FAM−) labeled universal M13 forward primer to allow subsequent quantification of the products [7, 44]. In each RT-PCR, 2 µl of cDNA (12.5 ng RNA-equivalents) was used, and the reactions were performed for 23–28 cycles, in log-linear range, with appropriate controls. The results were analyzed using capillary electrophoresis on an ABI3100 sequencer, and the measured peak heights for each isoform were added together to obtain the total signal. The percentage of each individual peak representing a unique isoform was then measured, and the statistical significance of the results were calculated using the Student´s t test. Primer sequences for RYR1 and ATP2A1 RT-PCR were as described before [20] with the addition of M13 tails (Supplemental Table 2).
Western blotting
To prepare samples for SDS-PAGE and western blotting of membrane-bound proteins RYR1, DHPR and SERCA1, ice-cold RIPA-buffer containing protease inhibitor cocktail (Complete®, Sigma-Aldrich) was added on top of the muscle biopsies, followed by mechanical homogenization Subsequently, the samples were sonicated on ice four times for 5 sec (40% amplitude), while cooled between cycles. The samples were then centrifuged at 13,000 rpm for 15 min, and the supernatant was recovered. One volume of (4×) reducing SDS-PAGE sample buffer containing beta-mercaptoethanol was added to three volumes of the supernatant, and samples were heated at 35°C for 15 min. For soluble proteins, a sample preparation method described previously was applied [45].
Different kinds of SDS-PAGE gels were used for proteins of distinct sizes: for CASQ1/2 and TRDN, 8% SDS-PAGE gels and Bio-Rad Mini Protean system (Bio-Rad Laboratories, Hercules, CA, USA) were used, whereas for RYR1, DHPR and SERCA1, 5% SDS-PAGE gels and Bio-Rad Protean System were used for better resolution. The following primary antibodies were used for detection: SERCA2 (sc-8095; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA); TRDN (sc-33390; Santa Cruz Biotechnology); JPH1 (H00056704-A01; Abnova Corporation, Taipei, Taiwan); calcineurin (clone CN-A1; Sigma-Aldrich, Saint Louis, MO, USA); NFATC3 (H00004775-B01; Abnova); RYR (clone 34C; Abcam, Cambridge, UK); DHPR (clone 1A; Novus Biologicals, Inc., Littleton, CA, USA); SERCA1 (clone VE121G9; Research Diagnostics, Flanders, NJ, USA); CASQ1 (SC-16571; Santa Cruz Biotechnology); CASQ2 (ab3516; Abcam). The HRP-conjugated secondary antibodies swine anti-rabbit (DAKO P399), rabbit anti-mouse (P260), and rabbit anti-goat (P449) were all obtained from DAKO (Glostrup, Denmark). ECL-detection was performed using the Super Signal Femto kit (Thermo Scientific, Rockford, IL, USA) at 1:10, diluted in PBS, pH 7,4. We confirmed that the molecular weight in kilodaltons (kD) for each protein detected agreed with published data and the information provided by the antibody manufacturer. Equal protein loading was monitored using Coomassie-stained myosin heavy chain (MyHC) band, and immunodetection of β-actin (ACTB) (latter not shown). The muscle fibre-specific MyHC provides a reliable control, especially in dystrophic muscles with potential fatty and connective tissue replacement. The ImageJ 1.43u free software (National Institutes of Health, USA) was used for quantification of WB bands. Integrated density of bands was determined, after which the ratio between the specific band and its corresponding loading control band (Coomassie-stained MyHC or immunoblotted ACTB) was measured. The average ratio of the healthy controls group was set to 1.0, and the relative protein quantities in the DM1 and DM2 groups were obtained by proportioning their average ratios relative to this. Microsoft® Excel Office 2007 (Microsoft Corporation, Redmond, WA, USA) was used to generate graphs, and to calculate p-values using Student´s t-test (with two-tail distribution and two-sample unequal variance).
Immunohistochemistry
Frozen muscle sections (6 µm) were used for immunohistochemical analyses. For immunofluorescence staining, the slides were fixed in 4% PFA for 10 min, permeabilized in 0.2% Triton X-100 for 10 min and blocked in 5% BSA for 30 min, after which primary and secondary antibody (Ab) incubations were performed at room temperature for 1 h. For nuclear counterstain, the slides were dipped in aquaeous DAPI solution before mounting in Gel/Mount (Sigma G0918). BSA (1%) was used as an Ab diluent, and PBS was used as a buffer throughout the procedure. Slides were rinsed in PBS four times for 5 min between each step. Double stainings were performed by pooling primary and secondary antibodies, except for the RYR/DHPR double staining, where RYR mAb was labeled using the Zenon mouse IgG labeling kit Alexa-546 (Molecular Probes, Inc., Eugene, OR) according to the manufacturer´s instructions. The primary antibodies used were CASQ2, TRDN, SERCA1, RYR, DHPR, MyHC-IIa/x (MHCf, clone MY-32; Sigma-Aldrich), CASQ1, and calcineurin. Fluorescently labelled secondary antibodies from Molecular Probes were used, including rabbit anti-mouse Alexa 488 (A-11059); goat anti-mouse Alexa 546 (A-11003); donkey anti-goat Alexa 546 (A-11056); and donkey anti-rabbit Alexa 488 (A-21206) (Invitrogen Corporation, Carlsbad, CA; USA).
Ventana Benchmark automated immunostainer (Ventana Medical Systems, Tucson, AZ) was used for DAB immunohistochemistry of frozen sections. iVIEW® DAB detection kit followed by Hematoxylin and Bluing reagent counterstain (Ventana). Partially the same primary antibodies as for IF were used for DAB stainings: MHCf, RYR, DHPR, SERCA1, JPH1, and CASQ2.
For comparison we also immunolabelled muscle membrane proteins dystrophin, sarcoglycans, caveolin 3, merosin and nNOS, the latter also being involved in calcium signaling. Beta-spectrin served as a membrane integrity control. Primary antibodies used were mouse mAbs beta-spectrin (clone RBC2/3D5; Leica Biosystems, Newcastle Upon Tyne, UK); C-terminal DYS (clone Dy8/6C5; Leica Biosystems); alpha-sarcoglycan (clone Ad1/20A6; Leica Biosystems); beta-sarcoglycan (clone Bsarc/5B1; Leica Biosystems) merosin (clone 2Q598; United States Biological, Swampscott, MA, USA); caveolin-3 (sc-5310; Santa Cruz Biotechnology); and rabbit pAb nNOS (sc-648; Santa Cruz Biotechnology). We carefully confirmed the subcellular distribution of all antibodies for immunohistochemistry using healthy control muscle material, and confirmed that the obtained results agreed with published information or that provided by manufacturer.
