Abstract
RATIONALE
The aim of this study was to demonstrate, and to characterize by high resolution mass spectrometry, that it is possible to preferentially induce covalent cross-links in peptides by using high energy femtosecond UV laser pulses. The cross-link is readily formed only when aromatic amino acids are present in the peptide sequence.
METHODS
Three peptides, xenopsin, angiotensin I, interleukin, individually or in combination, were exposed to high energy femtosecond UV laser pulses, either alone or in the presence of spin trapping molecules, the reaction products being characterized by high resolution mass spectrometry.
RESULTS
High resolution mass spectrometry and spin trapping strategies showed that cross-linking occurs readily, proceeds via a radical mechanism, and is the highly dominant reaction, proceeding without causing significant photo-damage in the investigated range of experimental parameters.
CONCLUSIONS
High energy femtosecond UV laser pulses can be used to induce covalent cross-links between aromatic amino acids in peptides, overcoming photo-oxidation processes, that predominate as the mean laser pulse intensity approaches illumination conditions achievable with conventional UV light sources.
Introduction
Cross-linking (CL) with pulsed UV lasers has been heralded as a revolutionary technique to increase the photochemical yield of protein-nucleic acid CL by one to two orders of magnitude,[1–9] and to significantly reduce the timescale of the reaction. Protein-DNA photochemical CL reactions proceed in two distinct steps[10, 11]: (i) biphotonic UV light absorption and excitation of the DNA bases, in the ns-, ps- or even fs-time scale, and (ii) CL with proteins interacting with the DNA excited site and therefore lying nearby (zero-length CL), which is completed in less than 1 μs.[12] Because conformational transitions of biomolecular complexes usually require more than 100 μs, a ns or ps UV laser pulse can freeze protein-DNA interactions in real time. The exploitation of UV laser-induced CL has allowed investigators to take snapshots at various steps during the assembly of the protein-DNA complexes.[13]
These studies place great emphasis on the nucleic acid side, neglecting the possibility that proteins could also be susceptible to photo-excitation by exposure to a UV laser and that CLs could be generated in proteins. In contrast to nucleic acid–protein cross-linking, peptide–peptide and, more generally, protein–protein cross-linking mediated by UV light has not been reported in the scientific literature to the best of our knowledge. Typically, photo–cross-linking is induced between proteins by replacing some amino acids (e.g. leucine and methionine) with their photo–sensitive analogs, which contain photo–sensitive diazirine rings, or by the addition of external reagents, [14–18] or it is considered as a side reaction that follows the exposure of peptides and proteins to reactive oxygen species, namely singlet oxygen 1O2 or hydroxyl radicals HO• on an amino acid side chain. [19–29]. We have addressed the feasibility of peptide–peptide photo–cross-linking without any external intervention other than the absorption of UV light, as a way of studying transient interactions between proteins in their most native biological background.
The working hypothesis is that, upon absorption of UV light, the side chains of aromatic amino acids produce radicals that are able to react and generate covalent bonds with nearby residue(s).
A UV femtosecond pulsed laser source was chosen from three different types of UV sources: cw-lamps, nanosecond, and femtosecond pulsed lasers, because of its capability to deliver higher radiation doses in shorter irradiation times, while transferring a smaller amount of heat to the irradiated target, thus inducing only minor damage to the biomolecule of interest (see for instance reference 30).
The experiments reported herein provide proof of concept that it is possible to introduce covalent cross-links in peptides by ultrashort UV laser pulses i) without any incorporation of unnatural aminoacids, reagents, ii) within seconds or even less, iii) with high efficiency, and iv) only with the presence of aromatic side chains. We have demonstrated that cross-links are readily formed and proceed via a radical mechanism without extensive photo-damage to the peptides.
EXPERIMENTAL
Xenopsin, angiotensin I, interleukin, Glu-1-Fibrinopeptide B, 5,5-dimethyl-1-pyrroline N-oxide (DMPO), 2-methyl-2-nitrosopropane (MNP), L-ascorbic acid, matrix-assisted laser desorption/ionization (MALDI) matrix α-cyano-4-hydroxycinnamic acid, and ammonium bicarbonate (AMBIC) were purchased from Sigma (St. Louis, MO, USA). Trifluoroacetic acid (TFA) and acetonitrile (ACN) were HPLC grade solvents obtained from Carlo Erba Reagenti SPA (Arese, Italy), and the other solvents were from Baker (Mallinckrodt Baker, Milan, Italy). The molecular weight standards for the calibration of the Voyager-DE STR system were calibration mixture 1 and calibration mixture 2 purchased from AB Sciex (Framingham, MA, USA).
UV laser peptide-peptide cross-linking
To induce the cross-link we used a powerful source of UV radiation, a custom-made version of the PHAROS laser system (Light Conversion Ltd, Vilnius, Lithuania) which is a very compact femtosecond amplified laser source - a single-unit integrated system, combining up-to-millijoule pulse energies and high average output power. This system, based on the new Yb:KGW lasing medium and on a very compact Chirped Pulse Amplification scheme, emits 1.3 mJ, 170 fs pulses, centred at 1030 nm, at a repetition rate of 2 kHz, corresponding to an average power of 2.5 – 2.6 W. The repetition rate can be increased up to 200 kHz, where the average output power reaches nearly 7 W. The IR pulse is then frequency up-converted into a harmonic generator stage (HIRO) where II (515 nm), III (343 nm), and VI (257 nm) harmonic pulses, lasting about 130 fs, are obtained. The system is equipped with a sophisticated pulse picker which allows one to separately select any possible repetition rate, from single-shot to 200 kHz.
