Abstract
Myocyte hypertrophy antecedent to heart failure involves changes in global gene expression, although the preceding mechanisms to coordinate DNA accessibility on a genomic scale are unknown. Chromatin-associated proteins alter chromatin structure by changing their association with DNA, thereby altering the gene expression profile. Little is known about the global changes in chromatin subproteomes that accompany heart failure, and the mechanisms by which these proteins alter chromatin structure. The present study tests the fundamental hypothesis that cardiac growth and plasticity in the setting of disease recapitulates conserved developmental chromatin remodeling events. We used quantitative proteomics to identify chromatin-associated proteins extracted via detergent and to quantify changes in their abundance during disease. Our study identified 321 proteins in this subproteome, demonstrating it to have modest conservation (37%) with that revealed using strong acid. Of these proteins, 176 exhibited altered expression during cardiac hypertrophy and failure; we conducted extensive functional characterization of one of these proteins, Nucleolin. Morpholino-based knockdown of nucleolin nearly abolished protein expression but surprisingly had little impact on gross morphological development. However, hearts of fish lacking Nucleolin displayed severe developmental impairment, abnormal chamber patterning and functional deficits, ostensibly due to defects in cardiac looping and myocyte differentiation. The mechanisms underlying these defects involve perturbed bone morphogenetic protein 4 expression, decreased rRNA transcription, and a shift to more heterochromatic chromatin. This study reports the quantitative analysis of a new chromatin subproteome in the normal and diseased mouse heart. Validation studies in the complementary model system of zebrafish examine the role of Nucleolin to orchestrate genomic reprogramming events shared between development and disease.
Keywords: proteomics, chromatin, nucleolin, cardiac hypertrophy, BMP4
during the development of hypertrophy and failure in the mammalian heart, adult cardiomyocytes undergo extensive transcriptional reprogramming, becoming more plastic in response to stress. Myocytes increase their mass and adopt a gene expression profile, as well as phenotypic features, normally associated with fetal myocardium (36). These gross changes in transcription may initially promote functional stability in the heart by allowing cells to compensate for an increased workload; however, studies from humans and animal models have consistently shown that hypertrophic remodeling leads to cardiac dysfunction and failure. While the transcription factors responsible for cardiac hypertrophy have been extensively studied (10, 35), the global chromatin remodeling events are less well understood. In the past few years, the field has gained a new appreciation for the global changes in chromatin features responsible for cardiovascular phenotypes in development and disease (3, 31), including from studies on chromatin remodeling complexes (20). However, large changes in gene expression must be preceded by coordinated alterations in chromatin structure to allow or deny accessibility of transcriptional machinery to specific genomic regions. This global process has local ramifications, in which the functional unit of chromatin—the nucleosome (the DNA-protein complex comprised of two copies each of four histone proteins)—alters its accessibility between structurally compact and transcriptionally inactive heterochromatin and structurally loose and transcriptionally active euchromatin. Therefore, knowledge of the constitutive protein occupants of chromatin, as well as the remodelers and other structural proteins that alter their association with the genome during development and disease, is an important step toward understanding how global changes in chromatin accessibility are coordinated.
To address this question, we sought to quantify a subproteome of chromatin during the development of hypertrophy and failure following pressure overload in the mouse. Of the 321 (698 total, 321 by 2 or more peptides) proteins measured, 176 (338 total, 176 by 2 or more peptides) changed with heart disease in the mouse. We then employed a complementary model system (zebrafish development) to investigate the role of one chromatin-bound protein, Nucleolin.
Nucleolin is commonly characterized as a nucleolar protein, although it has been observed throughout the nucleus, in the cytoplasm, and on the plasma membrane. Nucleolin is highly expressed in proliferating cells and has been proposed to regulate cell growth and apoptosis in noncardiac systems. It is also involved in several aspects of gene expression, including chromatin remodeling, RNA transcription, rRNA processing, nucleo-cytoplasmic transport, and mRNA stabilization (11, 17–19, 22). Much research has focused on Nucleolin's ability to regulate ribosome biogenesis, because protein synthesis is a key element for controlling growth, making this process a key readout for endogenous Nucleolin function. Nucleolin has also been shown to modulate the expression of p53 both negatively and positively, depending on the cell type. Robust cardiac Nucleolin expression has been observed during murine embryogenesis, followed by a precipitous decline during the first week of life, concomitant with decreased myocyte growth and cell cycle exit, with relatively low levels of Nucleolin detected in the normal adult heart (2). However, the functional role of Nucleolin in cardiac phenotype during development and disease is unknown in any species. Our findings implicate Nucleolin in cardiomyocyte differentiation and heart formation, providing evidence for this protein in regulation of chromatin structure and ribosome biogenesis.
MATERIALS AND METHODS
Mouse model of cardiac hypertrophy and failure and echocardiographic determination of cardiac function.
All protocols involving animals conform to the NIH Guide for the Care and Use of Laboratory Animals and were approved by the UCLA Animal Research Committee. Adult male balb/c mice aged 8–12 wk (Charles River Laboratories) were used for this study. The murine model of transverse aortic banding (TAC)-induced cardiac hypertrophy was performed as described previously (13, 32). SHAM operated mice underwent the same procedure without placement of aortic clamp. Echocardiography (ECHO) was used to determine cardiac parameters in live mice as described (13, 32), including the following indexes: left ventricular (LV) size (end-diastolic and end-systolic dimension), wall thickness (intraventricular septum and posterior wall thickness), ventricular mass, ventricular function [ejection fraction (EF)], and blood flow. All mice underwent ECHO analyses once before TAC or SHAM surgery, once a day after, and then once every 5 days for the duration of the study. Animals were considered hypertrophic when their LV mass was greater than the mean of the control animal with no depression of LV function as measured by EF; animals were considered in heart failure when the EF was significantly decreased below the mean of the control animals. These phenotypes, hypertrophy and failure, corresponded to ∼2 and ∼4 wk after TAC surgery, although animals were euthanized based on echo parameters and not solely on time.
Nuclear isolation and fractionation.
All buffers used for cell isolation or fractionation in this study contained the following protease, phosphatase, and deacetylase inhibitors, respectively: 0.1 mM phenylmethanesulfonylflouride, protease inhibitor cocktail pellet (Roche), 0.2 mM sodium orthovanadate, 0.1 mM sodium flouride, and 10 mM sodium butyrate. Cardiac nuclei were isolated as previously described (14). We consistently achieve ≥80% purity of nuclei with this method as observed by electron microscopy and Western blotting analysis. Following isolation of nuclei, further fractionation was carried out to separate nucleoplasm from chromatin using detergent extraction. Briefly, isolated nuclei were resuspended in buffer [20 mM HEPES (pH 7.6), 7.5 mM MgCl2, 30 mM NaCl, 1 M urea, 1% NP-40] to solubilize the nuclear membrane and extract soluble proteins in the nucleoplasm. After solubilization, samples were centrifuged at 13,000 g for 10 min to pellet the insoluble chromatin and remove the nucleoplasm fraction. The chromatin pellet was washed with PBS, solubilized in 50 mM Tris (pH 8), 10 mM EDTA, 1% SDS, sonicated to shear the DNA, and centrifuged at 13,000 g to extract proteins (referred to as detergent-extracted fraction). This method is distinct from the low pH method of protein extraction (acid-extraction) and, as demonstrated throughout the current manuscript (see Fig. 1), reveals a biologically distinct subproteome of molecules.
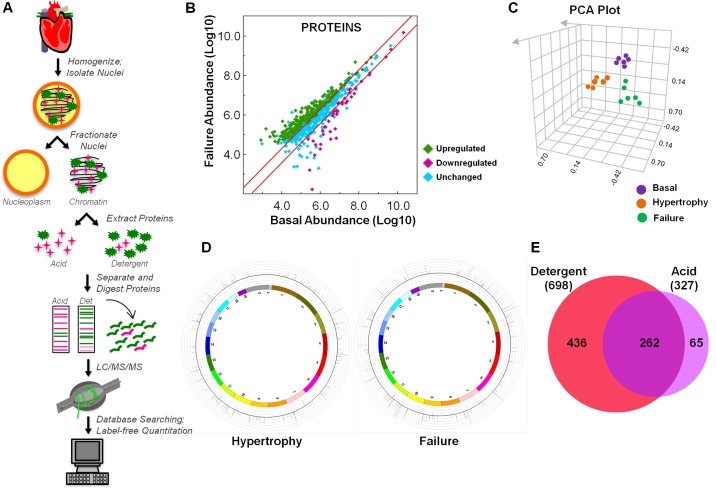
Fig. 1.