Results
Microarray expression profiling
To identify differentially expressed genes between DM1 and DM2, we performed global mRNA expression profiling of skeletal muscle biopsies in DM1 (n = 10) and DM2 (n = 20) patients and normal adults (n = 6). We previously reported preliminary results of muscle-specific genes, which identified numerous genes involved in Ca2+ metabolism (GEO accession number GSE7014; [17]). Analysis of all genes interrogated followed by Ingenuity Pathway Analysis of differentially expressed genes identified calcium signalling as the most significant canonical pathway with 29 of 204 genes (14%) in the curated pathway showing up-regulation and none showing down-regulation (Sirito et al., manuscript in preparation). Unsupervised clustering of all 2,490 probe sets representing 1,039 unique Ca2+ metabolism genes did not show separation by disease status. However, 200 probe sets (8%, representing 134 unique genes) showed differential expression in both DM1 and DM2 compared to normal samples (false-discovery rate 8%). Of these, 183 probe sets (91.5% or 121 unique genes) showed up-regulation, while only 17 probe sets (8.5%), representing 13 unique genes, showed down-regulation. When we compared DM2 to DM1 only 21 probe sets (16 unique genes) showed differential mRNA expression (false-discovery rate 4.8%).
This gene list included several Ca2+ transporting ATPases and other ion channels/pumps (K+, Na+, Cl−). Structurally, many of these proteins have one or several EF-hand motifs for Ca2+ binding, and many are identified as transmembrane proteins. Numerous myosin genes were also up-regulated, including unconventional/non-muscle myosins [17], regulated in a Ca2+-dependent fashion. In this study, we focused on a subset of genes known to play major roles in Ca2+ handling in muscle, including SERCA1 (ATP2A1) and SERCA2 (ATP2A2), both calsequestrin genes (CASQ1 and CASQ2), triadin (TRDN), junctophilin (JPH1) and calcineurin-α catalytic subunit (PPP3CA), all of which showed up-regulation (DM vs. N). In addition, ATP2A2, TRDN and JPH1 were differentially dysregulated between DM2 and DM1, the expression being higher in DM1. The results are summarized in Table 1.
Aberrant splicing of RYR1 and SERCA1 in DM1 and DM2
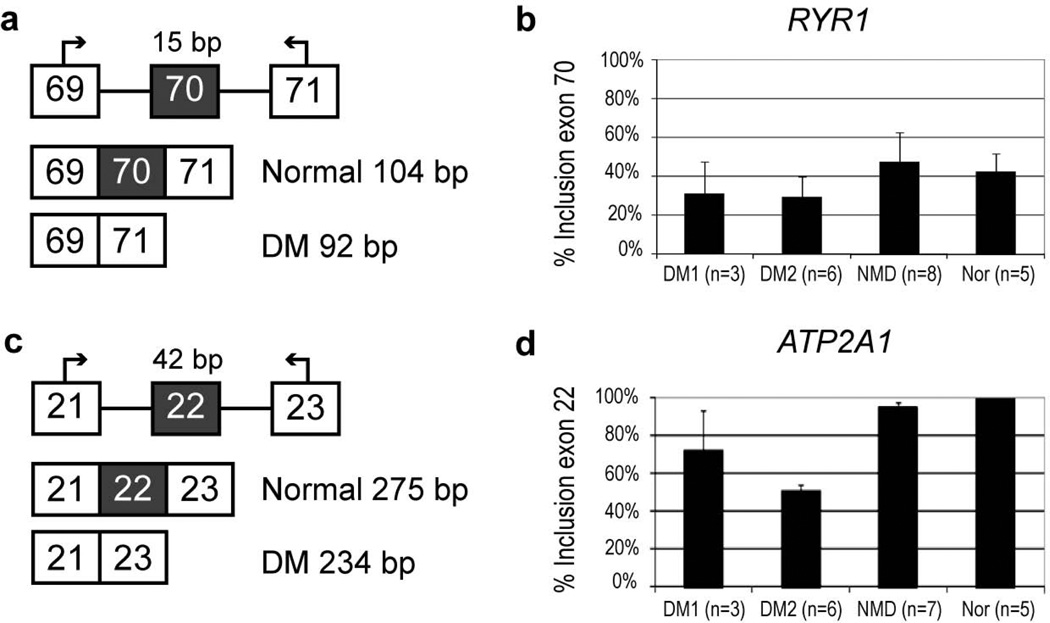
Splice variant analysis showed that the fetal/neonatal RYR1 isoform 2, [alternatively spliced sequence I, ASI(−)], lacking exon 70 (residues 3481–3485), was present in both DM1 and DM2 muscle specimens (Fig. 1b), previously reported only for DM1 [20]. Similarly, ATP2A1, encoding SERCA1, was aberrantly spliced in both DM2 and DM1, resulting in the fetal/neonatal isoform ATP2A1B, lacking exon 22 (Fig. 1d). The relative proportions of the RYR1 ASI(−) isoform were similar in DM1 and DM2, whereas the proportion of ATP2A1B was significantly higher in DM2 compared to DM1 (p-value = 0.031).
Figure 1.
Alternative isoforms of ATP2A1 and RYR1 in muscle biopsies from DM1, DM2 and non-DM dystrophy (NMD) patients compared to muscle of normal individuals. Panels a and c show design for RT-PCR assays. Panels b and d show results of quantitative fluorescent RT-PCR. Percent inclusion of the aberrantly spliced exon is shown on the Y axis. Error bars represent the standard deviation for each group. DM1 was not significantly different from normal for RYR1 (p-value = 0.251), while DM2 was (p-value = 0.0239). For ATP2A1 both DM1 and DM2 were significantly different from normal (p-value = 0.0211 and p-value = 2.68E-11, respectively)
Western blotting
The genes summarized in Table 2 were further evaluated by Western blotting to determine the total protein amounts in the samples in DM1 (n = 7) and DM2 (n = 5) (Fig. 2). RYR1 was included in the study to determine if aberrant splicing affects total protein levels, DHPR was included based on its crucial role in the excitation-contraction coupling with RYR1, although it did not show transcriptional dysregulation. In addition, NFATC3, a member of the NFAT family of transcription factors, the major downstream effectors of calcineurin, was included.
Table 2.
Summary of gene and protein expression in DM1 and DM2
| Gene | Protein | IHCa,b | EP | SVA | WB | |
|---|---|---|---|---|---|---|
| DM1 | DM2 | |||||
| RYR1 | RYR1 | - | - | no change | exon 70 exclusionc | no change |
| CACNA1S | DHPR | - | = | no change | na | no change |
| ATP2A1 | SERCA1 | = | = | ↑ | exon 22 exclusion | no change |
| ATP2A2 | SERCA2 | na | = | ↑ | na | N > DM2 |
| CASQ1 | CASQ1 | na | = | ↑ | na | no change |
| CASQ2 | CASQ2 | - | -- | ↑ | na | no change |
| TRDN | TRDN | na | = | ↑ | na | DM2 > DM1 |
| JPH1 | JPH1 | - | = | ↑ | na | N > DM |
| PPP3CA | Calcineurin | na | = | ↑ | na | DM2 > DM1 |
| NFATC3 | NFATC3 | na | na | no change | na | DM2 > DM1 |
IHC, immunohistochemistry; EP microarray expression profiling; SVA, splice variant analysis; WB, western blotting; na, not assessed.