Standard peptides (10 nmol of angiotensin I, xenopsin, interleukin), individually or mixed together, were dissolved in 6 μl of ammonium bicarbonate buffer (10 mM pH 7.0) and irradiated with a laser energy of 110 μJ/pulse, at a frequency of 2 kHz and a carrier λ of 257 nm, at room temperature for different time intervals from 0.01 sec to 3 min. The reaction was stopped by adding 4 μL of ascorbic acid to achieve a final concentration of 20 mM, dissolved in the same buffer, immediately before use.
The same conditions were used to irradiate the individual peptides in the presence of 5,5-dimethyl-1-pyrroline N-oxide (DMPO) (100 mM) or 2-methyl-2-nitrosopropane (MNP) (10 mM). Sample concentration and desalting were performed using C18 reversed phase ZipTip™ pipette tips (Millipore Corp., Billerica, MA USA). The peptides were eluted with 20 μL of a solution containing 50% acetonitrile, 0.5% formic acid in Milli-Q water at a final concentration of 25 μM.
MALDI-TOF MS analysis
MALDI-TOF mass spectra were recorded in positive ion mode using an Applied Biosystems (San Jose, CA, USA) Voyager STR instrument equipped with a nitrogen laser (337 nm, 3 nsec pulse width). The analytes were mixed (1/1, v/v) with a 10 mg/mL solution of α-cyano-hydroxycinnamic acid in acetonitrile/50 mM citrate buffer (7/3, v/v); for each analysis, 2 μL of this mixture was applied to the metallic sample plate and dried at room temperature. The acceleration and reflector conditions were set as follows: target voltage at 20 kV, grid voltage at 66% of the target voltage, and delayed extraction at 150 ns, to obtain the best signal-to-noise ratios and the best possible isotopic resolution. Mass calibration was performed using external peptide standards purchased from Applied Biosystems. Raw data were analyzed as monoisotopic masses, using the software provided by the manufacturer.
For semiquantitative measurements of the cross-linked peptides, a reference peptide, Glu-1-Fibrinopeptide B, EGVNDNEEGFFSAR ([MGlu + H]+ m/z 1570.7), was added to the matrix in a concentration (10 μM) that yielded peak intensities of the order of those observed for the abundant analytes. The addition of a reference peptide, whose signal does not overlap with those of the peptides contained in the samples, allows correction for crystallization variability inherent to MALDI sample preparations [31]. Moreover, to average out microheterogeneity in the matrix crystals the spectra were automatically acquired using uniformly random laser shot pattern with fixed intensity from all over the crystal rim of the matrix-analyte preparation (25 spectra per sample, 200 shots/spectrum).
MS/MS analysis
The cross-linked products were analyzed in both MS and MS/MS modes by high resolution mass spectrometry using a hybrid Quadrupole-hexapole/Fourier transform ion cyclotron resonance mass spectrometer (Qh/FTICR (SolariX)) equipped with a 12-T actively shielded magnet (Bruker Daltonics, Billerica, MA, USA). This instrument, equipped with a nano-spray source, was operated in positive ion mode. The high voltage used for ionization was between 1,000 and 1,500 V and nitrogen was used as a countercurrent drying gas with its temperature maintained at 180 °C. To record the spectra, an electron capture dissociation (ECD) current of 1.6 A, an ECD bias of 1.5 V and an electron pulse length of 0.07 s were employed. For collision-induced dissociation (CID) spectra, the collision voltage was set between 8 and 15 V. The collision gas was argon at a pressure of 6 × 10−6 mbar. In the electron transfer dissociation (ETD) mode, radical negative ions of fluoranthene, produced in the chemical ionization (CI) source of the SolariX mass spectrometer, were used as the reagent ions. During the ETD experiments, the crucible was heated to about 60 °C to sublimate the fluoranthene. The fluoranthene vapor then passed into the CI chamber where it was ionized via chemical ionization with methane. The methane tank was connected to the CI source with stainless steel tubing. Negatively charged fluoranthene ions were extracted from the CI source via a set of lenses. The filament was operated with a current of 3 μA. The acceleration time for the reagent was 50–100 ms and the reaction time was 20 ms.
Mass spectra were acquired in the positive-ion mode over the range m/z 200–2000 at a mass resolution of 60,000 at m/z 400. The mass accuracy was under 1 ppm. Compass Data analysis software (Bruker Daltonics) was used for data analysis; peptide sequencing and cross-linking site assignments were conducted manually employing a ±1 ppm error limit on the product ions.
RESULTS AND DISCUSSION
Three peptides, xenopsin (pyroEGKRPWIL), angiotensin I (DRVYIHPFHL), and interleukin (VQGEESNDK), alone or in combination, were exposed to the UV laser under various irradiation conditions. The first two peptides were selected because they contain one or more aromatic amino acids, whereas interleukin was included because it is devoid of aromatic moieties. Moreover, all of them fall within a molecular weight range adequate to allow direct MS and MS/MS analyses, even when cross-linked, without further manipulation of the sample. The laser setups were inspired by Fecko et al., [32] who studied CL in vitro between oligonucleotides and proteins with a femtosecond laser system, although with a much higher repetition rate and lower-energy pulses than those used in our experiments. Our laser system allowed us to span the pulse energy in the range 10–160 μJ and laser pulse repetition rate in the range 30 Hz – 200 kHz, the irradiation time being only a fraction of a second in some cases. The sample solution was introduced as 6-μL drops with peptide concentrations ranging from 0.5 mM to 5 mM.