Proteomic quantification of chromatin proteins in murine heart. A: schematic workflow of mass spectrometric identification of mouse chromatin proteins and label-free quantitation. In this study, loosely associated chromatin proteins were isolated using detergent (as opposed to tightly bound proteins which can be isolated only in the presence of low pH, referred to as “acid-extracted proteins” in the figure and text) to investigate proteins capable of transient regulation of the genome during stages of cardiac hypertrophy and failure. B: peptides identified by mass spectrometry were mapped to proteins and relative quantitation determined using the Elucidator software program. Proteins increasing in abundance during the failure stage are shown as green diamonds, those decreasing as purple diamonds, and those unchanged (or not statistically significant) as blue diamonds. Red lines indicate 2-fold change. C: the reproducibility of peptide abundance changes from basal, hypertrophied, and failing hearts was calculated using ANOVA on the 6 replicates (in each of the 3 conditions) and principal component analysis (PCA) performed. D: all proteins whose abundance was found to change in stages of hypertrophy or failure by 2-fold or greater were mapped to their chromosomal location, with relative change in abundance indicated by the inflection of the lines (toward the center being downregulated, and toward the outside being upregulated), each of which corresponds to a single protein. E: all proteins identified in this study were compared to proteins identified from our recent analysis of acid-extracted chromatin (13) using a Venn diagram to display overlap in the two datasets; note that the majority of proteins identified in the present study are distinct from our previous analysis, supporting this fractionation approach as having revealed a biologically distinct pool of proteins. F: to identify groups of proteins with similar changes in abundance, we converted relative abundance values to Z-scores, with each protein displayed as a single line, and performed unsupervised clustering (G) based on similar quantitative behavior during disease. H: the nine resulting modules contain proteins with corresponding changes in abundance. I: gene ontology (GO) analysis of each module—rendered in this panel as a heat map—highlights biological processes and molecular functions enriched in each module. J: changes in protein abundance across disease states were compared between the two chromatin compartments [detergent (this study) vs. acid-extraction (13)], with some proteins (Histone H1.1) showing the same pattern in both fractions, while others (Ruvb like-1) behaved differently. K: we also performed GO term enrichment analysis in which proteins differentially regulated in hypertrophy or failure chromatin were compared to proteins differentially regulated in published datasets from more or less differentiated cells or cancer or healthy tissue to determine if biological processes involved in heart disease are more similar to either cancer or development (see Supplemental Table 4, available with the online version of this article). To compare these subproteomes, we examined the “biological process” term set, with that grouping being the broadest categorization; we subdivided other more discriminating terms into “intermediate” or “specific” as informed by their tier in the ontological tree. While processes enriched in either hypertrophic or failing chromatin had greater overlap with healthy processes, as opposed to cancer (right panels), when comparing the most specific processes (red boxes), we observed that hypertrophy shares more common processes with less differentiated cells while failure better matches more differentiated cells (left panels).
Enzyme digestion.
Detergent-extracted proteins isolated from chromatin were separated by SDS-PAGE. Each gel lane was cut into 25 slices (∼2 mm each) for protein identification by mass spectrometry (MS). Gel plugs were dehydrated in acetonitrile and dried in a Speedvac. Samples were reduced and alkylated with 10 mM dithiotreitol and 10 mM tris(2-carboxyethyl)phosphine solution in 50 mM ammonium bicarbonate (30 min at 56°C) and 100 mM iodoacetamide (25 min in dark), respectively. Gels were washed with 50 mM ammonium bicarbonate, dehydrated with acetonitrile, and dried in a Speedvac. Gel pieces were then swollen in digestion buffer containing 50 mM ammonium bicarbonate, and 20.0 ng/μl of trypsin (37°C, overnight). Peptides were extracted with 0.1% formic acid in 50% acetonitrile solution, dried down, and resuspended in 0.1% formic acid, 2% acetonitrile. For each condition (basal, hypertrophy, and failure) three biological (de novo preparation of samples from different animals) and two technical (multiple LC/MS/MS experiments on the same preparation) replicates were analyzed by mass spectrometry.
Mass spectrometry analyses and database searching.
Extracted peptides were analyzed by nano-flow LC/MS/MS on a Thermo Orbitrap with dedicated Eksigent nanopump using a reversed-phase column (75 μm ID, 10 cm, BioBasic C18 5-μm particle size, New Objective) and a flow rate of 200 nl/min. For peptide separation a linear gradient was utilized from 95% buffer A (0.1% formic acid, 2% acetonitrile) and 5% buffer B (0.1% formic acid, 20% water in acetonitrile) to 50% buffer A and 50% buffer B over 60 min. Spectra were acquired in data-dependent mode with dynamic exclusion where the instrument selects the top six most abundant ions in the parent spectra for fragmentation. Data were searched against the Uniprot database (version 03.2011) using the SEQUEST algorithm in the BioWorks software program version 3.3.1 SP1 and through the Rosetta Elucidator software (Microsoft). False discovery rate, which was calculated on several independent datasets within this study by reverse database searching, ranged from 1.4 to 1.7%. All spectra used for identification had deltaCN>0.1, consensus score ≥20, and met the following Xcorr criteria: >3 (+2), >4 (+3), and >5 (+4). Searches required full tryptic cleavage, ≤3 missed cleavages and were performed with the differential modifications of carbamidomethylation on cysteine and methionine oxidation. Mass tolerance was 0.5 Da for precursor and 1 Da for product ions. All proteins were identified on the basis of two or more unique peptides.
Bioinformatics and protein annotation.
Label-free quantitation of peptide/protein expression was accomplished using the Rosetta Elucidator software (Microsoft). For Elucidator analyses, peptides across the entire chromatographic run for each sample were aligned between MS runs and between conditions (basal, hypertrophy, failure). The peak intensity for each eluting peptide was calculated as area under the extracted ion chromatographic curve. To determine protein abundance, intensity data for all peptides mapping to a protein were combined, and data from three biological and two technical replicates was averaged for each of the three conditions. Proteins whose intensity changed ≥2 fold between conditions with a P value ≤ 0.01 were considered to be statistically significant. To identify modules of proteins with similar expression behavior, intensity values were converted to Z-scores. Proteins were then clustered using a self-organizing map with x and y nodes of 3,3 and cosine correlation. Intensity data were coupled to peptide identification, which was determined using the SEQUEST algorithm described above.
Protein expression plots were generated as described (27). Redundancy in proteins was eliminated at the primary sequence level by manual inspection using CLUSTAL to compare the sequences in UniProt. For genome analysis (i.e., for determining from where in the genome mRNAs for the detected proteins were transcribed) UniProt IDs, from MS data, were converted into Ensembl gene IDs using the UniProt ID Mapping tool (http://www.uniprot.org/?tab=mapping), and gene annotations were obtained from Ensembl data (version 65). GO annotation enrichment analysis was performed using the DAVID Bioinformatics Resource (v6.7) developed by the NIAID (NIH). The Interpro and KEGG analysis functions of DAVID were utilized to determine enrichment in protein domains and functional pathways, respectively.
Gene ontology (GO) term enrichment analyses of proteins differentially regulated in hypertrophy or failure chromatin were compared to GO terms of proteins differentially regulated in published datasets from more or less differentiated cells or cancer or healthy tissue to determine if biological processes involved in heart disease are more similar to either cancer or development. To compare these subproteomes, we examined the “biological process” term set, with that grouping being the broadest categorization; we subdivided other more discriminating terms into “intermediate” or “specific” as informed by their hierarchical level in the ontological tree. PMID references to each dataset can be found in Supplemental Table 4 (all supplemental tables cited are available with the online version of this article).
Zebrafish studies.
Zebrafish colonies were cared for and bred under standard conditions (7, 39a). Developmental stages of embryos were determined using standard morphological features of fish raised at 28.5°C (39a).
Three of the proteins identified from our mass spectrometry data (with known or hypothesized roles in regulating chromatin structure and gene expression) were initially screened in zebrafish. Nucleolin was one of these three proteins and gave the most promising phenotype after knockdown and overexpression; we therefore chose to examine it in further detail.
For overexpression experiments, the full-length cDNA for zebrafish nucleolin was amplified with KOD polymerase (Novagen) and cloned into pCS2 + 3XFLAG. Plasmids were cut with SalI, and SP6 RNA polymerase was used to generate mRNA for injection. Nucleolin mRNA was injected alone or coinjected with a p53 MO. Control injections performed using p53 MO alone have no effect on zebrafish development (data not shown). For Morpholino-based knockdown experiments a Morpholino oligonucleotide targeting the translation initiation site of nucleolin (Ncl MO) was purchased from Gene Tools. Nucleolin MO was coinjected with a p53 Morpholino to prevent nonspecific cell death. The sequences of the Morpholinos used are as follows (from 5′ to 3′): Nucleolin MO: TAGCTGCCTTAGCGAGCTTTACCAT; p53 MO: GCGCCATTGCTTTGCAAGAATTG. Morpholino efficacy was tested by Western blotting to detect levels of Nucleolin protein in 24 and 72 hpf embryo lysates. Embryos were deyolked in calcium-free Ringer's solution with 1 mM EDTA and then lysed in buffer containing 50 mM Tris (pH 8), 10 mM EDTA, and 1% SDS, with protease [0.1 mM phenylmethanesulfonylflouride, protease inhibitor cocktail pellet (Roche)], phosphatase (0.2 mM sodium orthovanadate, 0.1 mM sodium flouride), and deacetylase (10 mM sodium butyrate) inhibitors, sonicated to shear the DNA and centrifuged at 13,000 g to extract proteins.
Whole mount in situ hybridization.
Embryos for in situ hybridization were raised in embryo medium supplemented with 0.2 mM 1-phenyl-2 thiourea to maintain optical transparency (39a). Whole mount in situ hybridization was performed as described previously (4). The antisense RNA probes used in this study include nucleolin (ncl), ventricular myosin heavy chain (vmhc), atrial myosin heavy chain (amhc, also known as myh6), bone morphogenetic protein 4 (bmp4), and versican.
Whole mount immunohistochemistry and cell size analysis.