Fibres > 30–40 um in diameter, most of which presumably contractile.
Evaluation of protein expression: =, staining similar to healthy control; -, moderately reduced; --, severely reduced.
Statistically significant only in DM2
Figure 2.
Quantification of proteins after western blotting showing differential expression in DM muscle biopsies with significant p-values (p < 0.05). The average of healthy controls (N, normal) in each group was set to 1.00. The rows are labeled as A immunoblotted protein band; B coomassie brilliant blue -stained MyHC band as loading control. Each band is labeled with a patient identifier (X = outlier omitted from analysis). Standard deviation is indicated by error bars. a, SERCA2 (observed band 115 kD) was decreased in DM2 vs. N (p-value = 0.0074). b, TRDN (100 kD) showed higher expression in DM2 vs. DM1 (p-value = 0.0012). c, JPH1 (80 kD) was reduced in DMs as a group (p-value = 0.0256), however, only DM2 showed significant decrease alone vs. N (p-value = 0.0147). d, Calcineurin (60 kD) showed higher expression in DM2 and lower expression in DM1 compared to N, however, only comparing DM1 vs. DM2 showed significant difference (p-value = 0.0423). e, NFATC3 (115 kD) appeared increased in DM2, but the p-value DM2 vs. N was not quite significant (p-value = 0.0590); comparing DM1 vs. DM2 showed that NFATC3 was more expressed in DM2 than DM1 (p-value = 0.0359).
The proteins showing differential expression included SERCA2, which was decreased approximately 25% in DM2 vs. N (p-value = 0.0074) and TRDN, which was increased in DM2 and decreased in DM1 (DM1 vs. DM2, p-value = 0.0012) (Fig. 2a and 2b). JPH1 was decreased in both DM1 and DM2 as a group (p-value = 0.0256), and in DM2 vs. N (p-value = 0.0147) (Fig. 2c). PPP3CA, the catalytic α subunit of calcineurin and NFATC3 both showed a similar trend: decreased in DM1 and increased in DM2 compared to N, however, only DM1 vs. DM2 was significant (PPP3CA p-value = 0.0423; NFATC3 p-value = 0.0359; Fig. 2d and 2e). Of the proteins studied, not all showed dysregulation at the total protein level, when assessed by western blotting (Table 2; figure not shown). RYR1 and SERCA1 were assayed because both have shown aberrant splicing in DM. However, they did not show abnormal expression by western blotting, and the fetal/neonatal isoforms could not be distinguished due to methodological limitations of immunoblotting to identify small size variations of these large proteins.
Immunohistochemistry and immunofluorescence analysis
In DAB immunohistochemistry, RYR1 was detected in the SR of all fibres, with fast fibres showing slightly more intense labeling (Fig. 3). In DM1 and DM2 specimens, RYR1 protein expression was slightly decreased in all fibres, except for the very atrophic fibres, where there was accumulation of RYR1 (Fig. 3). The dihydropyridine receptor (DHPR) alpha-subunit (CACNA1S) appeared intact in DM2, whereas it showed reduced immunolabeling in the more severely affected tibialis anterior (TA) muscles in DM1 (Fig. 3). SERCA1 (ATP2A1) immunohistochemistry did not show major differences between control specimens and DM1/DM2 patients (Fig. 3). All fast fibres expressed SERCA1 in their SR. However, SERCA1 expression was not restricted to fast type 2 fibres, but was also expressed in about 10–20 % of slow type 1 fibres. Also junctophilin (JPH1) immunolabeling was slightly more intense in fast fibres. While no abnormality was observed in DM2, DM1 samples showed generally more variable and weak JPH1 labeling (Fig. 3). CASQ2 expression was higher in slow compared to fast fibres (Fig. 3). However, in DM2 CASQ2 immunolabeling was clearly weaker in all fibre types, with the most severely affected muscles showing even more reduced staining. DM1 biopsies showed less prominent reduction in CASQ2 staining intensity, even in the most severely affected biopsies (Fig. 3).
Figure 3.
Representative immunohistochemical stainings of Ca2+ handling proteins in DM. In DAB immunohistochemistry (a–k, m, n, p, q), the specific signal is dark brown, and the background staining (H&E) is blue. In the upper panel, in control both RYR1 (b) and DHPR (c) are slightly more intensely expressed in fast fibres, when compared to MyHC-II (a) (detecting fast fibres). Reduction of RYR1 immunoreactivity in DM2 (e), and reduction of both RYR1 and DHPR in DM1 (h, i) is evident. In the lower panel, normal SERCA1 staining, similar to control (j) is seen in both DM2 (m) and DM1 (p). Reduction of JPH1 is seen in DM1 (q), but not in DM2 (n). The last panel with immunofluorescent staining shows drastic reduction of CASQ2 (green) in DM2 (o), and a less extensive reduction in DM1 (r) compared to healthy control (l). (a–c, j, k) N_??_VL_03; (L) N_M40_G_004; (d–f, m–o) DM2_M38_VL_001; (g, h, p) DM1_M48_TA_004; (i) DM1_M55_TA_010; (q) DM1_M48_VL_005; (r) DM1_M46_TA_001
When analyzed by confocal microscopy, no major abnormality in the organization of RYR1 and DHPR on the SR and T-tubule membranes was observed in functional fibres, whereas in the highly atrophic fibres, they accumulated in part independently (Supplemental Fig. 1). RYR1 was found to co-localize with TRDN in DM2 specimens, forming a similar doublet band staining pattern as in control tissue (Supplemental Fig. 1). Similarly, CASQ2 co-localized with TRDN, also in DM2 muscle samples even though CASQ2 labeling was very weak. In contrast to the findings above CASQ2 was more highly accumulated in the fast type 2 nuclear clump fibres in DM2, where it still colocalized with accumulated TRDN (Supplemental Fig. 1). This was also observed at later stages with severe pathology in DM1.
In DM2 muscle, neuronal nitric oxide synthase (nNOS) was severely reduced on the sarcolemma of very atrophic fibres (Supplemental Fig. 2), whereas sarcoglycans and dystrophin, integral members of the dystrophin associated glycoprotein complex (DAG), showed decreased labeling. Merosin (laminin-2) and caveolin-3 were unaffected.