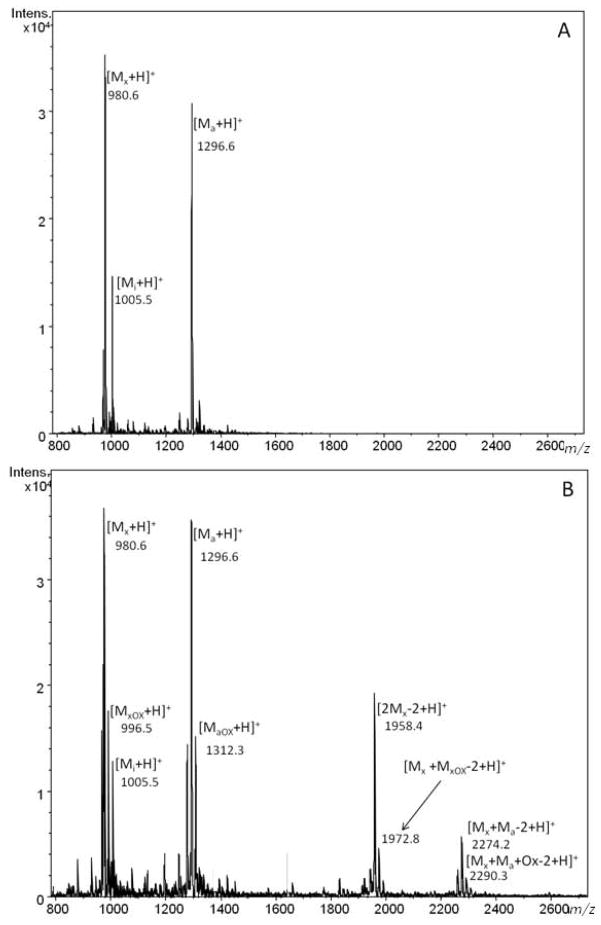
Figure 1 presents the MALDI-TOF mass spectrum of a mixture of xenopsin, interleukin and angiotensin (1.6 mM each) before exposure to the laser (Fig. 1a), and after exposure of the mixture to UV laser pulses of 110 μJ for 10 sec at a repetition rate of 2 kHz (Fig. 1b). The three signals in the spectrum in Fig. 1a correspond to the peptide standards (xenopsin [Mx + H]+ m/z 980.6; interleukin [Mi + H]+ m/z 1005.5; angiotensin I [Ma + H]+m/z 1296.6). In Fig. 1b, there are two new signals, generated after UV-laser exposure: the peak at m/z 1958.4 that could be tentatively assigned as [(2Mx − 2H) + H]+ for two molecules of xenopsin cross-linked to one another, and the signal at m/z 2274.2 that could correspond to one molecule of xenopsin cross-linked to one molecule of angiotensin I, [(Mx + Ma − 2H) + H]+. Signals that can be ascribed to modified products resulting from oxidation of the peptides can be also observed in Fig. 1b.
Figure 1.
Positive-ion MALDI-TOF mass spectra of a mixture of xenopsin (Mx), interleukin (Mi) and angiotensin I (Ma) not irradiated (panel A) and irradiated for 10 sec (panel B).
It should be noted that interleukin (that has no aromatic residue) does not appear to be involved in any of the species generated upon UV exposure. Each of the three peptides was separately exposed to the UV laser (see Fig. 6 for xenopsin as an example, and Figs S1 and S2 in the Supporting Information for angiotensin and interleukin, respectively), and the results confirmed that only peptides containing an aromatic side chain generated multimeric species upon exposure to UV laser light, thereby demonstrating that the CL reaction requires the presence of aromatic amino acids. These results suggest a reaction mechanism that is triggered by exposure to UV laser light and generates a CL of two aromatic ring centered radicals, analogous to the formation of bis-tyrosine and adducts of two tryptophans as observed in oxidative processes. [27, 28, 33]
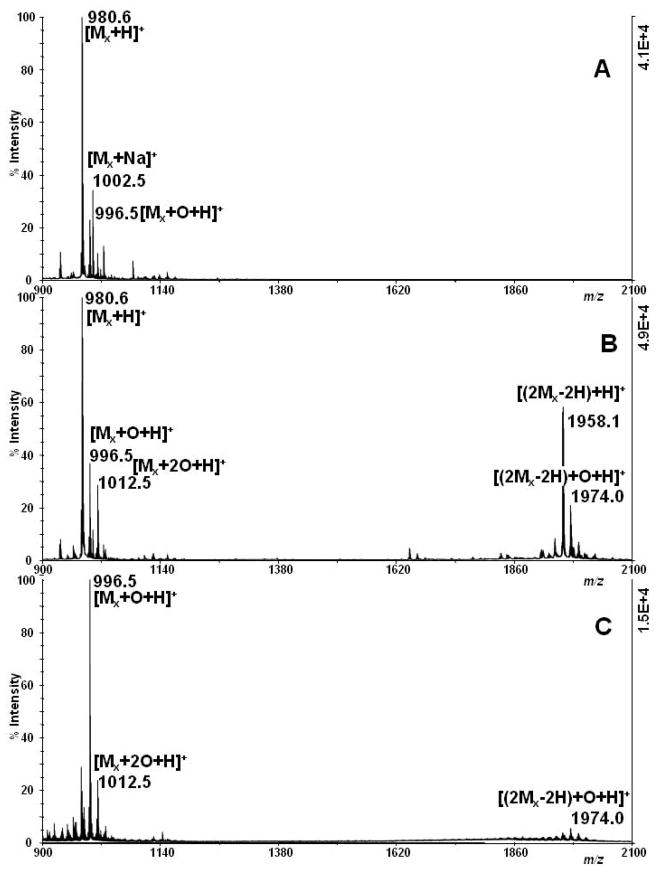
Figure 6.