Embryos expressing mCherry protein from the cmlc2 promoter (also known as myl7) were injected with Nucleolin Morpholino at the one-cell stage and assessed at 48 hpf for a cardiac phenotype. Uninjected embryos were also maintained for control. At 72 hpf, embryos were fixed overnight in 4% paraformaldehyde in phosphate-buffered saline (PBS) at 4°C. Embryos were then rinsed two times in PBS with 0.1% Tween 20 (PBST) and left in methanol overnight at 4°C. The pericardia were manually removed and the embryos left in acetone at −20°C for 15 min. They were then rinsed two times in water and once in PBS and then washed two times, 5 min each, in PBST. They were blocked 1 h in blocking buffer [0.2 g/ml Roche Blocking Reagent (cat. no. 11 096 176 001), and 5% goat serum in PBST] at room temperature and left overnight at 4°C in a 1:100 primary antibody dilution in blocking buffer [anti-Neurolin Zn8 (Zn-8-s, Developmental Studies Hybridoma Bank)]. Embryos were washed 4 times 15 min each in PBST and left overnight at 4°C in a 1:150 secondary antibody dilution in blocking buffer [goat anti-mouse FITC (1070–02, Southern Biotech)]. Embryos were again washed 4 times, 15 min each, in PBST and mounted using low-melt agarose. Imaging was performed on a Leica TCS-SP1 upright confocal microscope (in the CNSI Advanced Microscopy/Spectroscopy Core at UCLA) using a water-immersion objective (HCX APO L 40× 0.8W). FITC was visualized with an argon laser (488 nm), and mCherry with a diode laser (561 nm). The ventricle was optically sectioned into 3.012-μm sections and the maximum intensity projection used for analysis in Photoshop. Anti-Zn8 antibody demarcates the cell boundary by labeling the cell membrane protein Neurolin. Laser power was optimized for each fish to enable discrimination of the cell borders in the maximum intensity projection. Any cell with its complete border clearly visible was traced in Photoshop, with roughly 12 cells analyzed per fish.
For imaging Nucleolin's localization in the zebrafish heart, whole mount immunohistochemistry was performed on uninjected, 48 hpf embryos expressing cmlc2-driven mCherry following the same protocol as for cell size analysis with the exception of the antibodies [primary: anti-Nucleolin (ab22758, abcam); secondary: goat anti-rabbit FITC (4050-02, Southern Biotech and A11008, Invitrogen)]. Embryos were soaked overnight in PBS containing DAPI and rinsed three times in PBST prior to imaging. Images were acquired on a Zeiss LSM 510 with a water immersion objective (63×/0.9W Achroplan).
Cardiac output.
End-diastolic volume (EDV), end-systolic volume (ESV), stroke volume, and cardiac output (product of heart rate and stroke volume) were determined at 48 hpf from time-lapse recordings of a lateral view of the beating zebrafish heart. To calculate stroke volume we separately averaged EDV and then ESV for a given fish across three consecutive beats. Volume was calculated using the Simpson method of stacked discs (8, 37). Briefly, two-dimensional cross sectional images of the ventricle were obtained (end diastole and end systole determined as the largest and smallest 2D sections, respectively), and the ventricular space was divided into a series of bands representing the three-dimensional chamber. We set the thickness of each disc (or band) to be one pixel (1.2 μm) and measured the length of the band (diameter of the disc) in pixels as well. We assumed that the height of the disc is the same as length and from there calculated volume. Finally, the volumes of each disc were summed to give chamber volume at end diastole or end systole.
Isolated rat cardiomyocytes.
Neonatal rat ventricular myocytes (NRVMs) were obtained by enzymatic dissociation from 1-day-old litters and plated in DMEM media (Invitrogen, no. 11965) containing 1% penicillin, 1% streptomycin, 1% insulin-transferrin-sodium selenite supplement, and 10% fetal bovine serum for the first 24 h, after which the cells are cultured in serum- and antibiotic-free media. NRVMs were treated with 50 nM siRNA targeted to nucleolin (25 nM each SI00252476 and SI03107776; Qiagen) or scrambled siRNA (cat. no. 1027280; Qiagen) for 72 h. Transfections were performed with Lipofectamine RNAiMax Transfection Reagent (Invitrogen). To induce hypertrophy in isolated NRVMs, cells were treated with one of the following three hypertrophic agonists—isoproterenol (ISO, 1 μM), phenylephrine (PE, 10 μM), or endothelin-1 (ET-1, 1 nM)—for 48 h.
Electrophoresis and western blotting.
Proteins were separated by standard SDS-PAGE using Laemmli buffer. Gels (12%) were stained with Oriole (Bio-Rad). For Western blotting, proteins were transferred to nitrocellulose, membranes blocked with milk, and protein signals detected by enzyme linked chemiluminescence (GE Biosciences). Ponceau staining of membranes was used to confirm transfer and protein loading. Antibodies used in this study were as follows, including sources: Histone H3 (Abcam, ab1791, 1:10,000 dilution); Gapdh (Santa Cruz, sc-20357, 1:1,000 dilution); Histone H3-trimethylated-K9 (Abcam, ab8898, 1:500 dilution); Nucleolin (Abcam, ab22758, 1:1,000 dilution); β-Actin (Sigma, A1978, 1:1,000 dilution); p53 [Santa Cruz, sc1313, 1:500 dilution (NRVMs) and Abcam, ab77813, 1:200 dilution (zebrafish)]; and FLAG (Sigma, F1804, 1:1,000 dilution).
Quantitative real-time PCR analysis.
Total RNA was isolated from the left ventricle of the heart, from cultured NRVMs, and from zebrafish embryos using TRIzol (Invitrogen) according to the manufacturer's protocol. Total RNA was transcribed using SuperScript First-Strand Synthesis system for RT-PCR (Invitrogen) according to the manufacturer's protocol to produce cDNA. cDNA transcripts were amplified on the iCycler iQ real-time PCR detection system with iQ SYBR Green Supermix (Bio-Rad). Expression levels were analyzed using the iQ5 Optical Systems software v2.0 and normalized against GAPDH by subtracting the mean cycle number for each experimental group from the mean cycle number for GAPDH from the same group. Fold change was calculated using the comparative ΔΔCt method. Primers [forward (F) and reverse (R)] used in this study are as follows: GAPDH F-5′- CCCACTAACATCAAATGGGG-3′ R-5′-CCTTCCACA ATGCCAAAGTT-3′; ANF F-5'-CTGATGGATTTCAAGAACCTGCT-3′ R-5′-CTCTGGGCTCCAATCCTGTC-3′; SERCA2a F-5′-CCTTCTACCAGCTGAGTCATTT-3′ R-5′-CAGATGGAGCCCACGACCCA-3′; α-MHC F-5′-GAACAGCTGGGAGAAGGGGG-3′ R-5′-GCCTCTGAGGCTATTCTATTGG-3′; and β-MHC F-5′-CTCAACTGGGAAGAGCATCCA-3′ R-5′-CCTTCAGCAAACTCTGGAGGC-3′; Ncl-Mouse F-5′-AAGCAGCACCTGGAAAACG-3′ R-5′-TCTGAGCCTTCTACTTTCTGTTTCTTG-3′; 18S-Mouse F-5′-CGAGCCGCCTGGATACC-3′ R-5′-CATGGCCTCAGTTCCGAAAA-3′; ITS-Mouse F-5′-TCCGTGTCTACGAGGGGCGG-3′ R-5′-GGGTGCCGGGAGAGCAAAGC-3′; 18S-Rat F-5′-CGAGCCGCCTGGATACC-3′ R-5′-CATGGCCTCAGTTCCGAAAA-3′; ITS-Rat F-5′-GGCGGAGGGGGTTTC-3′ R-5′-GAGCGAGAAAACGGAGGAG-3′; 18S-Zebrafish F-5′-GATTGATAGCTCTTTCTCGATTCTG-3′ R-5′-GTAACTATTTAGCATGCCGGAGTCT-3′; ITS-Zebrafish F-5′-GTTCAAAGACCTTCCCGTCTC-3′ R-5′-CGACACCACAAAGAGGATGTT-3′; Gapdh-Zebrafish F-5′-TGTGATGGGAGTCAACCAGGACAA-3′ R-5′-TTAGCCAGAGGAGCCAAGCAGTTA-3′.
RESULTS
Measurement of chromatin proteins with altered expression during disease.
To identify proteins regulating gene expression changes in the mouse heart during disease, we isolated chromatin-binding proteins from healthy animals as well as those in stages of cardiac hypertrophy or failure (Fig. 1A). In contrast to our recent report (13) using this same disease model, the present study used detergent to extract proteins, thereby selecting for a more loosely associated chromatin subproteome compared with the low-pH extraction (acid extraction) used in our previous paper. This technical change in the experimental workflow has biological implications, as shown in the Venn diagram comparing the two studies in Fig. 1E (this study vs. Ref. 13).