Discussion
Differences in the spatial and temporal expression of the mutant (CUG)DM1 and (CCUG)DM2 located in DMPK and ZNF9, combined with differences in multiple downstream effector genes likely underlie the different phenotypic manifestations seen between DM1 and DM2, including muscle weakness and differential fibre type involvement. Aberrant splicing with only minor qualitative and quantitative differences was previously reported in DM1 and DM2 [10, 46]. Nevertheless, we recently observed differences in abnormal splicing of the sarcomeric genes TNNT3 and LDB3 between the two diseases [17]. Here, we have focused on proteins involved in Ca2+ metabolism, as calcium ion has a key role in cell signalling and muscle contraction, and defects in Ca2+-regulated pathways have been associated to DM1 previously [20, 41, 43]. As a novel finding, we present aberrant splicing of RYR1 and ATP2A1 in DM2. More importantly, we observed abnormal expression of mRNA and protein of a larger range of molecules involved in muscle Ca2+ release and re-uptake, Ca2+ storage, and Ca2+-dependent signalling. While similar in many ways, we have identified novel differences between DM1 and DM2. Interestingly, in most cases increases in mRNA transcript levels were not consistently reflected in increased protein levels. Our findings are consistent with recent reports that have identified a translational block in DM as one of the underlying mechanisms [26, 31].
Pathway analysis of differentially expressed genes identified calcium signalling as the most significant canonical pathway with 29 of 204 genes (14%) showing up-regulation and no genes showing down-regulation. The proportion of dysregulated genes involved in Ca2+ signaling is similar to that which we previously reported for sarcomeric protein encoding genes and myogenic transcription factors [17].
Several genes encoding triad constituents were up-regulated in the DMs. Two of them, TRDN (triadin) and JPH1 (junctophilin), were differentially dysregulated between DM1 and DM2, with higher mRNA levels in DM1. TRDN, which has numerous alternatively spliced isoforms, provides a scaffold for EC coupling components. It binds RYR1 with its short cytoplasmic domain and anchors CASQ in the vicinity of RYR1, forming a quaternary complex for regulation of Ca2+ release [47]. The Ca2+ storage protein encoding genes CASQ1 and CASQ2, preferentially expressed in fast and slow fibres, respectively, both showed approximately 2-fold up-regulation in DM samples compared to controls. In addition, the Ca2+ re-uptake channel encoding genes, ATP2A1 (SERCA1) and ATP2A2 (SERCA2) were both up-regulated, ATP2A2 showing more increased expression in DM1 compared to DM2.
The catalytic alpha subunit PPP3CA of calcineurin also showed increased mRNA expression levels. Calcineurin, a Ca2+-dependent Ser/Thr protein phosphatase has a pivotal role in regulating fibre type plasticity in adult muscle, and it has been implicated in establishing the slow fibre phenotype [48]. Calcineurin regulates the activity of NFAT transcription factors by dephosphorylation [49].
It should be noted that not all mRNA expression changes or aberrant splicing events are DM-specific, but also occur in other muscular dystrophies as part of downstream molecular pathology [50]. In a recent study comparing mRNA expression in two mouse models for DM1, the HSALR and Clc1 knock-out mouse indicated that up to 25% of changes could be caused by secondary changes induced by myotonia [41]. Another comparative study between HSALR and Mbnl1 knock-out mice suggested that more than 80% of aberrant splice events are directly caused by Mbnl1 loss of function [51]. Similarly, aberrant splicing of two of the four MEF2 family members (MEF2A and MEF2C) was also observed in non-DM neuromuscular patients [42].
Three RYR genes (RYR1, -2 and -3) show tissue-specific expression patterns [52]. RYR1 is the main subtype in skeletal muscle and its expression is under developmental regulation: alternative splicing of two exons, 70 and 83, gives rise to fetal/neonatal isoforms 2 [ASI(−)] and 3 [ASII(−)], whereas the adult RYR1 isoform contains both exons [52]. Here we show that the neonatal ASI(−) isoform was expressed in both DM1 and DM2 muscle. We did not find RYR1 ASI(−) differentially expressed between DM1 and DM2 (p-value = 0.59). Surprisingly, we found relatively high ASI(−) expression rates in our controls. We do not have an explanation for this unexpected finding at the moment. This together with small number of DM1 samples likely resulted in non-significant p-value for DM1 vs. controls. However, it should be noted that RYR1 aberrant splicing was still significantly higher in DM2 samples, suggestive of more pronounced RYR1 aberrant splicing. As previously reported, we did not detect exclusion of RYR1 exon 83 [20]. The properties of the RYR1 splice variants ASI(+) and ASI(−) have been investigated, and the ASI domain has been shown to contribute to the inhibitory domain in RYR1 [53]. No difference was observed in Ca2+ dependence of the two isoforms [20]. RYR1 activity is modulated by posttranslational modifications, such as glycosylation and phosphorylation involving at least PKA and CAMKII, and by binding of several ligands, including calmodulin (CALM1). The DHPR beta-subunit binding site is adjacent to the ASI domain [54]. Recently, skipping of CACNA1S exon 29 was reported with both DM1 and DM2, and this aberrant splicing event was associated with increased channel conductance and voltage sensitivity of DHPR [23].
The Ca2+ re-uptake SERCA pumps are expressed by three highly conserved genes (ATP2A1, -2 and -3), which are alternatively spliced in both tissue-specific and developmentally regulated manner [55]. Of the ATP2A1 (SERCA1) isoforms, ATP2A1A (SERCA1a, including exon 22) is expressed in adult fast skeletal muscle fibres, and ATP2A1B (SERCA1b, excluding exon 22) in fetal/neonatal muscle fibres [56]. We found that ATP2A1B is expressed in DM1 and DM2 [10, 20], with the novel finding that the proportion of this fetal isoform is greater in DM2 compared to DM1 (p-value = 0.031). No functional differences between SERCA1 splice isoforms have been confirmed so far.
We previously reported proportionally more pronounced aberrant splicing of TNNT3 and LDB3 in DM2 [17], and here we show a similar trend for ATP2A1. Aberrant splicing events in DM are the result of MBNL1 sequestration in ribonuclear foci, resulting in MBNL1 loss-of-function, and the up-regulation of CUGBP1. SERCA1, RYR, LDB3, and TNNT3 are all MBNL1 targets, and it has been reported that higher levels of MBNL1 accumulate in DM2 ribonuclear foci than DM1, with concomitant depletion of free MBNL1 in nucleoplasm [10]. This could provide one explanation for our findings that aberrant splicing tends to be more pronounced in DM2. However, since the muscle pathology including myotonia is generally more severe in DM1, in spite of larger mutant expansions in DM2, aberrant splicing alone is unlikely to explain all manifestations leading to muscle weakness and atrophy in DM. Interestingly, in a study of developing mouse hearts in CUGBP1-overexpressing and Mbnl1-deficient mice, half of the aberrant splicing events were not regulated by either of them [57]. Differences in aberrant splicing patterns between DM1 and DM2 have been reported for GFAT1, which is not misspliced in DM2 [10].
Although splice variant analysis showed aberrant splicing of RYR1 and SERCA1, western blotting could not verify the existence of the abnormal fetal isoforms due to the lack of resolution for small differences in isoform sizes (RYR1, 5 residues; SERCA1, 7 residues). However, total protein amounts of both RYR1 and SERCA1 were similar between control and DM samples, even though ATP2A1 showed significant increase of the total mRNA level. SERCA1 is almost exclusively expressed in fast fibres, and given that the fast type 2 fibres are more affected in DM2 than in DM1, our observation that aberrant SERCA1 splice variants occur more frequently in DM2, suggests that it may have a role in DM2 differential muscle pathogenesis.