Positive-ion MALDI-TOF mass spectrum of xenopsin after exposure to the high intensity and low intensity UV laser. Xenopsin ([M + H]+ m/z 980.5) (A, control) was exposed to (B) high intensity UV light for 1.56 s (1.3×106 μW cm−2, 160 μJ/pulse, 2 kHz repetition rate, 1.56 s), so that an energy of 0.5 J was released to the solution. (C) The same energy (0.5 J) was released in 58 minutes (6×102 μW cm−2, 0.75×10−3 μJ/pulse, 200 kHz repetition rate, 58 min)
The systematic loss of 2 Da, with respect to the sum of the molecular masses of the two peptides, was observed, suggesting that two hydrogen atoms are lost in the formation of the cross-link, and this assignment was confirmed by high resolution accurate mass measurements: the mass difference was within 1 ppm of 2.015650, the accurate mass of 2H.
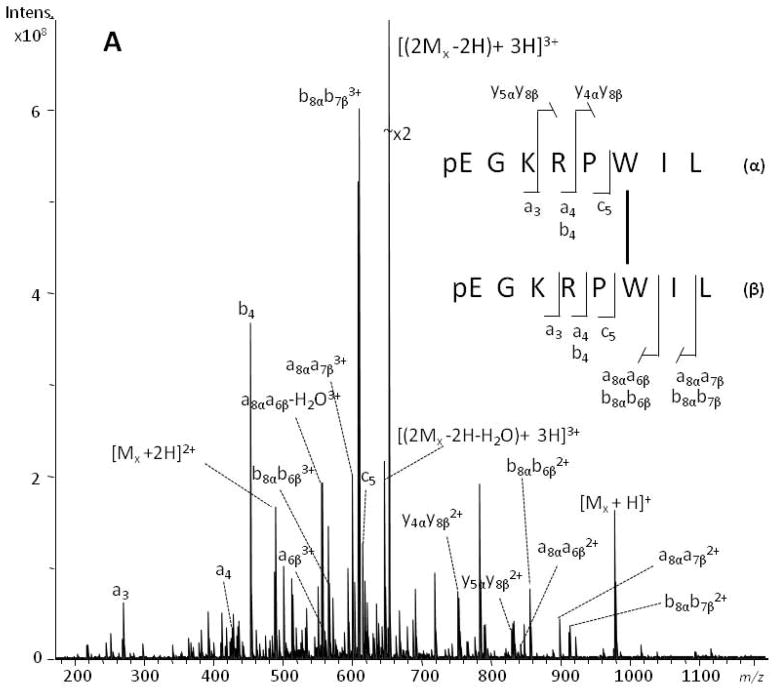
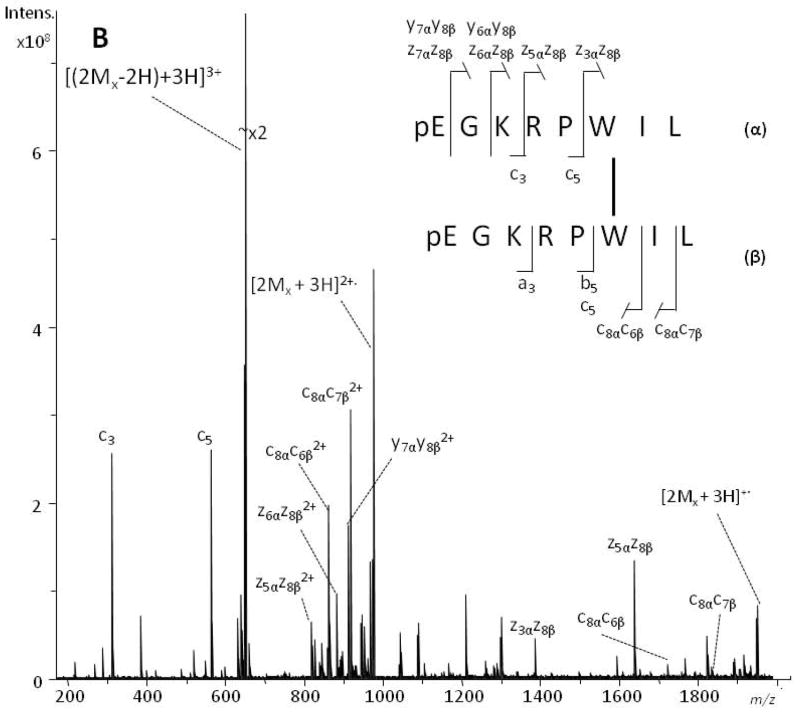
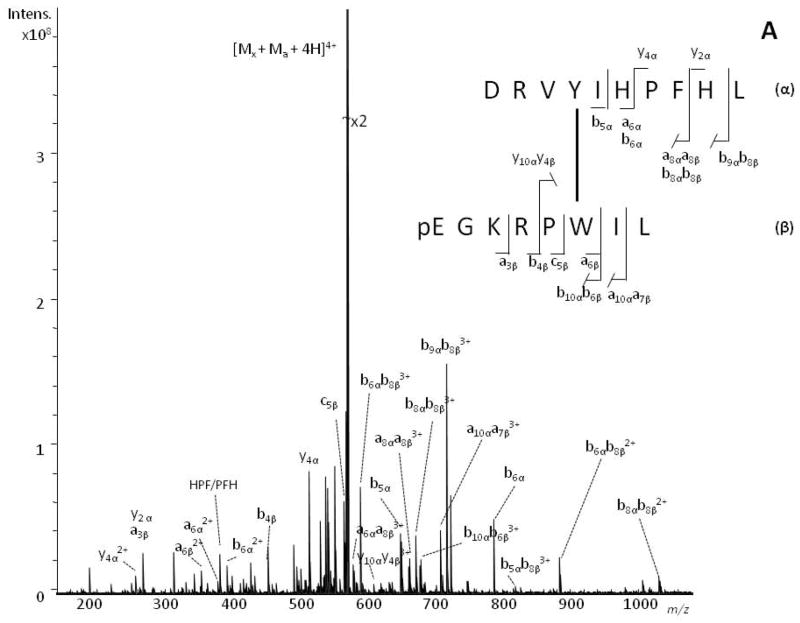
MS/MS spectra of the dimeric species generated upon irradiation with UV laser pulses were obtained on the SolariX 12-T FTICR mass spectrometer with collision-induced dissociation (CID), electron capture dissociation (ECD) and electron transfer dissociation (ETD) fragmentation modes. Displayed in Figs 2 and 3 are the CID (a) and ECD (b) MS/MS spectra generated from the [(2Mx − 2H) + 3H]3+ ion (m/z 653.3759 (CID), m/z 653.3757 (ECD); calc. m/z 653.3756, see Fig. S3, Supporting Information), and the [(Ma + Mx − 2H) + 4H]4+ ion (m/z 569.3127 (CID), m/z 569.3131 (ECD); calc. m/z 569.3128, see Fig. S4, Supporting Information), that had been tentatively assigned to the homodimer of xenopsin and the heterodimer of angiotensin I-xenopsin, respectively. Cleavage products originating from both peptide chains can be assigned, and product ions corresponding to the two peptides are, therefore, designated with either the α or the β subscript to indicate the peptide of origin.