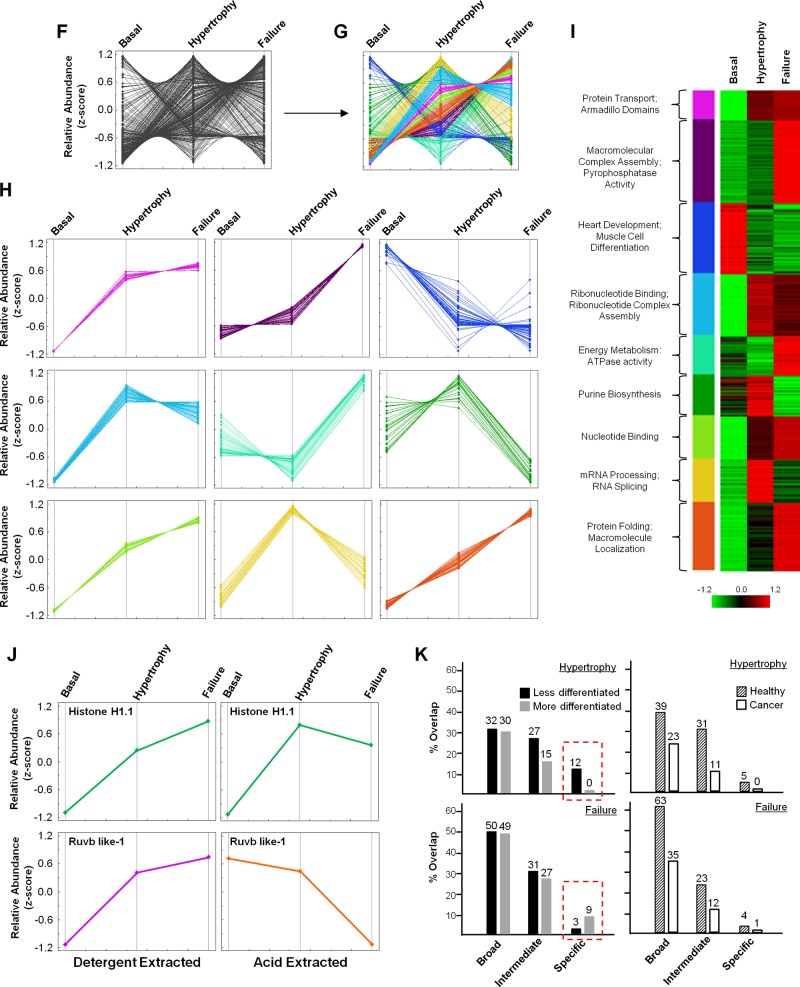
Proteins were separated by 1D electrophoresis, in-gel digested, and analyzed on an Orbri-trap mass spectrometer (Fig. 1A). Label-free quantitation of identified peptides was performed to elucidate abundance changes. In total, 2,068 peptides were identified and mapped to 698 proteins (377 identified by 1 peptide, 321 identified by 2 or more peptides, 338 of which displayed altered abundance by twofold or greater with a P value ≤ 0.01) (Fig. 1B, Supplemental Tables 1–3, available with the online version of this article). To confirm the reproducibility of the abundance changes, we performed statistical analysis (ANOVA) on all peptides identified as well as principal component analysis (PCA), which shows clustering of both biological and technical replicates (Fig. 1C). The 338 proteins whose abundance was modulated during disease (176 quantified by 2 or more peptides) were mapped to their chromosomal location to identify genomic regions altered during cardiac dysfunction in the mouse (Fig. 1D). We also performed unsupervised clustering of these 338 proteins to identify self-organizing trends, which revealed nine modules of proteins, the members of which have similar behavior in terms of abundance on chromatin during disease progression (Fig. 1, F–H). Gene ontology (GO) analysis of the proteins in each module revealed the molecular functions and biological processes enriched in each group (Fig. 1I). Proteins identified in this study were compared to our previous analysis of acid-extracted chromatin, which primarily enriches for histone proteins. This comparison revealed only 37% of the proteins in the detergent-extracted procedure (present study) were also present in the pool revealed following acid-extraction (previous study; Fig. 1E), with some proteins behaving differently during disease in the distinct fractions (Fig. 1J). We also performed GO term enrichment analysis in which proteins differentially regulated in hypertrophy or failure chromatin were compared to proteins differentially regulated in published datasets from more or less differentiated cells or cancer or healthy tissue to determine if biological processes involved in heart disease are more similar to either cancer or development (Supplemental Table 4). Grouping the biological terms by hierarchical level in the GO analysis tree revealed that the processes enriched in either hypertrophy or failure had greater overlap with those enriched in healthy, as opposed to those enriched in cancer, at all levels of the hierarchical tree (broad, intermediate, and specific) (Fig. 1K, right panel). However, when comparing the most specific processes of hypertrophy and failure to differentiation (red boxes), we observe distinct behavior: hypertrophy shares more processes with less differentiated cells, while failure better matches more differentiated cells (Fig. 1K, left panels).
Nucleolin knockdown in mammalian cardiomyocytes promotes fetal gene expression.
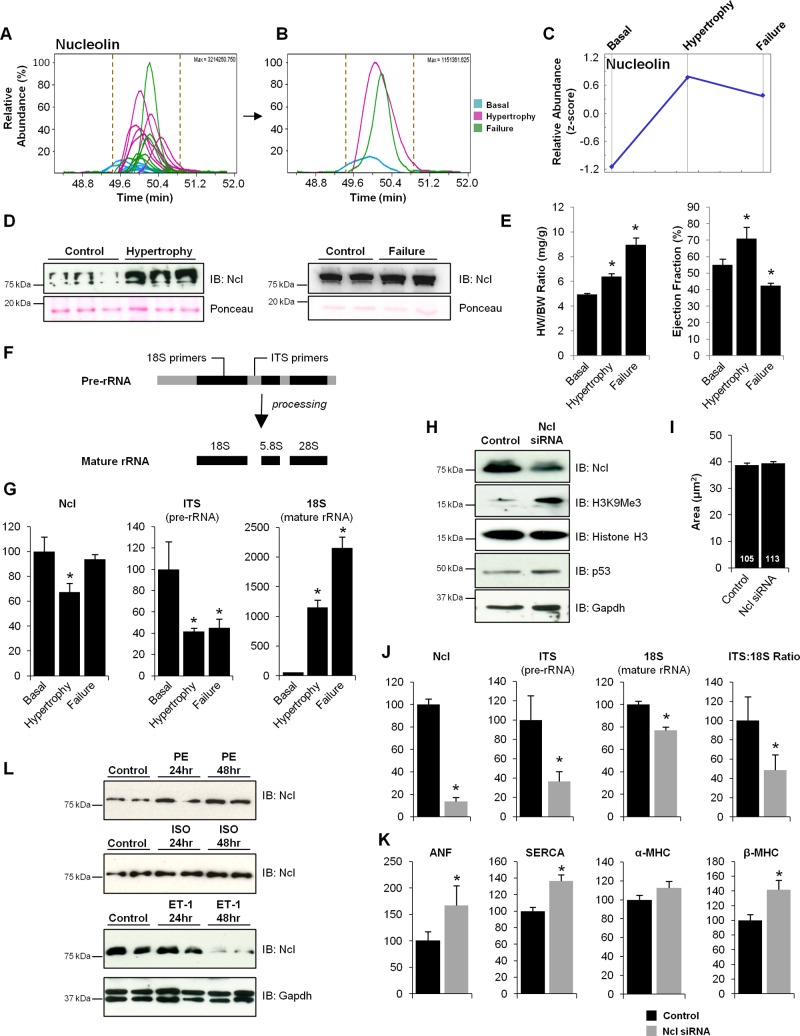
To examine the hypothesis that cardiac growth and plasticity in disease recapitulates conserved aspects of development, we explored the role of one protein—Nucleolin—whose abundance on chromatin increased in the mouse heart during hypertrophy and failure (Fig. 2, A–D). Mass spectrometry-based quantitation of one nucleolin peptide (Fig. 2, A–C), performed by determining the area under the curve for all 18 chromatographic peaks, identified by mass spectrometry from mouse hearts under basal, hypertrophy, or failure conditions (6 replicates per condition), was confirmed by Western blotting (Fig. 2D). Although nuclear Nucleolin was found to increase in this model, total cellular (nuclear and cytoplasmic) levels of Nucleolin, in mice euthanized in stages of hypertrophy or failure (Fig. 2E), decreased in the hypertrophic myocardium (Fig. 2G), accompanied by changes in pre-rRNA (ITS) and mature (18S) rRNA (Fig. 2, F and G).
Fig. 2.
Identification of Nucleolin as a candidate regulator of cardiac growth. A: abundance of chromatin-bound Nucleolin was quantified by calculating the area under the curve (from 18 replicates: 6 replicates in each of 3 conditions) for each of two Nucleolin peptides used for identification and quantitation. Shown are the extracted ion chromatograms of the Nucleolin peptide VEGSEPTTPFNLFIGNLNPNK; the x-axis indicates chromatographic time. B: replicates from the three biological conditions (basal, hypertrophy, and failure) were averaged to give average abundance values for each condition which, as shown in the trend plot (panel C; this plot includes mean value of two peptides used for detection/quantitation), demonstrate an increase in Nucleolin during hypertrophy that persists in the failing heart. D: increased levels of chromatin-bound Nucleolin (Ncl) from mice in cardiac hypertrophy and failure were confirmed via Western blotting, supporting mass spectrometry results. IB, immunoblot. Total cellular (nuclear and cytoplasmic) levels of Nucleolin was also examined in mice euthanized in stages of hypertrophy or failure (E; *P < 0.01.), as determined by heart weight (HW) to body weight (BW) ratio and ejection fraction (ECHO), by qPCR analysis showing a decrease in the hypertrophic myocardium, accompanied by changes in pre-rRNA (ITS) and mature (18S) rRNA using primers that uniquely detect these species (F and G). These results suggest that while chromatin bound Nucleolin increases during cardiac hypertrophy, total Nucleolin mRNA levels in the cell decrease. To examine the role of Nucleolin in the cardiomyocyte, isolated neonatal rat ventricular myocytes (NRVMs) were treated with Nucleolin siRNA or lipofectamine control. Western blot analysis confirmed that loss of Nucleolin results in an increase in the heterochromatin mark H3K9Me3 (blots are indicative of 3 independent experiments, n = 3 in each; H) but had no effect on cell size under basal conditions when quantified via phalloidin staining (I). Number of cells measured is indicated in each bar. F: rRNA transcription and maturation was examined using primers that detect pre-rRNA (ITS) and mature rRNA (18S) by qRT-PCR. J: loss of Nucleolin in NRVMs also led to a decrease in both rRNA transcription and maturation. n = 3/group. Asterisks indicate a P value < 0.05. K: strikingly, loss of Nucleolin in rat ventricular myocytes also led to an increase in the expression of β-MHC, ANF and Serca2a, suggesting that Nucleolin regulates some of the mRNA-encoding genes known to control pathological cardiac growth. L: additionally, while the fraction of Nucleolin bound to chromatin increases in a mouse model of pressure-overload hypertrophy, total cellular levels of Nucleolin were altered in an agonist-dependent manner in the setting of hypertrophy in NRVMs (n = 3/group). Error bars indicate SE. ISO, isoproterenol; ET-1, endothelin-1; PE, phenylephrine.