Calcineurin (PPP3CA) and TRDN behaved in a different way compared to most of the other studied molecules being increased both at the mRNA and protein level, with significantly higher increase in DM2 compared to DM1 (p-value = 0,0423 and p-value = 0,0012, respectively) when assessed by western blotting. On the contrary, ATP2A2 and JPH1 showed reduced total protein levels, even though mRNA was up-regulated. NFATC3, a downstream effector of calcineurin, also showed increased protein levels in DM2 vs. DM1 (p-value = 0,036), although without significant dysregulation at the mRNA level [17]. mRNA expression levels were increased for both CASQ1 and CASQ2, but the corresponding protein amounts were unaltered in DM1 and DM2. The observed discrepancy between mRNA and protein levels may be due to a translational block or abnormal post-transcriptional mRNA processing in the myotonic dystrophies [26, 27, 31]. It has recently been reported that the transcriptional up-regulation of several genes is not DM-specific [41]. Thus, a large proportion of the reported transcriptional changes may reflect an unspecific response to muscle injury, but since most mRNA expression changes do not result in any corresponding increase of the protein they probably represent indicators linked to the catabolic pathology of the disease.
Upon membrane depolarization, the voltage-gated L-type Ca2+ channel DHPR on T-tubules undergoes a conformational change and activates RYR1, the primary Ca2+ release channel in the terminal SR, thus initiating muscle contraction. When assessed by immunohistochemistry, RYR1 levels in SR were reduced in both DM1 and DM2, while CACNA1S levels were reduced in the severely affected DM1 muscle fibres only. The triad structure, as well as organization of RYR1 and DHPR in the excitable membranes appeared intact in DM2 biopsies in confocal fluorescent microscopy. The reduction of RYR1 and DHPR observed on IHC could not be replicated by western blotting. A similar pattern of expression was seen with CASQ2, showing severely reduced immunolabeling in DM2, and less prominent reduction in DM1, but without significantly reduced amounts in DM2 or DM1 specimens compared to healthy controls on western blotting. A slight reduction in JPH1 immunoreactivity was also observed in the severely affected DM1 muscles. Together with reduced DHPR levels in DM1, this is intriguing in the context of the more severe muscle phenotype in DM1. JPH1 facilitates the interaction of DHPR and RYR1, hence enhancing EC coupling [58]. In accordance with its function, JPH1 also was more intensely expressed in fast muscle fibres.
The observed discrepancy between IHC and western blotting results is challenging to explain. However, as the fibre type distributions can vary greatly, especially in diseased muscle, comparison of total expression of proteins which show intrinsic fibre type expression differences is not easy. In addition, several proteins, including RYR1, accumulated heavily in the highly atrophic fibres, which may contribute to these results. Moreover, immunohistochemistry in contrast to western blotting may have different affinity for posttranslational modifications and reveal functionally important subcellular localization of proteins, which, based on our data presented here, we believe is the case with RYR1 and CASQ2.
CASQ is the major Ca2+ binding protein in the SR lumen of both skeletal and cardiac muscle, with low affinity and high capacity for Ca2+ [59]. Calsequestrin binds directly to RYR1 and TRDN, and recruits Ca2+ to the junctional membrane of terminal cisternae, keeping free SR Ca2+ concentration low. In addition, CASQ modifies the activity of RYR1 in concert with triadin, in a [Ca2+] dependent manner [60]. At present the significance of CASQ2 reduction in DM muscle is not known, as CASQ1 expression appeared intact in both IHC and immunoblotting. However, the different CASQ isoforms regulate RYR in different ways, and it is possible that CASQ2 depletion may influence the Ca2+ handling capacity in slow fibres. Type I fibres are generally more affected in DM1 muscles.
nNOS is regulated by Ca2+/calmodulin, and decrease in sarcolemma-associated nNOS has been implicated in Duchenne muscular dystrophy and some other neuromuscular diseases [61, 62]. Given the atrophy of type 2 fibres in DM2, it is interesting that dystrophin-mediated signaling has been linked to regulation of muscle atrophy [63]. In addition, nitric oxide mediates epigenetic changes via regulating histone deacetylases (HDAC), thus affecting gene expression [61]. We found the sarcolemmal nNOS staining in the atrophic fibres in DM2 clearly decreased, with concomitant reduction in dystrophin and sarcoglycans, members of the DAG complex. Since overt fibre necrosis and highly elevated creatine kinase levels are not hallmarks of the phenotype, it is likely that these changes are secondary in DM2, even though dystrophin undergoes aberrant splicing in DM [21].
Several genes show differential expression patterns in different fibre types. Of the genes studied here, ATP2A1 (SERCA1) is predominantly expressed in fast fibres, while RYR1, CACNA1S (DHPR) and JPH1 are only slightly more expressed in fast fibres. On the other hand, CASQ2 shows higher expression in slow fibres. All human muscles are mixed populations of type 1 and 2 fibres, with significant variation between muscle types and individuals. DM2 biopsies in this study were obtained mostly from vastus lateralis muscle with fast fibre type predominance, whereas tibialis anterior muscle, which is generally more affected in DM1, usually contains slightly more slow fibres. Biceps brachii and gastrocnemius usually contain equal numbers of slow and fast fibres. We previously showed more pronounced aberrant splicing in DM2 for TNNT3 [17], and here for ATP2A1, both of which show predominant expression in type 2 fibres. Apart from this, however, results of gene and protein expression in this study do not allow direct explanation for the differential fibre and muscle type involvement in DM1 and DM2, which are likely due to more complex pathways.
As with the previously reported sarcomeric proteins [17], we have been able to identify specific molecular differences in the extent of aberrant splicing, total mRNA, and protein expression for a set of Ca2+ handling proteins. For some of the studied proteins these changes are not extensive, but considering the absolute requirement of tight Ca2+ control in the muscle cell, they may still be of importance for muscle pathology and some of the muscle phenotypic differences between DM1 and DM2.
Supplementary Material
Acknowledgments
We are grateful to the participating patients for their cooperation. This study has been accomplished through the active collaboration and sharing of patient samples within the European Neuromuscular Centre (ENMC) consortium on DM2 and Other Myotonic Dystrophies by the following members: Josep Gamez, Jerry Mendell, Guillaume Bassez, Bruno Eymard, Tetsuo Ashizawa and Lubov Timchenko. We thank Shohrae Hajibashi, Helena Luque, Valerie L. Neubauer and Tamara J. Nixon for expert technical assistance. RK was supported by grants from the National Institutes of Health, NIH (AR48171), Muscular Dystrophy Association USA and the Kleberg Foundation. BU was supported by funding from the Folkhälsan Research Foundation, and grants from the Liv&Hälsa Foundation, the Vasa Central Hospital District Medical Research funds and Kung Gustav V Adolfs och Drottning Victorias minnesfond Foundation.