Figure 2.
Positive-ion ESI-CID (panel A) and ESI-ECD (panel B) MS/MS spectra of xenopsin homodimer. The [(2Mx −2H) + 3H]3+ peaks at m/z 653.3759 and m/z 653.3757 were selected as the precursor ions in ESI-CID and ESI-ECD MS/MS, respectively. The cross-linked sites are indicated with a line connecting the linked residues.
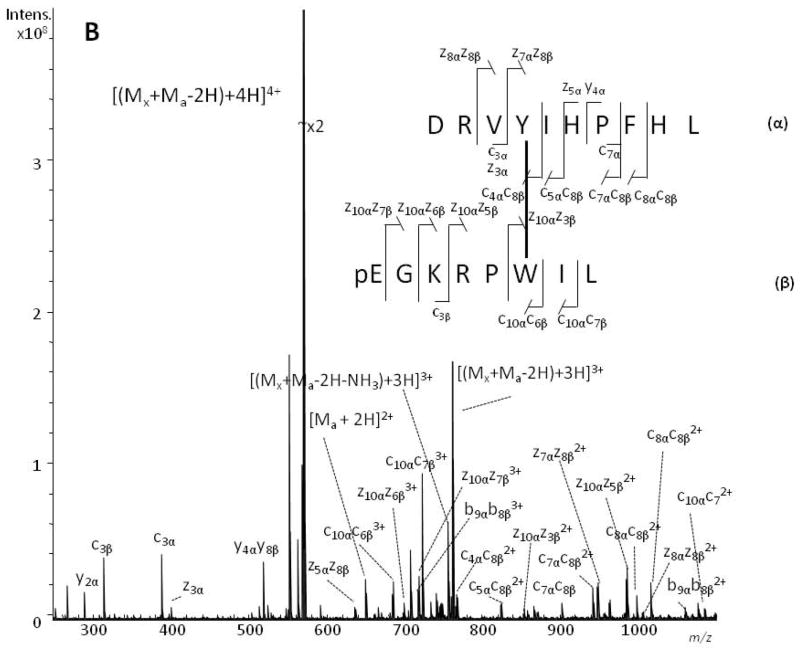
Figure 3.
Positive-ion ESI-CID (panel A) and ESI-ECD (panel B) MS/MS spectra of the angiotensin I/xenopsin heterodimer. The [(Mx + Ma − 2H) + 4H]4+ peaks at m/z 569.3127 and m/z 569.3131 were selected as the precursor ions in ESI-CID and ESI-ECD MS/MS, respectively. Product ions arising from angiotensin I (DRVYIHPFHL) are labelled with an α subscript, and those from xenopsin (pEGKRPWIL) with a β subscript. The cross-linked sites are indicated with a line between the sequences.
The fragmentation observed in the low-energy CID spectrum of the homodimer (Fig. 2a) is dominated by ions corresponding to a-, b- and y- ions whereas, as expected, c and z• ion series dominate the corresponding ECD spectrum (Fig. 2b). In addition to short z- and a-, b-, c-ion series, it is possible to observe several yαyβ, zαzβ, bαbβ, cαcβ-ions detected in their singly, doubly and/or triply charged states, that facilitate assessment of the CL sites as detailed in Tables S1 and S2 (Supporting Information). The z3αz8β and c8αc6β2+ ions are key signals to univocally determine that Trp-6 is involved in the cross-link.