Having observed alterations of chromatin-associated Nucleolin in pressure-overload hypertrophy, we sought to explore the actions of Nucleolin in mammalian cardiomyocytes. Knockdown was carried out in isolated neonatal rat ventricular myocytes (NRVMs) using siRNA, leading to an ∼80% decrease in Nucleolin protein level, concomitant with an increase in the heterochromatin mark, H3K9Me3 (Fig. 2H), supporting Nucleolin's role in decondensing chromatin. While Nucleolin KD did not have an effect on cell size under basal conditions (Fig. 2I), the addition of hypertrophic agonist in the presence of Ncl siRNA was lethal to these cells (data not shown). To evaluate the effect of Nucleolin knockdown on gene transcription we quantified levels of pre-rRNA (ITS) and mature rRNA (18S) in the absence or presence of siRNA (Fig. 2F). Nucleolin KD in NRVMs resulted in decreased rRNA transcription (ITS) and subsequent maturation (18S) (Fig. 2J), providing a functional readout for Nucleolin knockdown and suggesting that Nucleolin plays a role in regulating ribosomal transcription in the cardiomyocyte. Surprisingly, we observed increases in the fetal isoform (beta) of myosin heavy chain, ANF, and the calcium handling protein SERCA (Fig. 2K), suggesting that Nucleolin may regulate transcription of other (nonribosomal) genes in the cardiomyocyte (although whether this regulation involves direct binding of Nucleolin to these genes remains to be determined). Interestingly, NRVMs treated with hypertrophic agonists [endothelin-1 (ET-1), isoproterenol (ISO) or phenylephrine (PE)] displayed differential regulation of Nucleolin protein in an agonist-dependent manner with increases in protein observed after ISO and PE treatment and a decrease in protein after ET-1 treatment (Fig. 2L).
Nucleolin knockdown and overexpression in zebrafish leads to cardiac dysfunction.
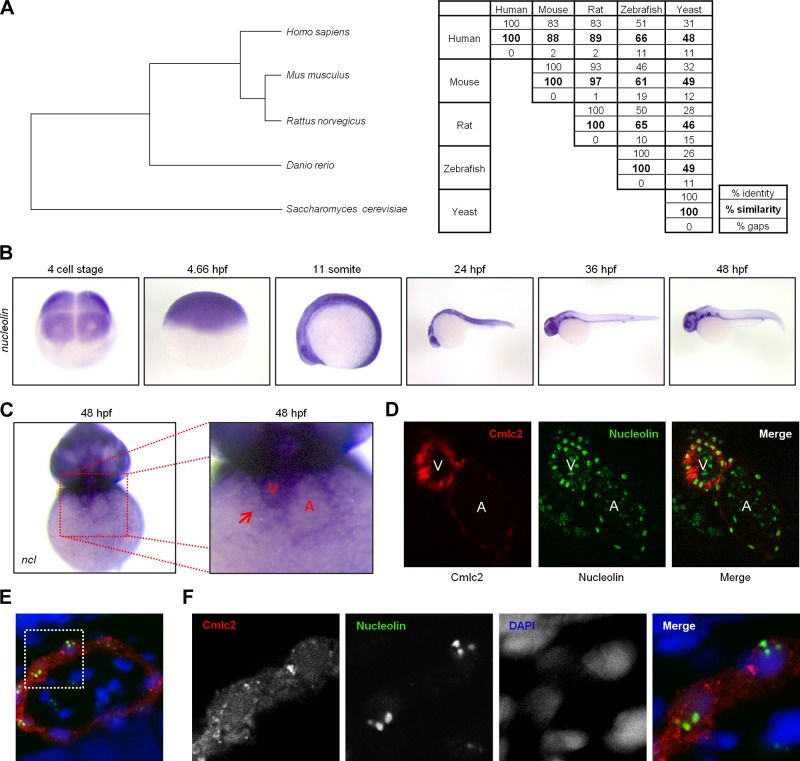
To further evaluate our hypothesis we employed a new model system to examine development in the zebrafish (Danio rerio), whose translucent and relatively porous viscera respectively facilitate microscopic observation and studies of cardiac development (because morphogenic abnormalities in the heart are often not embryonically lethal, in that the animal can absorb oxygen directly from the water through early maturation). Nucleolin is a highly conserved protein (Fig. 3A) whose mRNA is ubiquitously expressed during development (Fig. 3B), including in both chambers of the heart (Fig. 3, C and D), where it is most highly expressed in the nucleus (Fig. 3, E and F).
Fig. 3.
Nucleolin expression in the developing zebrafish embryo. A, left: phylogenetic analysis of Nucleolin protein sequence demonstrates degree of conservation amongst vertebrates and between model animals used in this study and a representative of the kingdom fungi (source blastP, MultiAlin, GeneBee). A, right: numbers highlight the percent similarity between two species (source: blastP and EBI). B: to explore the role of Nucleolin in cardiac function, endogenous nucleolin expression was visualized by in situ hybridization at the indicated stages following fertilization (hours postfertilization, hpf) in the developing zebrafish embryo. C: at 48 hpf the zebrafish ventricle (indicated by red arrow, V) expresses higher levels of nucleolin than the atria (A). D: embryos expressing the mCherry fluorescent protein under the cmlc2 promoter to demarcate cardiac cells (red) were fixed at 48 hpf and stained via whole mount immunohistochemistry to visualize Nucleolin protein expression (green) in the ventricle and atria. E: the nuclear marker DAPI (blue) confirms nuclear localization of Nucleolin (green) in cardiac cells expressing cmlc2 driven mCherry (red). White box indicates area enlarged in F.
Because of its high conservation, ubiquitous (and early, relative to fertilization) expression and known role in ribosomal biogenesis, we anticipated that loss of Nucleolin from the single-cell stage would have drastic effects on multiple aspects of the zebrafish body plan. While MO-induced knockdown was very effective in reducing Nucleolin protein levels (Fig. 4A) and increasing the heterochromatic mark histone H3 lysine 9 trimethylation (H3K9Me3) and p53 (Fig. 4B), the morphant embryos appeared grossly normal, with the exception of some thoracic edema (Fig. 4C). In some cell types Nucleolin knockdown (KD) has been shown to increase p53 expression, so coinjection of nucleolin and p53 MO was performed to eliminate cell death-specific phenotypes resulting from increased p53. While all nucleolin MO-based experiments were performed in the presence and absence of p53 MO, no differences were observed between these two conditions; therefore data obtained with p53 MO are presented, as it is the more rigorous control.
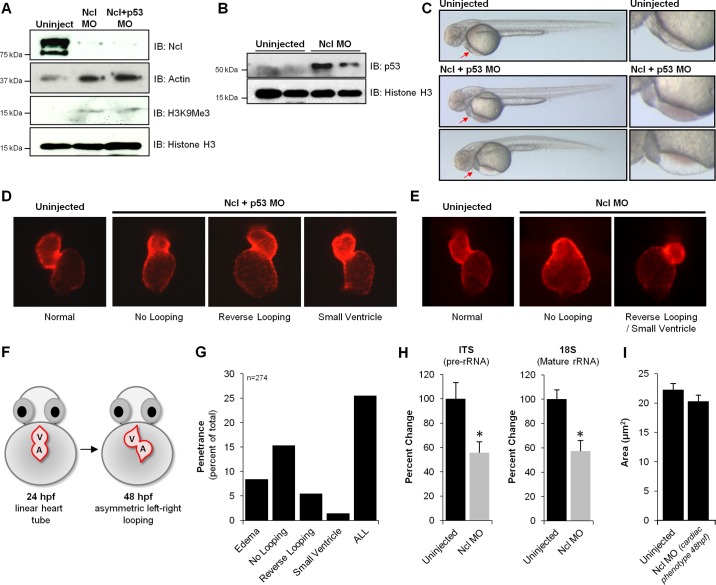
Fig. 4.
Loss of nucleolin in zebrafish results in cardiac morphological defects. To examine the role of Nucleolin on zebrafish development, Morpholino-based knockdown was performed. Embryos were dechorinated and injected at the one cell stage with 3 ng of Nucleolin (Ncl) Mopholino (MO) in the presence or absence of p53 MO (3 ng). Note: in this experiment, the coinjection with p53 is a control to rule out generalized cell death phenotypes induced by the Nucleolin MO; in all experiments performed, the coinjected Ncl+p53 was indistinguishable from the Ncl alone, and so the former is shown as this is the more rigorous control. A: Western blot analysis of 24 hpf embryos reveals near complete loss of Nucleolin protein, concomitant with significant increases in Histone H3 trimethylation on lysine K9 (H3K9Me3) and p53 (B), supporting an endogenous role for Nucleolin to maintain euchromatin (Actin and total Histone H3 shown as controls). C: depletion of Nucleolin protein had minimal effects on gross development in zebrafish, as observed 48 hpf (a time point at which Nucleolin protein levels are still significantly diminished), with the striking exception of the heart, in which a significant population of embryos displayed blood pooling (middle panels) and/or edema (fluid accumulation in pericardial space; bottom panels), both symptoms of cardiac dysfunction. Red arrows highlight regions of defect, expanded in right column. To further investigate this localized cardiac phenotype, we examined heart development after Ncl KD in the presence (D) or absence (E) of p53 MO by fluorescence imaging. Embryos expressing mCherry fluorescent protein under control of the cmlc2 promoter enabled live imaging of heart morphology (images taken at 48 hpf). F: normal cardiac development involves looping of the linear heart tube between 24 and 36 hpf, eventually exhibiting proper orientation by 48 hpf; however, three prominent defects in chamber patterning and morphology were observed after Nucleolin knockdown: no looping, reverse looping and decreased ventricular size (small ventricle). G: penetrance of phenotypes observed after Nucleolin knockdown indicates that ∼25% of zebrafish displayed a cardiac defect, with delayed looping being the most prominent phenotype (15%). H: to investigate the mechanistic basis for these cardiac defects, rRNA transcription and maturation was examined using primers that detect pre-rRNA (ITS) and mature rRNA (18S) by qRT-PCR, demonstrating that loss of Nucleolin impaired both rRNA transcription and processing (n = 3/group). Asterisks indicate a P value < 0.01. I: embryos expressing cardiac restricted mCherry fluorescent protein (red) 72 hpf were fixed and stained via whole mount immunohistochemistry against Zn8, a cell membrane marker (green). Confocal microscopy was used to optically section the heart and generate a 2-dimensional maximum intensity projection of several images acquired serially in the z-dimension. Zn8 staining in ventricular cardiomyocytes enabled quantification of cell size in embryos with normal or reduced Nucleolin levels (displaying a cardiac phenotype at 48 hpf).