Footnotes
Supplemental Data Supplemental data will be posted on the journal web page (if accepted for publication).
Data Deposition GEO series numbers GSE7014; www.ncbi.nlm.nih.gov/geo/.
Author contributions
Conceived and designed the project and experiments: AV, RK, and BU. Performed the experiments: AV, MS, LLB, OR, MS, and TS. Analyzed the data: AV, MS, LLB, OR, MS, TS, RK, and BU. Wrote and revised the manuscript: AV, MS, LLB, RK, and BU.
Conflict of Interest The authors declare that they have no conflict of interest.
Contributor Information
Anna Vihola, Folkhälsan Institute of Genetics and Department of Medical Genetics, Haartman Institute, University of Helsinki, 00014 Helsinki, Finland.
Mario Sirito, Department of Genetics, University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Linda L. Bachinski, Department of Genetics, University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA
Olayinka Raheem, Neuromuscular Research Unit, University and University Hospital of Tampere, 33520 Tampere, Finland.
Mark Screen, Folkhälsan Institute of Genetics and Department of Medical Genetics, Haartman Institute, University of Helsinki, 00014 Helsinki, Finland.
Tiina Suominen, Neuromuscular Research Unit, University and University Hospital of Tampere, 33520 Tampere, Finland.
Ralf Krahe, Department of Genetics, University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA; Graduate Programs in Human and Molecular Genetics and Genes and Development, University of Texas at Houston Graduate School of Biomedical Sciences, Houston, TX 77030, USA.
Bjarne Udd, Folkhälsan Institute of Genetics and Department of Medical Genetics, Haartman Institute, University of Helsinki, 00014 Helsinki, Finland; Neuromuscular Research Unit, University and University Hospital of Tampere, 33520 Tampere, Finland; Department of Neurology, Vaasa Central Hospital, 65100 Vaasa, Finland.
References
- 1.Harper PS. Myotonic Dystrophy. 3rd edn. London: Saunders; 2001. [Google Scholar]
- 2.Udd B, Meola G, Krahe R, Thornton C, Ranum L, Day J, Bassez G, Ricker K. Report of the 115th ENMC workshop: DM2/PROMM and other myotonic dystrophies. 3rd workshop, 14–16 February 2003, Naarden, the Netherlands. Neuromuscul Disord. 2003;13:589–596. doi: 10.1016/s0960-8966(03)00092-0. [DOI] [PubMed] [Google Scholar]
- 3.Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Hudson T. Molecular basis of myotonic dystrophy: Expansion of a trinucleotide (CTG) repeat at the 3' end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 4.Fu YH, Pizzuti A, Fenwick RG, Jr, King J, Rajnarayan S, Dunne PW, Dubel J, Nasser GA, Ashizawa T, de Jong P. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science. 1992;255:1256–1258. doi: 10.1126/science.1546326. [DOI] [PubMed] [Google Scholar]
- 5.Mahadevan M, Tsilfidis C, Sabourin L, Shutler G, Amemiya C, Jansen G, Neville C, Narang M, Barcelo J, O'Hoy K. Myotonic dystrophy mutation: An unstable CTG repeat in the 3' untranslated region of the gene. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- 6.Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LP. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 7.Bachinski LL, Udd B, Meola G, Sansone V, Bassez G, Eymard B, Thornton CA, Moxley RT, Harper PS, Rogers MT, Jurkat-Rott K, Lehmann-Horn F, Wieser T, Gamez J, Navarro C, Bottani A, Kohler A, Shriver MD, Sallinen R, Wessman M, Zhang S, Wright FA, Krahe R. Confirmation of the type 2 myotonic dystrophy (CCTG)n expansion mutation in patients with proximal myotonic myopathy/proximal myotonic dystrophy of different european origins: A single shared haplotype indicates an ancestral founder effect. Am J Hum Genet. 2003;73:835–848. doi: 10.1086/378566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider C, Ziegler A, Ricker K, Grimm T, Kress W, Reimers CD, Meinck H, Reiners K, Toyka KV. Proximal myotonic myopathy: Evidence for anticipation in families with linkage to chromosome 3q. Neurology. 2000;55:383–388. doi: 10.1212/wnl.55.3.383. [DOI] [PubMed] [Google Scholar]
- 9.Lee JE, Cooper TA. Pathogenic mechanisms of myotonic dystrophy. Biochem Soc Trans. 2009;37(Pt 6):1281–1286. doi: 10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin X, Miller JW, Mankodi A, Kanadia RN, Yuan Y, Moxley RT, Swanson MS, Thornton CA. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum Mol Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 11.Yuan Y, Compton SA, Sobczak K, Stenberg MG, Thornton CA, Griffith JD, Swanson MS. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 2007;35:5474–5486. doi: 10.1093/nar/gkm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osborne RJ, Thornton CA. RNA-dominant diseases. Hum Mol Genet. 2006;15 Spec No 2:R162–R169. doi: 10.1093/hmg/ddl181. [DOI] [PubMed] [Google Scholar]
- 14.Mankodi A, Takahashi MP, Jiang H, Beck CL, Bowers WJ, Moxley RT, Cannon SC, Thornton CA. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 15.Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 16.Savkur RS, Philips AV, Cooper TA, Dalton JC, Moseley ML, Ranum LP, Day JW. Insulin receptor splicing alteration in myotonic dystrophy type 2. Am J Hum Genet. 2004;74:1309–1313. doi: 10.1086/421528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vihola A, Bachinski LL, Sirito M, Olufemi SE, Hajibashi S, Baggerly KA, Raheem O, Haapasalo H, Suominen T, Holmlund-Hampf J, Paetau A, Cardani R, Meola G, Kalimo H, Edstrom L, Krahe R, Udd B. Differences in aberrant expression and splicing of sarcomeric proteins in the myotonic dystrophies DM1 and DM2. Acta Neuropathol. 2010;119:465–479. doi: 10.1007/s00401-010-0637-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 19.Buj-Bello A, Furling D, Tronchere H, Laporte J, Lerouge T, Butler-Browne GS, Mandel JL. Muscle-specific alternative splicing of myotubularin-related 1 gene is impaired in DM1 muscle cells. Hum Mol Genet. 2002;11:2297–2307. doi: 10.1093/hmg/11.19.2297. [DOI] [PubMed] [Google Scholar]