Similar considerations allowed the interpretation of the MS/MS spectra (Fig. 3) generated from the [(Mx + Ma − 2H) + 4H]4+ ions tentatively assigned to the heterodimer of angiotensin I (α-chain) and xenopsin (β-chain). The MS/MS spectra of the quadruply charged ion at m/z 569.3127 consist of dominant b- and y-series ions for the CID spectrum (Fig. 3a) while c,z-series ions dominate the ECD MS/MS spectrum (Fig. 3b) generated from the ion m/z 569.3131, as detailed in Tables S3 and S4 (Supporting Information). Several ions that can be interpreted as cross-linked species allowed definition of the location of the CL site within the peptide sequences. The c10αc6β3+, z10αz3β2+, z7αz8β2+, and c4αc8β2+ ions are key signals to define that Tyr-4 and Trp-6 are involved in the CL. It should be noted that a few cross-link-free product ions were observed in the CID spectrum and one in the ECD spectrum. To check whether this phenomenon was due to the existence of another cross-linking radical site, we attempted to assign these few product ions in the CID and ECD spectra by considering other radical sites. No ions in the MS/MS spectra could be assigned as having been formed from a differently cross-linked peptide. Moreover, the cross-link-free product ions were formed when the CID mode was employed, whereas the occurrence of such ions was minimal in the ECD spectra; it has been widely established that ECD is a more gentle activation mode than CID. Thus, we suggest that the formation of some low abundant cross-link-free product ions results from a competition between cleavages of the cross-link and cleavage along the peptide backbone, indicating that the relative stability of the cross-link is comparable with that of the bonds along the peptide backbone. ETD (spectra not shown) confirmed this view by leading to assignments similar to those from ECD.
To test the hypothesis that UV-induced CL is a radical reaction, exposure to the laser was carried out in the presence of spin trap molecules. Spin traps are often used to allow the visualization of transient free radical populations by reacting with short-lived radicals to produce persistent spin adduct radicals that can be studied by electron paramagnetic resonance (EPR) [34, 35] or MS since the spin trap molecules covalently label the radical site in the molecule. [36–41]
Therefore, the peptides were exposed to the UV laser as described above, for either 10 sec or 1 min, but in the presence of either 5,5-dimethyl-1-pyrroline N-oxide (DMPO), or 2-methyl-2-nitrosopropane (MNP).
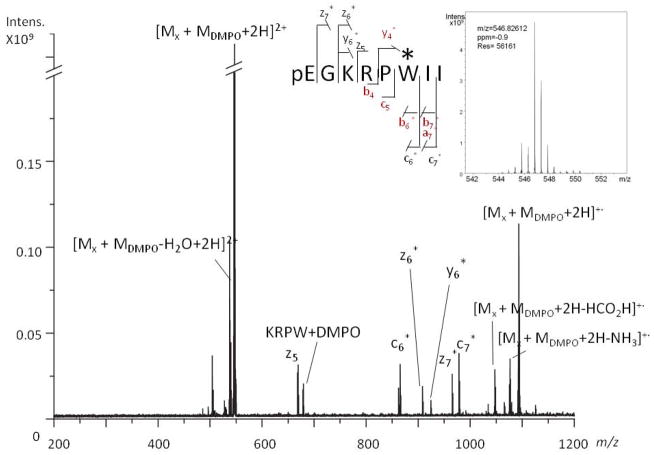
MALDI-TOF MS analysis of xenopsin irradiated in the presence of 10 mM DMPO (Fig. 4b) yielded a signal at m/z 1091.6, which is 111.1 m/z units higher than the mass of protonated xenopsin ([Mx + H]+ m/z 980.5, Fig. 4a), thus suggesting that a single DMPO molecule was trapped on the peptide. The ETD spectrum of the [Mx + MDMPO + 2H]2+ ion (m/z 546.8261; calc. m/z 546.8256), presented in Fig. 5, yielded the charge-reduced ion [Mx + MDMPO + 2H]+· at m/z 1093.6499 (calc. m/z 1093.6517) and, more interestingly, c-, z- y- and b-series product ions bearing the DMPO modification. For example, the ion at m/z 924.5906 (labelled as y6*) (calc. m/z 924.5903) corresponds in mass to a y6 ion plus DMPO (+112.0762 m/z units) and the ion at m/z 865.4914 (labelled as c6*) (calc. m/z 865.4917) corresponds in mass to a c6 ion containing DMPO (+112.0762 m/z units). Moreover, different internal product ions of the peptide also show the addition of DMPO (e.g., [Mx + MDMPO − H2O + H]+ at m/z 537.8209 (calc. m/z 537.8203)). The CID spectrum showed complementary ions such as some corresponding to cleavages of amino acids from the N-terminus of the peptide backbone with loss of DMPO (b4 and c5) and some corresponding to cleavages of amino acids from the N-terminus of the peptide backbone bearing DMPO (y4*, b6*, b7*, a7*). The concomitant presence of the ions y4*, c6* and c5 indicates that the DMPO adduct is located on Trp-6 in xenopsin. The ECD spectrum (Fig. S5, Supplementary Information) further confirms the interpretation, showing several internal product ions bearing the DMPO moiety.