Although the hearts in Nucleolin MO-injected zebrafish appeared normal at 24 hpf, by 48 hpf a significant proportion exhibited defects in chamber morphology and patterning. Epifluorescence imaging of the heart revealed a number of severe cardiac abnormalities, including small ventricles (2%) and no (15%) or reverse (5%) looping (Fig. 4, D–F). Overall, we observed a cardiac phenotype in ∼25% of injected embryos (Fig. 4G), consistent with the penetrance commonly observed in Morpholino-based screens (9, 23, 28). Interestingly, the most common abnormality was an inability of the linear heart tube to loop, a process that normally begins by ∼36 hpf and is complete by 48 hpf (Fig. 4F), leading to an underdeveloped cardiac structure in these animals. These defects in chamber looping, as well as the presence of reverse-looped hearts, suggests abnormal left-right asymmetry, most likely resulting from the disruption of proper signaling responses (24). To evaluate the effect of Nucleolin knockdown on gene transcription we quantified levels of pre-rRNA (ITS) and mature rRNA (18S) in the absence or presence of nucleolin MO injection (Fig. 4H). Loss of Nucleolin led to a ∼40% reduction in rRNA transcription and maturation in these fish, suggesting that Nucleolin plays a role in regulating ribosomal transcription in zebrafish as it does in mammals. No change in cardiomyocyte size was detected in zebrafish displaying a cardiac phenotype after loss of Nucleolin (Fig. 4I).
To characterize the effects of Nucleolin knockdown on cardiac parameters, we analyzed live videos of zebrafish hearts imaged by virtue of cmlc2-driven, cardiac-specific expression of the mCherry fluorescent protein (Fig. 5A). Line-scanning software enabled quantitation of heart rate and fractional shortening in MO-injected and control fish (34). The heart rate of embryos exhibiting edema or no looping decreased ∼10% compared with uninjected controls, while the fractional shortening increased 15–30% and decreased 10% in fish with no looping/small ventricles and reverse-looped hearts, respectively (Fig. 5B). To assess whether changes in fractional shortening were compensated by inverse changes in heart rate, we calculated the cardiac output (Fig. 5C). Zebrafish with small ventricular chambers or edema displayed a significant decrease in overall cardiac output (Fig. 5D).
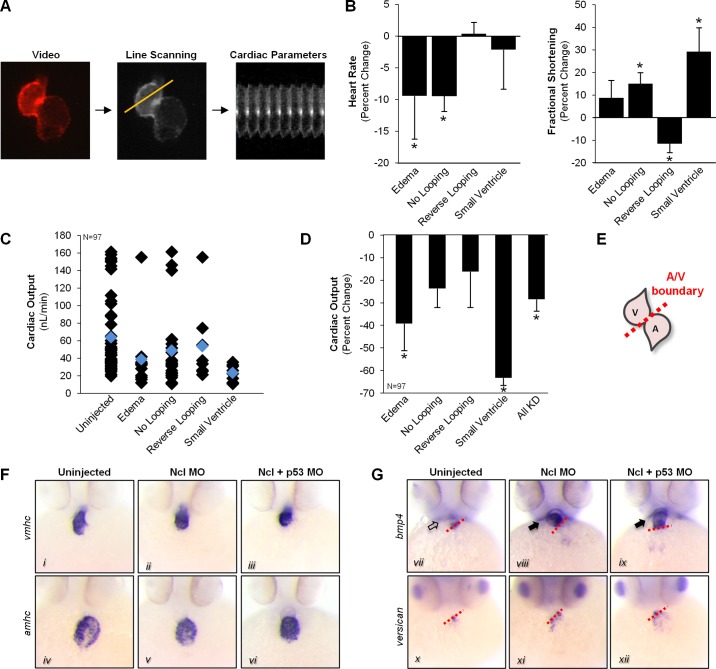
Fig. 5.
Nucleolin is essential for proper cardiac function and myocyte differentiation. A: embryos were injected with Nucleolin MO and real-time movies of heart muscle recorded at 48 hpf via fluorescence video illumination of the cardiac-specific mCherry fluorescent protein. Line scanning software was used to analyze videos of beating hearts to calculate heart rate and fractional shortening in Nucleolin knockdown embryos (see methods and Ref. 29). B: animals with pericardial edema or no heart looping displayed decreased heart rate, while significant changes in fractional shortening were observed in no looping, reverse looping, and small ventricle fish. C: to assess whether changes in fractional shortening were compensated by inverse changes in heart rate, we calculated the cardiac output for each fish (left panel, black diamond indicates individual fish, blue indicates group average) and the percent change in the cardiac output compared uninjected controls (D). E: in situ hybridization was performed in 48 hpf embryos in the presence or absence of Nucleolin MO to examine markers of chamber specification and myocyte differentiation; schematic illustrates normal zebrafish heart chambers at 48 hpf. A, atrium; V, ventricle. F: endogenous expression and distribution of atrial (amhc; top panels) and ventricular (vmhc, bottom panels) specific myosin heavy chain were normal in Nucleolin KD embryos. G: however, staining for bmp4 (top panels), a myocardial lineage marker normally restricted to the AV boundary (red dashed line), displayed intense expression throughout the ventricle of hearts lacking nucleolin, while A/V boundary expression of versican (bottom panels), which is an endocardial marker, appeared normal. The expression of bmp4 in the ventricle at this late stage suggests that loss of Nucleolin perturbs bmp4 expression leading to a delay in myocyte differentiation and provides a mechanistic explanation for the defects in chamber morphology and patterning, given BMP4's role in regulating muscle development and cardiac left-right asymmetry.
Analysis of chamber specification and differentiation in zebrafish hearts was accomplished by in situ hybridization using markers of the atrium (amhc), the ventricles (vmhc), and the A/V boundary (versican and bmp4) in 48-hpf embryos (Fig. 5, E–G). Chamber-specific expression of atrial (amhc) and ventricular (vmhc) myosin heavy chain appeared normal after Nucleolin MO injection (Fig. 5F). However, analysis of the myocardial A/V boundary marker bmp4 (myocardial specific) displayed aberrant staining throughout the ventricular chamber in Nucleolin deficient animals, in contrast to the A/V boundary restriction observed with the endocardial marker versican (Fig. 5G), which was indistinguishable in localization between normal and MO-treated animals. While ventricular expression of bmp4 is normal in 24-hpf embryos, these cells typically silence bmp4 transcription prior to 48 hpf as they continue differentiating into cardiomyocytes, suggesting that Nucleolin knockdown inhibits differentiation of ventricular cells. In addition, abnormal signaling by bmp4, a known regulator of left-right cardiac asymmetry during development (5, 6, 26), provides a mechanistic explanation for the morphological defects in chamber looping.
To examine the effects of Nucleolin overexpression, flag-tagged nucleolin RNA was injected into one-cell-stage embryos. Western blotting for total (Nucleolin antibody) and exogenous Nucleolin (FLAG antibody) in 24-hpf embryos confirmed an increase of ∼3-fold in injected animals (Fig. 6A) and increased p53. Embryos began exhibiting gross morphological defects at 24 hpf, with ∼60% of embryos displaying severe defects at 48 hpf (Fig. 6, B and C) including edema and abnormal dorsal ventral patterning indicative of a bent or truncated body axis (short, curled tail) (15, 39). Analysis of the hearts in these fish revealed defects in chamber morphology and patterning similar to those observed after Nucleolin KD, which were mildly attenuated with coinjection of the p53 MO (Fig. 6, D and E). Staining for bmp4 in embryos with elevated Nucleolin levels revealed aberrant expression seen by intense ventricular staining in contrast to the A/V restricted expression in uninjected embryos (Fig. 6F). These impairments in dorsal ventral axis formation (Fig. 6G) and cardiac looping (Fig. 6H) upon Nucleolin overexpression mirror the phenotypes observed after perturbation of BMP4 signaling in the developing embryo as reported previously (5, 6, 12, 15, 26, 38, 39).
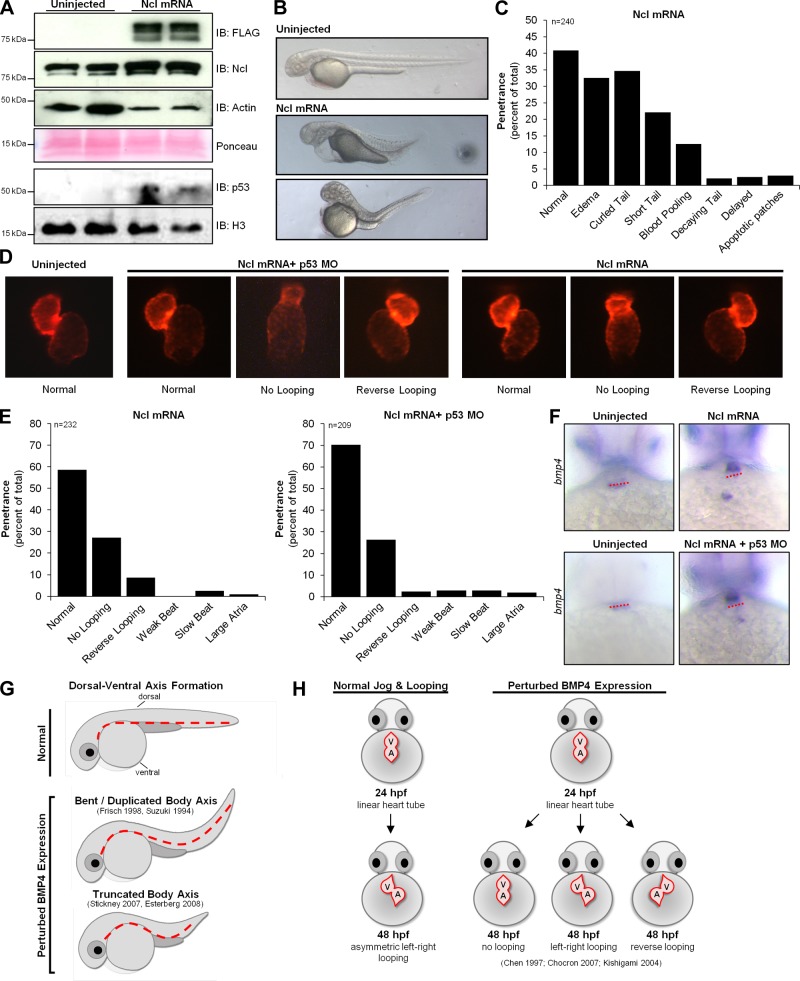
Fig. 6.