- 20.Kimura T, Nakamori M, Lueck JD, Pouliquin P, Aoike F, Fujimura H, Dirksen RT, Takahashi MP, Dulhunty AF, Sakoda S. Altered mRNA splicing of the skeletal muscle ryanodine receptor and sarcoplasmic/endoplasmic reticulum Ca2+-ATPase in myotonic dystrophy type 1. Hum Mol Genet. 2005;14:2189–2200. doi: 10.1093/hmg/ddi223. [DOI] [PubMed] [Google Scholar]
- 21.Nakamori M, Kimura T, Fujimura H, Takahashi MP, Sakoda S. Altered mRNA splicing of dystrophin in type 1 myotonic dystrophy. Muscle Nerve. 2007;36:251–257. doi: 10.1002/mus.20809. [DOI] [PubMed] [Google Scholar]
- 22.Nakamori M, Kimura T, Kubota T, Matsumura T, Sumi H, Fujimura H, Takahashi MP, Sakoda S. Aberrantly spliced alpha-dystrobrevin alters alpha-syntrophin binding in myotonic dystrophy type 1. Neurology. 2008;70:677–685. doi: 10.1212/01.wnl.0000302174.08951.cf. [DOI] [PubMed] [Google Scholar]
- 23.Tang ZZ, Yarotskyy V, Wei L, Sobczak K, Nakamori M, Eichinger K, Moxley RT, Dirksen RT, Thornton CA. Muscle weakness in myotonic dystrophy associated with misregulated splicing and altered gating of Cav1.1 calcium channel. Hum Mol Genet. 2011 Dec 2; doi: 10.1093/hmg/ddr568. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Timchenko NA, Patel R, Iakova P, Cai ZJ, Quan L, Timchenko LT. Overexpression of CUG triplet repeat-binding protein, CUGBP1, in mice inhibits myogenesis. J Biol Chem. 2004;279:13129–13139. doi: 10.1074/jbc.M312923200. [DOI] [PubMed] [Google Scholar]
- 25.Vlasova IA, Tahoe NM, Fan D, Larsson O, Rattenbacher B, Sternjohn JR, Vasdewani J, Karypis G, Reilly CS, Bitterman PB, Bohjanen PR. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol Cell. 2008;29:263–270. doi: 10.1016/j.molcel.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huichalaf C, Schoser B, Schneider-Gold C, Jin B, Sarkar P, Timchenko L. Reduction of the rate of protein translation in patients with myotonic dystrophy 2. J Neurosci. 2009;29:9042–9049. doi: 10.1523/JNEUROSCI.1983-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salisbury E, Schoser B, Schneider-Gold C, Wang GL, Huichalaf C, Jin B, Sirito M, Sarkar P, Krahe R, Timchenko NA, Timchenko LT. Expression of RNA CCUG repeats dysregulates translation and degradation of proteins in myotonic dystrophy 2 patients. Am J Pathol. 2009;175:748–762. doi: 10.2353/ajpath.2009.090047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furling D, Lemieux D, Taneja K, Puymirat J. Decreased levels of myotonic dystrophy protein kinase (DMPK) and delayed differentiation in human myotonic dystrophy myoblasts. Neuromuscul Disord. 2001;11:728–735. doi: 10.1016/s0960-8966(01)00226-7. [DOI] [PubMed] [Google Scholar]
- 29.Chen W, Wang Y, Abe Y, Cheney L, Udd B, Li YP. Haploinsuffciency for Znf9 in Znf9+/− mice is associated with multiorgan abnormalities resembling myotonic dystrophy. J Mol Biol. 2007;368:8–17. doi: 10.1016/j.jmb.2007.01.088. [DOI] [PubMed] [Google Scholar]
- 30.Pelletier R, Hamel F, Beaulieu D, Patry L, Haineault C, Tarnopolsky M, Schoser B, Puymirat J. Absence of a differentiation defect in muscle satellite cells from DM2 patients. Neurobiol Dis. 2009;36:181–190. doi: 10.1016/j.nbd.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 31.Sammons MA, Antons AK, Bendjennat M, Udd B, Krahe R, Link AJ. ZNF9 activation of IRES-mediated translation of the human ODC mRNA is decreased in myotonic dystrophy type 2. PLoS One. 2010;5:e9301. doi: 10.1371/journal.pone.0009301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raheem O, Olufemi SE, Bachinski LL, Vihola A, Sirito M, Holmlund-Hampf J, Haapasalo H, Li YP, Udd B, Krahe R. Mutant (CCTG)n expansion causes abnormal expression of zinc finger protein 9 in myotonic dystrophy type 2. Am J Pathol. 2010;177:3025–3036. doi: 10.2353/ajpath.2010.100179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Udd B, Krahe R, Wallgren-Pettersson C, Falck B, Kalimo H. Proximal myotonic dystrophy--a family with autosomal dominant muscular dystrophy, cataracts, hearing loss and hypogonadism: Heterogeneity of proximal myotonic syndromes? Neuromuscul Disord. 1997;7:217–228. doi: 10.1016/s0960-8966(97)00041-2. [DOI] [PubMed] [Google Scholar]
- 34.Udd B, Meola G, Krahe R, Thornton C, Ranum LP, Bassez G, Kress W, Schoser B, Moxley R. 140th ENMC international workshop: Myotonic dystrophy DM2/PROMM and other myotonic dystrophies with guidelines on management. Neuromuscul Disord. 2006;16:403–413. doi: 10.1016/j.nmd.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 35.Vihola A, Bassez G, Meola G, Zhang S, Haapasalo H, Paetau A, Mancinelli E, Rouche A, Hogrel JY, Laforet P, Maisonobe T, Pellissier JF, Krahe R, Eymard B, Udd B. Histopathological differences of myotonic dystrophy type 1 (DM1) and PROMM/DM2. Neurology. 2003;60:1854–1857. doi: 10.1212/01.wnl.0000065898.61358.09. [DOI] [PubMed] [Google Scholar]
- 36.Schoser BG, Schneider-Gold C, Kress W, Goebel HH, Reilich P, Koch MC, Pongratz DE, Toyka KV, Lochmuller H, Ricker K. Muscle pathology in 57 patients with myotonic dystrophy type 2. Muscle Nerve. 2004;29:275–281. doi: 10.1002/mus.10545. [DOI] [PubMed] [Google Scholar]
- 37.Auvinen S, Suominen T, Hannonen P, Bachinski LL, Krahe R, Udd B. Myotonic dystrophy type 2 found in two of sixty-three persons diagnosed as having fibromyalgia. Arthritis Rheum. 2008;58:3627–3631. doi: 10.1002/art.24037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rossi AE, Dirksen RT. Sarcoplasmic reticulum: The dynamic calcium governor of muscle. Muscle Nerve. 2006;33:715–731. doi: 10.1002/mus.20512. [DOI] [PubMed] [Google Scholar]
- 39.Berchtold MW, Brinkmeier H, Muntener M. Calcium ion in skeletal muscle: Its crucial role for muscle function, plasticity, and disease. Physiol Rev. 2000;80:1215–1265. doi: 10.1152/physrev.2000.80.3.1215. [DOI] [PubMed] [Google Scholar]
