To read out information from its genome, a cell needs the appropriate molecular machinery, RNA polymerases (RNAPs), ribosomes, and associated factors. Thus, the quantitative level of gene expression is dependent on the availability of that machinery, which in turn depends on the external conditions (1). In bacteria, the concentrations of free (i.e., available) RNAPs and ribosomes depend on the growth conditions and may (for example) change during stress responses, affecting the patterns of gene expression. Moreover, different genes may compete for these molecular machines. Then, the expression level of a gene can also depend on what other genes are expressed simultaneously, because expression of a gene reduces the pool of free RNAPs and ribosomes (at least transiently, if there is feedback to keeps these pools constant). Such competition is well established for ribosomes (2–4), but appears to be less pronounced for RNAPs (5).
Recent years have seen tremendous progress in microscopy techniques that can be used to image the localization and the dynamics of RNAPs and ribosomes in bacterial cells (6–10). In this issue of the Biophysical Journal, Bakshi et al. (11) report a study that uses superresolution microscopy to look at RNAPs one-by-one to obtain a global picture of their cellular activities and to determine the fraction of free RNAPs. These questions could so far only be addressed indirectly through integrative modeling approaches that inferred free RNAP concentrations from the integration of large amounts of experimental data into a mathematical model (5,12,13) and by analyzing chromosome-free minicells (14). Bakshi et al. (11) take advantage of the fact that RNAPs performing different functions (transcribing; free in the cytoplasm; nonspecifically bound to DNA) exhibit different patterns of mobility in the cell. Thus, they determine diffusion coefficients from individual RNAP trajectories and study their distributions (Fig. 1) .
Figure 1.
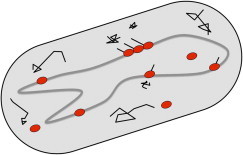
Fluorescently labeled RNA polymerases exhibiting different functional activities (specifically bound to DNA/transcribing; nonspecifically bound to DNA; free in the cytoplasm) are distinguished by their different diffusion behavior.
The distributions of these diffusion coefficients can be fitted with a model for several distinct populations that can be identified with RNAPs in different functional states. Diffusion measurements with low temporal resolution (0.1 s) show two main populations of RNAPs, somewhat rapidly diffusing ones with D ∼ 0.2 μm2/s, and very slow ones with a nominal diffusion coefficient of ∼0.003 μm2/s (but which likely exhibit subdiffusive behavior). Diffusion of the slow RNAPs closely resembles the motion of fluorescently labeled chromosome loci, therefore these RNAPs are interpreted as RNAPs specifically bound to DNA, and are most likely transcribing. Switching between the two populations is seen in longer individual trajectories. Based on measurements with higher temporal resolution (20 ms), the more rapid population is split further into freely diffusing, cytoplasmic RNAPs (with diffusion coefficient ∼0.7 μm2/s) and RNAPs nonspecifically bound to DNA. These subpopulations interconvert rapidly, so an average diffusion coefficient is seen in the lower time-resolution measurements. A small fourth population is given by RNAPs that do not bind DNA. These appear as very rapid even in the lower time-resolution experiments; their physical interpretation remains to be determined.
By quantifying these fractions, Bakshi et al. (11) obtain a global picture of the activities of RNAPs in the cell. Only approximately one-half of the RNAPs are found engaged in active transcription, but a large fraction is bound to DNA, either specifically or nonspecifically (82%). The free fraction is 12%, which corresponds to ∼700 RNAPs or a concentration of free RNAPs of ∼1 μM, much higher than an old estimate (30 nM) that has been widely used in models of gene regulation (15), but similar to a more recent estimate (0.5−1.1 μM, depending on growth conditions (5)).
The overall picture drawn from these results is similar to the one obtained from the integrative modeling studies, in particular that a substantial fraction of RNAPs is not busy transcribing and that most RNAPs are bound to DNA. However, there is pronounced quantitative disagreement with all models proposed earlier (which all relied on data from minicells as input to determine the free RNAP concentration). The biggest difference is in the fraction of specifically bound or transcribing RNAPs, which is smaller in the models than in the new data. This fraction, however, could easily be underestimated in the models if transcription was slower than generally believed; therefore, it would be interesting to measure the speed of transcription under the conditions of the new experiment.
