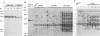Abstract
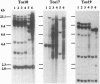
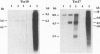
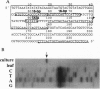
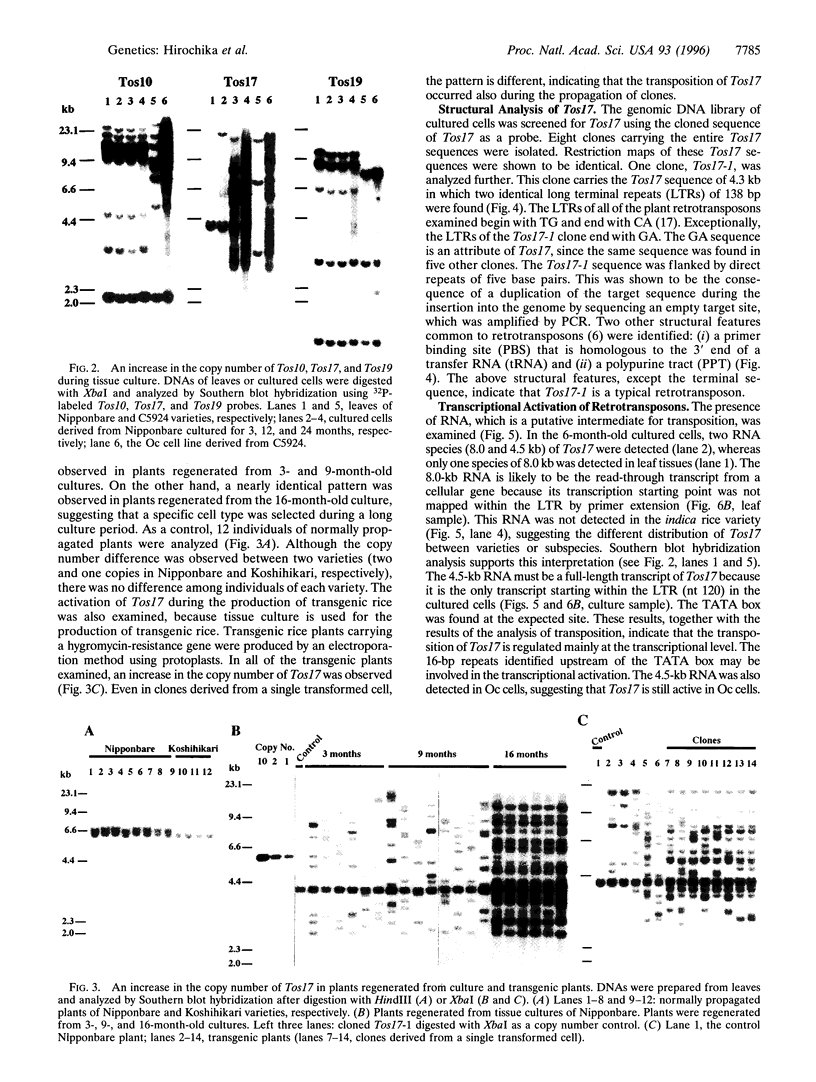
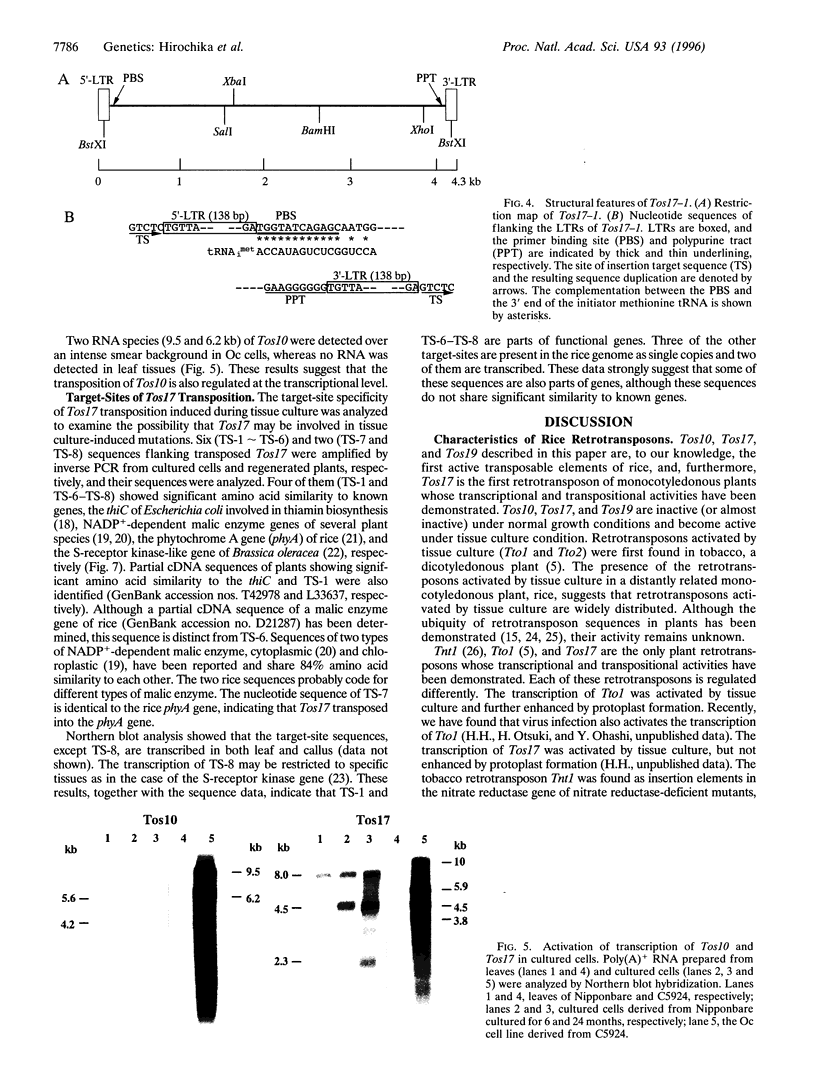
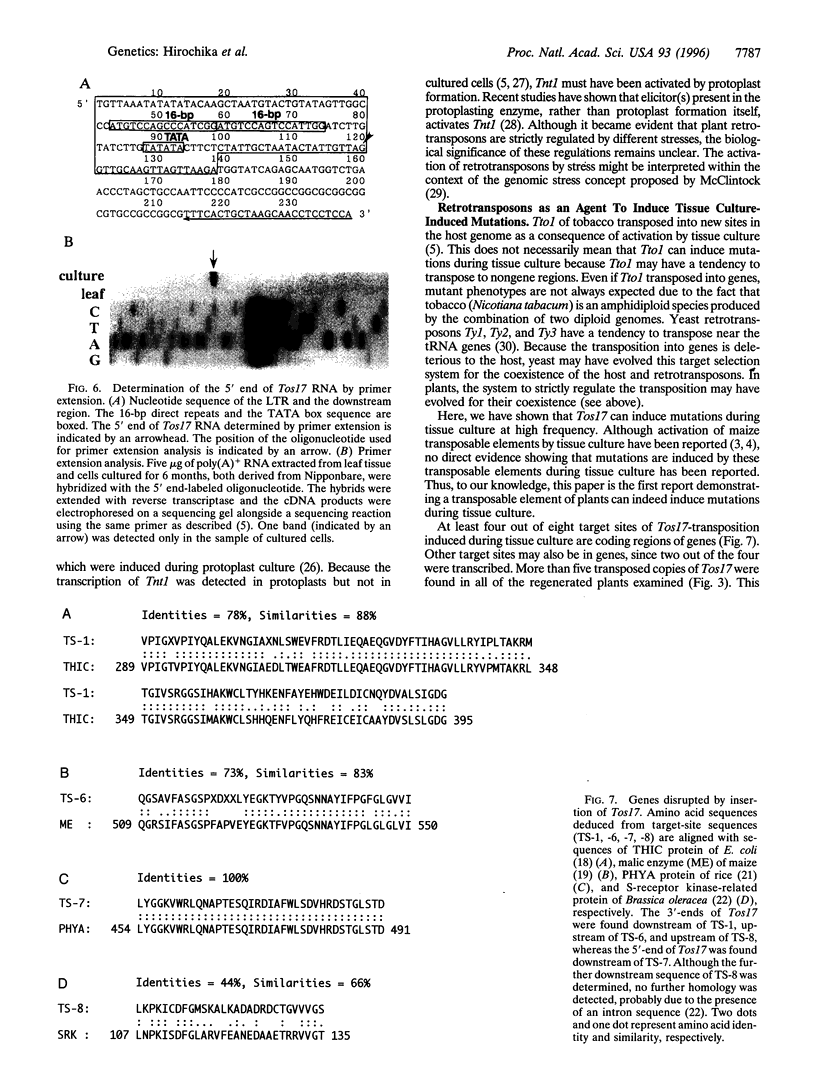
Five retrotransposon families of rice (Tos1-Tos5) have been reported previously. Here we report 15 new retrotransposon families of rice (Tos6-Tos20). In contrast to yeast and Drosophila retrotransposons, all of the rice retrotransposons examined appear inactive (or almost inactive) under normal growth conditions. Three of the rice retrotransposons (Tos10, Tos17, and Tos19) are activated under tissue culture conditions. The most active one, Tos17, was studied in detail. The copy number of Tos17 increased with prolonged culture period. In all of the plants regenerated from tissue cultures, including transgenic plants, 5 to 30 transposed Tos17 copies were detected. The transcript of Tos17 was only detected under tissue culture conditions, indicating that the transposition of Tos17 is mainly regulated at the transcriptional level. To examine the target-site specificity of Tos17 transposition, sequences flanking transposed Tos17 copies were analyzed. At least four out of eight target sites examined are coding regions. Other target sites may also be in genes because two out of four were transcribed. The regenerated plants with Tos17-insertions in the phytochrome A gene and the S-receptor kinase-related gene were identified. These results indicate that activation of Tos17 is an important cause of tissue culture-induced mutations. Tissue culture-induced activation of Tos17 may be a useful tool for insertional mutagenesis and functional analysis of genes.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ballinger D. G., Benzer S. Targeted gene mutations in Drosophila. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9402–9406. doi: 10.1073/pnas.86.23.