- 40.Petersen OH, Michalak M, Verkhratsky A. Calcium signalling: Past, present and future. Cell Calcium. 2005;38:161–169. doi: 10.1016/j.ceca.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 41.Osborne RJ, Lin X, Welle S, Sobczak K, O'Rourke JR, Swanson MS, Thornton CA. Transcriptional and post-transcriptional impact of toxic RNA in myotonic dystrophy. Hum Mol Genet. 2009;18:1471–1481. doi: 10.1093/hmg/ddp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bachinski LL, Sirito M, Bohme M, Baggerly KA, Udd B, Krahe R. Altered MEF2 isoforms in myotonic dystrophy and other neuromuscular disorders. Muscle Nerve. 2010;42:856–863. doi: 10.1002/mus.21789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kimura T, Lueck JD, Harvey PJ, Pace SM, Ikemoto N, Casarotto MG, Dirksen RT, Dulhunty AF. Alternative splicing of RyR1 alters the efficacy of skeletal EC coupling. Cell Calcium. 2009;45:264–274. doi: 10.1016/j.ceca.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sallinen R, Vihola A, Bachinski LL, Huoponen K, Haapasalo H, Hackman P, Zhang S, Sirito M, Kalimo H, Meola G, Horelli-Kuitunen N, Wessman M, Krahe R, Udd B. New methods for molecular diagnosis and demonstration of the (CCTG)n mutation in myotonic dystrophy type 2 (DM2) Neuromuscul Disord. 2004;14:274–283. doi: 10.1016/j.nmd.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 45.Haravuori H, Vihola A, Straub V, Auranen M, Richard I, Marchand S, Voit T, Labeit S, Somer H, Peltonen L, Beckmann JS, Udd B. Secondary calpain3 deficiency in 2q-linked muscular dystrophy: Titin is the candidate gene. Neurology. 2001;56:869–877. doi: 10.1212/wnl.56.7.869. [DOI] [PubMed] [Google Scholar]
- 46.Salvatori S, Furlan S, Fanin M, Picard A, Pastorello E, Romeo V, Trevisan CP, Angelini C. Comparative transcriptional and biochemical studies in muscle of myotonic dystrophies (DM1 and DM2) Neurol Sci. 2009;30:185–192. doi: 10.1007/s10072-009-0048-4. [DOI] [PubMed] [Google Scholar]
- 47.Zhang L, Kelley J, Schmeisser G, Kobayashi YM, Jones LR. Complex formation between junctin, triadin, calsequestrin, and the ryanodine receptor, proteins of the cardiac junctional sarcoplasmic reticulum membrane. J Biol Chem. 1997;272:23389–23397. doi: 10.1074/jbc.272.37.23389. [DOI] [PubMed] [Google Scholar]
- 48.Mu X, Brown LD, Liu Y, Schneider MF. Roles of the calcineurin and CaMK signaling pathways in fast-to-slow fiber type transformation of cultured adult mouse skeletal muscle fibers. Physiol Genomics. 2007;30:300–312. doi: 10.1152/physiolgenomics.00286.2006. [DOI] [PubMed] [Google Scholar]
- 49.Calabria E, Ciciliot S, Moretti I, Garcia M, Picard A, Dyar KA, Pallafacchina G, Tothova J, Schiaffino S, Murgia M. NFAT isoforms control activity-dependent muscle fiber type specification. Proc Natl Acad Sci U S A. 2009;106:13335–13340. doi: 10.1073/pnas.0812911106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orengo JP, Ward AJ, Cooper TA. Alternative splicing dysregulation secondary to skeletal muscle regeneration. Ann Neurol. 2011;69:681–690. doi: 10.1002/ana.22278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Du H, Cline MS, Osborne RJ, Tuttle DL, Clark TA, Donohue JP, Hall MP, Shiue L, Swanson MS, Thornton CA, Ares M., Jr Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol. 2010;17:187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Futatsugi A, Kuwajima G, Mikoshiba K. Tissue-specific and developmentally regulated alternative splicing in mouse skeletal muscle ryanodine receptor mRNA. Biochem J. 1995;305:373–378. doi: 10.1042/bj3050373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kimura T, Pace SM, Wei L, Beard NA, Dirksen RT, Dulhunty AF. A variably spliced region in the type 1 ryanodine receptor may participate in an inter-domain interaction. Biochem J. 2007;401:317–324. doi: 10.1042/BJ20060686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng W, Altafaj X, Ronjat M, Coronado R. Interaction between the dihydropyridine receptor Ca2+ channel beta-subunit and ryanodine receptor type 1 strengthens excitation-contraction coupling. Proc Natl Acad Sci U S A. 2005;102:19225–19230. doi: 10.1073/pnas.0504334102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burk SE, Lytton J, MacLennan DH, Shull GE. cDNA cloning, functional expression, and mRNA tissue distribution of a third organellar Ca2+ pump. J Biol Chem. 1989;264:18561–18568. [PubMed] [Google Scholar]
- 56.Brandl CJ, deLeon S, Martin DR, MacLennan DH. Adult forms of the Ca2+ATPase of sarcoplasmic reticulum. Expression in developing skeletal muscle. J Biol Chem. 1987;262:3768–3774. [PubMed] [Google Scholar]
- 57.Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci U S A. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garbino A, van Oort RJ, Dixit SS, Landstrom AP, Ackerman MJ, Wehrens XH. Molecular evolution of the junctophilin gene family. Physiol Genomics. 2009;37:175–186. doi: 10.1152/physiolgenomics.00017.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beard NA, Laver DR, Dulhunty AF. Calsequestrin and the calcium release channel of skeletal and cardiac muscle. Prog Biophys Mol Biol. 2004;85:33–69. doi: 10.1016/j.pbiomolbio.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 60.Beard NA, Wei L, Dulhunty AF. Ca(2+) signaling in striated muscle: The elusive roles of triadin, junctin, and calsequestrin. Eur Biophys J. 2009;39:27–36. doi: 10.1007/s00249-009-0449-6. [DOI] [PubMed] [Google Scholar]
- 61.Cacchiarelli D, Martone J, Girardi E, Cesana M, Incitti T, Morlando M, Nicoletti C, Santini T, Sthandier O, Barberi L, Auricchio A, Musaro A, Bozzoni I. MicroRNAs involved in molecular circuitries relevant for the duchenne muscular dystrophy pathogenesis are controlled by the dystrophin/nNOS pathway. Cell Metab. 2010;12:341–351. doi: 10.1016/j.cmet.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 62.Suzuki N, Mizuno H, Warita H, Takeda S, Itoyama Y, Aoki M. Neuronal NOS is dislocated during muscle atrophy in amyotrophic lateral sclerosis. J Neurol Sci. 2010;294:95–101. doi: 10.1016/j.jns.2010.03.022. [DOI] [PubMed] [Google Scholar]
- 63.Acharyya S, Butchbach ME, Sahenk Z, Wang H, Saji M, Carathers M, Ringel MD, Skipworth RJ, Fearon KC, Hollingsworth MA, Muscarella P, Burghes AH, Rafael-Fortney JA, Guttridge DC. Dystrophin glycoprotein complex dysfunction: A regulatory link between muscular dystrophy and cancer cachexia. Cancer Cell. 2005;8:421–432. doi: 10.1016/j.ccr.2005.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.