So what’s next? Besides demonstrating a beautiful application of imaging techniques beyond imaging, the study of Bakshi et al. (11) opens the door to addressing the following questions:
Does the free RNAP concentration change when the conditions are changed?
If yes, is this change reflected in changing transcription rates of constitutively expressed genes?
The technique could be used to study dependencies on growth rate or other parameters (temperature, osmotic pressure, etc.) for cells in steady-state growth, as well as to investigate transient dynamics during growth shifts or in stress responses. In iteration with improved models, these experiments could provide a rather comprehensive picture of what RNAPs are doing in cells. Moreover, the method may also be applicable to other components of the gene expression apparatus for both native and synthetic (specifically orthogonal) expression systems.
References
- 1.Klumpp S., Zhang Z., Hwa T. Growth rate-dependent global effects on gene expression in bacteria. Cell. 2009;139:1366–1375. doi: 10.1016/j.cell.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vind J., Sørensen M.A., Pedersen S. Synthesis of proteins in Escherichia coli is limited by the concentration of free ribosomes. Expression from reporter genes does not always reflect functional mRNA levels. J. Mol. Biol. 1993;231:678–688. doi: 10.1006/jmbi.1993.1319. [DOI] [PubMed] [Google Scholar]
- 3.Scott M., Gunderson C.W., Hwa T. Interdependence of cell growth and gene expression: origins and consequences. Science. 2010;330:1099–1102. doi: 10.1126/science.1192588. [DOI] [PubMed] [Google Scholar]
- 4.Mather W.H., Hasty J., Williams R.J. Translational cross talk in gene networks. Biophys. J. 2013;104:2564–2572. doi: 10.1016/j.bpj.2013.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klumpp S., Hwa T. Growth-rate-dependent partitioning of RNA polymerases in bacteria. Proc. Natl. Acad. Sci. USA. 2008;105:20245–20250. doi: 10.1073/pnas.0804953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin D.J., Cabrera J.E. Coupling the distribution of RNA polymerase to global gene regulation and the dynamic structure of the bacterial nucleoid in Escherichia coli. J. Struct. Biol. 2006;156:284–291. doi: 10.1016/j.jsb.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Bakshi S., Siryaporn A., Weisshaar J.C. Superresolution imaging of ribosomes and RNA polymerase in live Escherichia coli cells. Mol. Microbiol. 2012;85:21–38. doi: 10.1111/j.1365-2958.2012.08081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montero Llopis P., Sliusarenko O., Jacobs-Wagner C. In vivo biochemistry in bacterial cells using FRAP: insight into the translation cycle. Biophys. J. 2012;103:1848–1859. doi: 10.1016/j.bpj.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.English B.P., Hauryliuk V., Elf J. Single-molecule investigations of the stringent response machinery in living bacterial cells. Proc. Natl. Acad. Sci. USA. 2011;108:E365–E373. doi: 10.1073/pnas.1102255108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Endesfelder U., Finan K., Heilemann M. Multiscale spatial organization of RNA polymerase in Escherichia coli. Biophys. J. 2013;105:172–181. doi: 10.1016/j.bpj.2013.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bakshi S., Dalrymple R.M., Weisshaar J.C. Partitioning of RNA polymerase activity in live E. coli from analysis of single-molecule diffusive trajectories. Biophys. J. 2013;105:2676–2686. doi: 10.1016/j.bpj.2013.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bremer H., Dennis P., Ehrenberg M. Free RNA polymerase and modeling global transcription in Escherichia coli. Biochimie. 2003;85:597–609. doi: 10.1016/s0300-9084(03)00105-6. [DOI] [PubMed] [Google Scholar]
- 13.Tadmor A.D., Tlusty T. A coarse-grained biophysical model of E. coli and its application to perturbation of the rRNA operon copy number. PLOS Comput. Biol. 2008;4:e1000038. doi: 10.1371/journal.pcbi.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shepherd N., Dennis P., Bremer H. Cytoplasmic RNA polymerase in Escherichia coli. J. Bacteriol. 2001;183:2527–2534. doi: 10.1128/JB.183.8.2527-2534.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McClure W.R. A biochemical analysis of the effect of RNA polymerase concentration on the in vivo control of RNA chain initiation frequency. In: Lennon D.L.F., Stratman F.W., Zahlten R.N., editors. Biochemistry of Metabolic Processes. Elsevier; New York: 1983. pp. 207–217. [Google Scholar]