Figure 4.
Positive-ion MALDI-TOF mass spectra of xenopsin (A) and xenopsin after exposure to the UV laser in the presence of DMPO (B). The peak at m/z 1091.6 corresponds to the [M + H]+ ion of the adduct formed between one molecule of DMPO and one molecule of xenopsin. In the inset, a zoom of the selected m/z range.
Figure 5.
Positive-ion ESI-ETD mass spectrum of xenopsin, exposed to the UV laser in the presence of DMPO. The [Mx + MDMPO + 2H]2+ peak at m/z 546.8261 was selected as the precursor ion. The asterisk indicates the residue at the adduct site. The members of the ion series detected in the CID spectrum are indicated in red. In the inset, the ESI-FTICR mass spectrum of the precursor ion.
Similar analyses were performed for the same peptide irradiated in the presence of 10 mM MNP (see Fig. S6, Supplementary Information, and relative comments).
The interpretation of these data was severely complicated by a significant loss of DMPO and MNP molecules during fragmentation. Possibly the bond between DMPO or MNP and the residue in the peptide is weak and thus susceptible to facile cleavage during MS/MS. To determine the adduct sites, we used CID, ETD and ECD activation modes. The ETD and ECD activation modes are both gentler than CID activation. In the ECD and ETD spectra, the most abundant product ions preserved the spin trap moieties, and a loss of MNP or DMPO could be observed only for a few, usually less abundant, product ions.
The product ions assigned in the CID spectra are indicated in red on the peptide sequence in the ETD or ECD spectra. We can still observe several ions bearing MNP or DMPO although some significant product ions can be attributed to losses of MNP or DMPO. This has been already reported in other cases, [36, 42], and interpreted as a cleavage competition between the covalent bond with the spin trap on the side chain and the peptide backbone.
Nevertheless, the presence of product ions that retained the DMPO moiety allowed us to suggest the assignment of the DMPO and MNP linkages to the Trp-6 residue of xenopsin, as also validated in the ECD analysis (Fig. S5, Supporting Information).
The same experiments with angiotensin I and DMPO and MNP (see Figs S7 and S8, Supporting Information), suggest DMPO and MNP linkages to the Tyr-4 residue of angiotensin I.
CL has already been reported as a possible type of UV-induced damage in polypeptides. [19–29] It is, however, important to stress that all the photochemical reactions previously described are reported to follow upon the initial addition of singlet oxygen 1O2 or hydroxyl radical HO• to an amino acid side chain. The intermediates then undergo a variety of further reactions which can result in radical formation and ring-opening reactions, including cross-links in proteins.
We then considered comparing the results obtained with our femtosecond laser source with those recorded after exposure of the samples to conventional UV light sources such as a UV lamp. Releasing the same dose (0.5 J) to the target with a typical UV lamp would, however, have taken a very long time, between several hours and two days of irradiation, and such a long time would have resulted in deterioration of our sample (even evaporation would no longer be non-negligible). Therefore, we compared our result with those obtained with the femtosecond laser system operated in very different conditions, to make it as close as possible to a standard UV cw-lamp. Xenopsin was exposed to UV light at reduced pulse intensity while extending the exposure time, so as to keep the same total energy amount (0.5 J): thus, the comparison was carried out under the two conditions 6×102 μW cm−2 and 1.3×106 μW cm−2 for 58 min (0.75×10−3 μJ/pulse, 200 kHz repetition rate) and 1.56 s (160 μJ/pulse, 2 kHz repetition rate). The former are the conditions employed for the result shown in Fig. 6c, where the same dose was released as in Fig. 6b, but with laser nano–pulses (carrying very low energy), delivered in a much larger number and for a much longer irradiation time than in the experiment shown in Fig. 6b.
For the low energy/pulse sample (Fig. 6c), ions arising from side chain photo-oxidation are the main components of the mass spectrum. The most abundant ion corresponds to the addition of one oxygen atom, probably due to the formation of hydroxytryptophan (Mx + O, Δ +16 Da), in agreement with data reported by Grosvenor et al., [19] where a mean intensity of the order of few mW cm−2 was used. The signal at m/z 1012.5 (Mx + 2O, Δ +32 m/z units) can be accordingly attributed to N-formylkynurenine (NFK)/dihydroxy- tryptophan. Further signals that can be ascribed to some hydroxy-formylkynurenine (M + 3O, Δ +48 Da) can also be detected, as well as a minor dimeric species. In the MS analysis of the high energy/pulse sample (Fig. 6b), the main reaction product is the dimeric form of the peptide (2M – 2H, Δ −2 Da) without notable increase in the signals of the photo-oxidation products with respect to the reference, non-irradiated sample (Fig. 6a).
Moreover, when using the high pulse energy irradiation, no notable difference was observed in the mass spectra of xenopsin samples, analyzed immediately or left in the open air for 58 min after irradiation (see Fig. S9, Supplementary Information). This suggests that the oxidation products detected in the low energy/pulse sample are a consequence of exposure to the UV laser.