Nucleolin overexpression causes defects in heart chamber looping and dorsal-ventral axis formation. Embryos were dechorinated and injected at the one-cell stage with 300 pg of flag-tagged Nucleolin RNA in the presence or absence of p53 MO. A: Western blot analysis of 24 hpf embryos confirms modest overexpression of Nucleolin (actin and Ponceau shown as controls; we observed a consistent increase in actin levels concomitant with Nucleolin overexpression, the origin of which is unknown at this time) and increased p53 (Histone H3 shown as control). B: analysis of gross morphology 48 hpf demonstrates severe developmental abnormalities indicative of a truncated or bent body axis. C: sixty percent of embryos displayed at least one defect, including edema, shortened yolk extension, and curled tail. D: the myocardia were visualized in these embryos at 48 hpf by imaging the cmlc2 driven mCherry fluorescent protein. E: quantification of cardiac phenotypes seen after Nucleolin overexpression; greater penetrance was observed in the embryos lacking the p53 MO. F: in situ hybridization for bmp4 in 48 hpf embryos in the presence of Ncl mRNA (with or without p53 MO) revealed ventricular expression as opposed to the AV boundary (red dashed line) restricted expression in uninjected controls. G: defects in dorsal-ventral axis formation (dotted red line) and cardiac chamber looping (H) observed in zebrafish embryos after Nucleolin overexpression mimic results from other investigators (5, 6, 26) after perturbation of bmp4 expression during development suggesting that Nucleolin regulates bone morphometric protein 4 (BMP4) signaling.
Taken together, these results demonstrate that Nucleolin decondenses chromatin and facilitates normal rRNA transcription, processing, and ribosomal biogenesis in the heart. Loss of Nucleolin perturbs gene expression (BMP4 and β-MHC) and impairs cardiac chamber looping and function, whereas upregulation affects dorsal-ventral axis formation (Fig. 7).
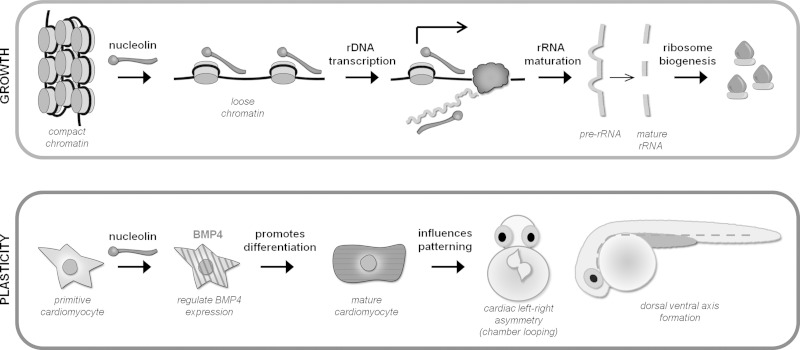
Fig. 7.
Nucleolin regulates growth and plasticity in cardiomyocytes. Top: nucleolin binds to and decondenses chromatin in the heart, thereby enhancing rDNA transcription and maturation to support hypertrophic growth in the cardiomyocyte during disease (loss of Nucleolin in NRVMs counteracts this process). Bottom: additionally, Nucleolin regulates BMP4 expression (and other fetal genes in mice) necessary for proper cardiomyocyte differentiation and patterning (cardiac looping and axis formation). The molecular mechanisms by which this regulation occurs and whether this is a property of nuclear or nonnuclear Nucleolin is unknown.
DISCUSSION
The hypothesis that cardiac hypertrophy and failure involves recapitulation of fetal gene expression is widely accepted; however, it lacks a fundamental mechanism to explain how such a genomewide change in DNA accessibility would occur. Genes whose expression is altered in heart disease are structurally distributed across the genome, so a global act of chromatin remodeling, that enables rapid, systematic, and reproducible reprogramming of genomic structure, would be a desirable feature of the above-referenced “mechanism” for fetal gene reprogramming. To address this knowledge gap, detergent-extracted chromatin proteins were identified and quantified in the healthy mouse heart and comparison made with the pressure-overloaded heart in stages of hypertrophy and failure. The first component to this study was the determination of the chromatin-associated proteins in the diseased heart, which enhances our understanding of the raw materials for genome packing and modification. The second major component of the present work involved testing the hypothesis that chromatin-associated proteins activated during disease are responsible for gene expression and phenotypic events normally operative in cardiac development.
To test this hypothesis, we performed gain- and loss-of-function studies in zebrafish, demonstrating that Nucleolin regulates chamber patterning and heart function. While additional studies will be necessary to elucidate the roles of all the individual proteins measured in the first phase of this study, our findings demonstrate the power of systems proteomics to investigate the relationship between chromatin subproteomes in the diseased and developing heart by combining unbiased screening and hypothesis-driven experiments.
Previous studies had implied a role for nucleolar stress in cardiac disease (1, 2), although the role of individual proteins, including Nucleolin, was unknown. While nuclear Nucleolin was found to increase its association with chromatin during hypertrophy, total cellular levels of Nucleolin fluctuated (increased or decreased) in the presence of different agonists. This observation is consistent with previous reports, which have shown that cytoplasmic Nucleolin is differentially expressed compared with its nuclear counterpart, each having substantially different half-lives of 45 min (21) and >24 h (25), respectively. Consistent with this, Hovanessian et al. (21) reported reduced levels of Nucleolin mRNA were accompanied by markedly diminished cytoplasmic but not nuclear Nucleolin protein.
Our results indicate that Nucleolin is necessary for normal cardiomyocyte differentiation during development and that loss of Nucleolin promotes the expression of fetal markers in zebrafish (Bmp4) and isolated cardiomyocytes (ANF, β-MHC). This phenotype is similar to that reported by Yang and colleagues (40), who observed abnormal differentiation of embryonic stem cells (ESCs) after Nucleolin knockdown. Specifically, ESCs displayed properties of differentiation but maintained the expression of pluripotency genes after loss of Nucleolin in their study. These effects were partially attributed to p53, which was found to be upregulated after Nucleolin knockdown, consistent with our observations in isolated myocytes.
The disruption in nucleolin expression involved misregulation of bmp4 expression and ultimately led to defects in patterning, specifically left-right cardiac asymmetry and dorsal-ventral axis formation. These results are consistent with perturbations in BMP signaling during development, which have shown that BMP4 is required for left-right patterning in the mouse (16, 26), chick (33) and zebrafish (5, 6) embryos. In addition, inhibition of BMP-4 signaling in Xenopus (15, 39) and zebrafish (12, 38) embryos leads to duplication, curving, or truncation of the dorsal-ventral body axis, resulting in embryos with short curled tails (similar to our results after Nucleolin overexpression). In these models, inhibition of BMP signaling altered the proliferation and differentiation of cells in the developing embryos. These data suggest that Nucleolin may influence myocyte plasticity via BMP4 signaling and provides a mechanistic explanation for the defects in chamber morphology and patterning, given BMP4's role in regulating muscle development, left-right asymmetry, and dorsal-ventral specification.
Overall, these data support the hypothesis that some regulators of chromatin structure in the developing heart are reactivated during heart disease to mobilize conserved aspects of chromatin remodeling and gene expression.
GRANTS
This study was supported by National Institutes of Health (NIH) Grants to T. M. Vondriska (HL-105699, HL-115238, and S10-RR-022371) and J.-N. Chen (R01 HL096980) and the Laubisch Endowment at UCLA (to T. M. Vondriska). Mass spectrometry instrument contract support came from an NHLBI Proteomics Contract (HHSN268201000035C). S. Franklin is the recipient of a K99/R00 Award from the NIH (K99 HL107674); E. Monte is the recipient of the Jennifer Buchwald Fellowship in Physiology from UCLA and an American Heart Association Predoctoral Fellowship; and H. Chen is the recipient of an American Heart Association Predoctoral Fellowship.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: E.M., K.M., Y.W., J.-N.C., T.M.V., and S.F. conception and design of research; E.M., K.M., T.K., S.R., and S.F. performed experiments; E.M., K.M., H.C., T.M.V., and S.F. analyzed data; E.M., K.M., H.C., J.-N.C., T.M.V., and S.F. interpreted results of experiments; E.M., H.C., T.K., T.M.V., and S.F. prepared figures; E.M., T.M.V., and S.F. drafted manuscript; E.M., K.M., H.C., T.K., J.-N.C., T.M.V., and S.F. edited and revised manuscript; E.M., K.M., H.C., T.K., S.R., Y.W., J.-N.C., T.M.V., and S.F. approved final version of manuscript.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. J. Zhang, A. K. Paulsson and M. Kolmakova for assistance with early stages of this work, T. Tran for technical assistance, and members of the J.-N. Chen and T. M. Vondriska laboratories for helpful feedback and suggestions.