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft I., Dean C. Transposition pattern of the maize element Ds in Arabidopsis thaliana. Genetics. 1993 Aug;134(4):1221–1229. doi: 10.1093/genetics/134.4.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft I., Jones J. D., Dean C. Heterologous transposon tagging of the DRL1 locus in Arabidopsis. Plant Cell. 1993 Jun;5(6):631–638. doi: 10.1105/tpc.5.6.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner F. R., Burland V., Plunkett G., 3rd, Sofia H. J., Daniels D. L. Analysis of the Escherichia coli genome. IV. DNA sequence of the region from 89.2 to 92.8 minutes. Nucleic Acids Res. 1993 Nov 25;21(23):5408–5417. doi: 10.1093/nar/21.23.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echalier G. Drosophila retrotransposons: interactions with genome. Adv Virus Res. 1989;36:33–105. doi: 10.1016/s0065-3527(08)60582-5. [DOI] [PubMed] [Google Scholar]
- Flavell A. J., Dunbar E., Anderson R., Pearce S. R., Hartley R., Kumar A. Ty1-copia group retrotransposons are ubiquitous and heterogeneous in higher plants. Nucleic Acids Res. 1992 Jul 25;20(14):3639–3644. doi: 10.1093/nar/20.14.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandbastien M. A. Retroelements in higher plants. Trends Genet. 1992 Mar;8(3):103–108. doi: 10.1016/0168-9525(92)90198-d. [DOI] [PubMed] [Google Scholar]
- Grandbastien M. A., Spielmann A., Caboche M. Tnt1, a mobile retroviral-like transposable element of tobacco isolated by plant cell genetics. Nature. 1989 Jan 26;337(6205):376–380. doi: 10.1038/337376a0. [DOI] [PubMed] [Google Scholar]
- Hirochika H. Activation of tobacco retrotransposons during tissue culture. EMBO J. 1993 Jun;12(6):2521–2528. doi: 10.1002/j.1460-2075.1993.tb05907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H., Fukuchi A., Kikuchi F. Retrotransposon families in rice. Mol Gen Genet. 1992 May;233(1-2):209–216. doi: 10.1007/BF00587581. [DOI] [PubMed] [Google Scholar]