REFERENCES
- 1.Avitabile D, Bailey B, Cottage CT, Sundararaman B, Joyo A, McGregor M, Gude N, Truffa S, Zarrabi A, Konstandin M, Khan M, Mohsin S, Volkers M, Toko H, Mason M, Cheng Z, Din S, Alvarez R, Jr, Fischer K, Sussman MA. Nucleolar stress is an early response to myocardial damage involving nucleolar proteins nucleostemin and nucleophosmin. Proc Natl Acad Sci USA 108: 6145–6150, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bicknell K, Brooks G, Kaiser P, Chen H, Dove BK, Hiscox JA. Nucleolin is regulated both at the level of transcription and translation. Biochem Biophys Res Commun 332: 817–822, 2005. [DOI] [PubMed] [Google Scholar]
- 3.Bruneau BG. Chromatin remodeling in heart development. Curr Opin Genet Dev 20: 505–511, 2010. [DOI] [PubMed] [Google Scholar]
- 4.Chen JN, Fishman MC. Zebrafish tinman homolog demarcates the heart field and initiates myocardial differentiation. Development 122: 3809–3816, 1996. [DOI] [PubMed] [Google Scholar]
- 5.Chen JN, van Eeden FJ, Warren KS, Chin A, Nusslein-Volhard C, Haffter P, Fishman MC. Left-right pattern of cardiac BMP4 may drive asymmetry of the heart in zebrafish. Development 124: 4373–4382, 1997. [DOI] [PubMed] [Google Scholar]
- 6.Chocron S, Verhoeven MC, Rentzsch F, Hammerschmidt M, Bakkers J. Zebrafish Bmp4 regulates left-right asymmetry at two distinct developmental time points. Dev Biol 305: 577–588, 2007. [DOI] [PubMed] [Google Scholar]
- 7.Choi J, Mouillesseaux K, Wang Z, Fiji HD, Kinderman SS, Otto GW, Geisler R, Kwon O, Chen JN. Aplexone targets the HMG-CoA reductase pathway and differentially regulates arteriovenous angiogenesis. Development 138: 1173–1181, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coucelo J, Joaquim N, Coucelo J. Calculation of volumes and systolic indices of heart ventricle from Halobatrachus didactylus: echocardiographic noninvasive method. J Exp Zool 286: 585–595, 2000. [DOI] [PubMed] [Google Scholar]
- 9.Delous M, Yin C, Shin D, Ninov N, Debrito Carten J, Pan L, Ma TP, Farber SA, Moens CB, Stainier DY. sox9b is a key regulator of pancreaticobiliary ductal system development. PLoS Gen 8: e1002754, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorn GW, 2nd, Molkentin JD. Manipulating cardiac contractility in heart failure: data from mice and men. Circulation 109: 150–158, 2004. [DOI] [PubMed] [Google Scholar]
- 11.Erard MS, Belenguer P, Caizergues-Ferrer M, Pantaloni A, Amalric F. A major nucleolar protein, nucleolin, induces chromatin decondensation by binding to histone H1. Eur J Biochem 175: 525–530, 1988. [DOI] [PubMed] [Google Scholar]
- 12.Esterberg R, Delalande JM, Fritz A. Tailbud-derived Bmp4 drives proliferation and inhibits maturation of zebrafish chordamesoderm. Development 135: 3891–3901, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franklin S, Chen H, Mitchell-Jordan SA, Ren S, Wang Y, Vondriska TM. Quantitative analysis of chromatin proteome reveals remodeling principles and identifies HMGB2 as a regulator of hypertrophic growth. Mol Cell Proteomics 11: M111.014258, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franklin S, Zhang MJ, Chen H, Paulsson AK, Mitchell-Jordan SA, Li Y, Ping P, Vondriska TM. Specialized compartments of cardiac nuclei exhibit distinct proteomic anatomy. Mol Cell Proteomics 10: 703, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frisch A, Wright CV. XBMPRII, a novel Xenopus type II receptor mediating BMP signaling in embryonic tissues. Development 125: 431–442, 1998. [DOI] [PubMed] [Google Scholar]
- 16.Fujiwara T, Dehart DB, Sulik KK, Hogan BL. Distinct requirements for extra-embryonic and embryonic bone morphogenetic protein 4 in the formation of the node and primitive streak and coordination of left-right asymmetry in the mouse. Development 129: 4685–4696, 2002. [DOI] [PubMed] [Google Scholar]
- 17.Ghisolfi-Nieto L, Joseph G, Puvion-Dutilleul F, Amalric F, Bouvet P. Nucleolin is a sequence-specific RNA-binding protein: characterization of targets on pre-ribosomal RNA. J Mol Biol 260: 34–53, 1996. [DOI] [PubMed] [Google Scholar]
- 18.Ginisty H, Amalric F, Bouvet P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J 17: 1476–1486, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goyette M, Petropoulos CJ, Shank PR, Fausto N. Regulated transcription of c-Ki-ras and c-myc during compensatory growth of rat liver. Mol Cell Biol 4: 1493–1498, 1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han P, Hang CT, Yang J, Chang CP. Chromatin remodeling in cardiovascular development and physiology. Circ Res 108: 378–396, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hovanessian AG, Soundaramourty C, El Khoury D, Nondier I, Svab J, Krust B. Surface expressed nucleolin is constantly induced in tumor cells to mediate calcium-dependent ligand internalization. PLoS One 5: e15787, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jordan G. At the heart of the nucleolus. Nature 329: 489–490, 1987. [DOI] [PubMed] [Google Scholar]
- 23.Just S, Meder B, Berger IM, Etard C, Trano N, Patzel E, Hassel D, Marquart S, Dahme T, Vogel B, Fishman MC, Katus HA, Strahle U, Rottbauer W. The myosin-interacting protein SMYD1 is essential for sarcomere organization. J Cell Sci 124: 3127–3136, 2011. [DOI] [PubMed] [Google Scholar]
- 24.Kathiriya IS, Srivastava D. Left-right asymmetry and cardiac looping: implications for cardiac development and congenital heart disease. Am J Med Genet 97: 271–279, 2000. [DOI] [PubMed] [Google Scholar]
- 25.Kim SK, Srivastava M. Stability of Nucleolin protein as the basis for the differential expression of Nucleolin mRNA and protein during serum starvation. DNA Cell Biol 22: 171–178, 2003. [DOI] [PubMed] [Google Scholar]
- 26.Kishigami S, Yoshikawa S, Castranio T, Okazaki K, Furuta Y, Mishina Y. BMP signaling through ACVRI is required for left-right patterning in the early mouse embryo. Dev Biol 276: 185–193, 2004. [DOI] [PubMed] [Google Scholar]
- 27.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langenbacher AD, Nguyen CT, Cavanaugh AM, Huang J, Lu F, Chen JN. The PAF1 complex differentially regulates cardiomyocyte specification. Dev Biol 353: 19–28, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu G, Ren S, Korge P, Choi J, Dong Y, Weiss J, Koehler C, Chen JN, Wang Y. A novel mitochondrial matrix serine/threonine protein phosphatase regulates the mitochondria permeability transition pore and is essential for cellular survival and development. Genes Dev 21: 784–796, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maunakea AK, Chepelev I, Zhao K. Epigenome mapping in normal and disease States. Circ Res 107: 327–339, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell-Jordan SA, Holopainen T, Ren S, Wang S, Warburton S, Zhang MJ, Alitalo K, Wang Y, Vondriska TM. Loss of Bmx nonreceptor tyrosine kinase prevents pressure overload-induced cardiac hypertrophy. Circ Res 103: 1359–1362, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monsoro-Burq A, Le Douarin NM. BMP4 plays a key role in left-right patterning in chick embryos by maintaining Sonic Hedgehog asymmetry. Mol Cell 7: 789–799, 2001. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen CT, Lu Q, Wang Y, Chen JN. Zebrafish as a model for cardiovascular development and disease. Drug Discov Today Dis Models 5: 135–140, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olson EN. A decade of discoveries in cardiac biology. Nat Med 10: 467–474, 2004. [DOI] [PubMed] [Google Scholar]
- 36.Razeghi P, Young ME, Alcorn JL, Moravec CS, Frazier OH, Taegtmeyer H. Metabolic gene expression in fetal and failing human heart. Circulation 104: 2923–2931, 2001. [DOI] [PubMed] [Google Scholar]
- 37.Schiller NB, Shah PM, Crawford M, DeMaria A, Devereux R, Feigenbaum H, Gutgesell H, Reichek N, Sahn D, Schnittger I. Recommendations for quantitation of the left ventricle by two-dimensional echocardiography American Society of Echocardiography Committee on Standards, Subcommittee on Quantitation of Two-Dimensional Echocardiograms. J Am Soc Echocardiogr 2: 358–367, 1989. [DOI] [PubMed] [Google Scholar]
- 38.Stickney HL, Imai Y, Draper B, Moens C, Talbot WS. Zebrafish bmp4 functions during late gastrulation to specify ventroposterior cell fates. Dev Biol 310: 71–84, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki A, Thies RS, Yamaji N, Song JJ, Wozney JM, Murakami K, Ueno N. A truncated bone morphogenetic protein receptor affects dorsal-ventral patterning in the early Xenopus embryo. Proc Natl Acad Sci USA 91: 10255–10259, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a.Westerfield M. The Zebrafish Book. Univ. of Oregon Press, 2000. [Google Scholar]
- 40.Yang A, Shi G, Zhou C, Lu R, Li H, Sun L, Jin Y. Nucleolin maintains embryonic stem cell self-renewal by suppression of p53 protein-dependent pathway. J Biol Chem 286: 43370–43382, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.