- Hirochika H., Hirochika R. Ty1-copia group retrotransposons as ubiquitous components of plant genomes. Jpn J Genet. 1993 Feb;68(1):35–46. doi: 10.1266/jjg.68.35. [DOI] [PubMed] [Google Scholar]
- Hirochika H., Otsuki H., Yoshikawa M., Otsuki Y., Sugimoto K., Takeda S. Autonomous transposition of the tobacco retrotransposon Tto1 in rice. Plant Cell. 1996 Apr;8(4):725–734. doi: 10.1105/tpc.8.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser K., Goodwin S. F. "Site-selected" transposon mutagenesis of Drosophila. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1686–1690. doi: 10.1073/pnas.87.5.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay S. A., Keith B., Shinozaki K., Chua N. H. The sequence of the rice phytochrome gene. Nucleic Acids Res. 1989 Apr 11;17(7):2865–2866. doi: 10.1093/nar/17.7.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Trick M. Sequence complexity of the S receptor kinase gene family in Brassica. Mol Gen Genet. 1993 Nov;241(3-4):440–446. doi: 10.1007/BF00284698. [DOI] [PubMed] [Google Scholar]
- Kurata N., Nagamura Y., Yamamoto K., Harushima Y., Sue N., Wu J., Antonio B. A., Shomura A., Shimizu T., Lin S. Y. A 300 kilobase interval genetic map of rice including 883 expressed sequences. Nat Genet. 1994 Dec;8(4):365–372. doi: 10.1038/ng1294-365. [DOI] [PubMed] [Google Scholar]
- McClintock B. The significance of responses of the genome to challenge. Science. 1984 Nov 16;226(4676):792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- Peschke V. M., Phillips R. L., Gengenbach B. G. Discovery of transposable element activity among progeny of tissue culture--derived maize plants. Science. 1987 Nov 6;238(4828):804–807. doi: 10.1126/science.238.4828.804. [DOI] [PubMed] [Google Scholar]
- Pouteau S., Huttner E., Grandbastien M. A., Caboche M. Specific expression of the tobacco Tnt1 retrotransposon in protoplasts. EMBO J. 1991 Jul;10(7):1911–1918. doi: 10.1002/j.1460-2075.1991.tb07717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothermel B. A., Nelson T. Primary structure of the maize NADP-dependent malic enzyme. J Biol Chem. 1989 Nov 25;264(33):19587–19592. [PubMed] [Google Scholar]
- Sasaki T., Song J., Koga-Ban Y., Matsui E., Fang F., Higo H., Nagasaki H., Hori M., Miya M., Murayama-Kayano E. Toward cataloguing all rice genes: large-scale sequencing of randomly chosen rice cDNAs from a callus cDNA library. Plant J. 1994 Oct;6(4):615–624. doi: 10.1046/j.1365-313x.1994.6040615.x. [DOI] [PubMed] [Google Scholar]
- Stein J. C., Howlett B., Boyes D. C., Nasrallah M. E., Nasrallah J. B. Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8816–8820. doi: 10.1073/pnas.88.19.8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Otsuki Y., Saji S., Hirochika H. Transposition of the maize Ds element from a viral vector to the rice genome. Plant J. 1994 Jun;5(6):863–871. doi: 10.1046/j.1365-313x.1994.5060863.x. [DOI] [PubMed] [Google Scholar]
- Van Doorsselaere J., Villarroel R., Van Montagu M., Inzé D. Nucleotide sequence of a cDNA encoding malic enzyme from poplar. Plant Physiol. 1991 Aug;96(4):1385–1386. doi: 10.1104/pp.96.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varagona M. J., Purugganan M., Wessler S. R. Alternative splicing induced by insertion of retrotransposons into the maize waxy gene. Plant Cell. 1992 Jul;4(7):811–820. doi: 10.1105/tpc.4.7.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas D. F., Boeke J. D. Yeast retrotransposons and tRNAs. Trends Genet. 1993 Dec;9(12):421–427. doi: 10.1016/0168-9525(93)90105-q. [DOI] [PubMed] [Google Scholar]
- Voytas D. F., Cummings M. P., Koniczny A., Ausubel F. M., Rodermel S. R. copia-like retrotransposons are ubiquitous among plants. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):7124–7128. doi: 10.1073/pnas.89.15.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]