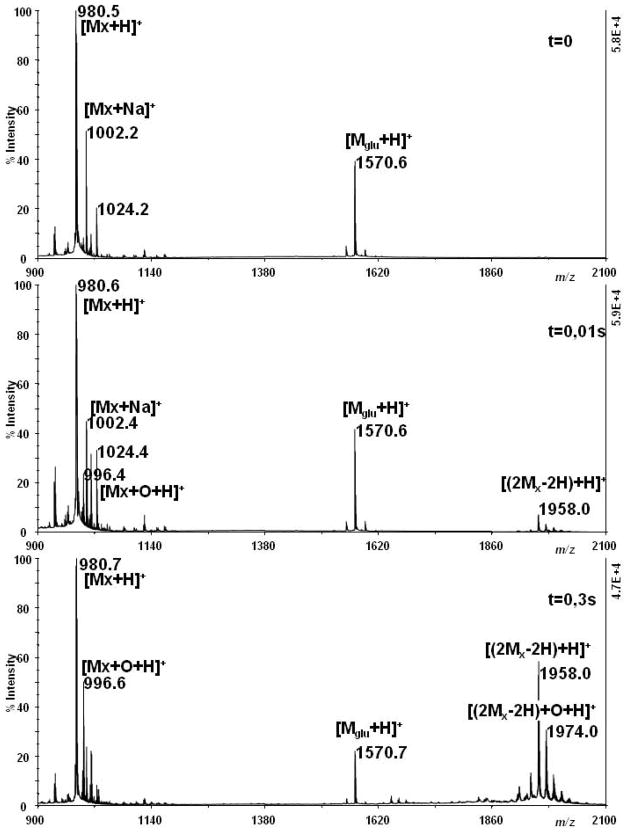
Figure 7 reports examples of a semi-quantitative analysis of the yield of cross-linking as a function of the exposure time. Xenopsin was irradiated with a UV laser (2 KHz, 160μJ/pulse) for different exposure times starting from 0.01 sec and analysed by MALDI-TOF, with the introduction of a reference peptide (Glu-1-Fibrinopeptide B, [MGlu + H]+ m/z 1570.6) in the matrix to provide a semi-quantitative evaluation of the yield of CL [31]. Even in the high energy/pulse regime, some photoxidation products could be observed, and these increased upon increasing the exposure time. However, some consistent cross-linking was achieved with exposure times as low as 0.01 sec (corresponding to only 20 laser pulses) and CL was consistently the predominant reaction.
Figure 7.
Positive-ion MALDI-TOF mass spectrum of xenopsin after exposure to the high intensity UV laser as a function of exposure time. Xenopsin ([M + H]+ m/z 980.5) (A, control) was exposed to high intensity UV light (2 KHz, 160μJ/pulse) for 0.01 s (B) and 0.3 s. (C). Glu-1-Fibrinopeptide B, [MGlu + H]+ m/z 1570.7, was introduced as reference peptide in the matrix to provide a semi-quantitative evaluation of the yield of CL.
These results suggest that a conventional UV cw - lamp would probably fail to induce the consistent amount of peptide–peptide cross-linking, observed with the irradiation conditions of Fig. 6b, typical of femtosecond laser sources. As a consequence, we can also conclude that the key parameter in obtaining the observed cross-linking is the peak power of the delivered laser pulses rather than the integrated dose of radiation energy, indicating a possible nonlinear excitation mechanism of the involved molecules at a microscopic level, that will be addressed in forthcoming studies.
CONCLUSIONS
We have demonstrated herein that, upon exposure to pulsed UV laser light of wavelength near 260 nm, a “zero-length” covalent bond between the aromatic side chains of amino acids in different peptide molecules can be formed with good efficiency on an extremely rapid time scale, probably in the pico- or even femtosecond range. We have determined that photochemical CL is by far the predominant reaction, and that it requires the light intensity that can be generated with pulsed laser sources since, as the average laser intensity is reduced, down to that od conventional UV lamps, photo-damage is observed, similar to damage occurring with conventional UV light sources. [20–28] We defined a molecular basis for the exploitation of UV pulsed laser sources as a powerful CL agent, that would certainly have a strong impact on the possibility of studying transient interactions among proteins, and the dynamics of the contacts within multi-protein complexes, and to discover transient interactions which have so far escaped observation in “molecular sociology of the cell” studies. [43] Although a demonstration that our initial observations of efficient cross-links generated between aromatic amino acids in peptides can be extended to proteins, to freeze biologically significant interactions, will demand further experiments. The results presented here offer the first indications of the feasibility of the development of such an approach.
The extremely fast kinetics of the reactions, the almost instantaneous diffusion of light within the cell, and the absence of exogenous chemical reactants, suggest that, once established, UV laser CL will represent an innovative and important tool well tailored for in vivo applications.
Supplementary Material
Acknowledgments
CA, RE, and RV wish to acknowledge the European Community (ATLAS contract n. 221952) for the support to set up the laser source experimental apparatus. The BUSM Mass Spectrometry Resource is supported by NIH grant P41 RR10888/GM104603; the SolariX FTMS was acquired with NIH grant S10 RR025082. LB and GL wish to aknowledge the “Programma per la Breve Mobilità” of the Università di Napoli Federico II, for the financial support to GL in the visit to CEC laboratory. We thank Prof. Orlando Crescenzi, Diparti di Scienze Chimiche, Napoli, for critical discussion.
Footnotes
Electronic Supplementary Information (ESI) available: assignments of the reported MS/MS spectra, additional MS/MS spectra and the experiments with the spin traps.
References
- 1.Harrison CA, Turner DH, Hinkle DC. Laser crosslinking of E. coli RNA polymerase and T7 DNA. Nucleic Acids Res. 1982;10:2399. doi: 10.1093/nar/10.7.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hockensmith JW, Kubasek WL, Vorachek WR, Von Hippel PH. Laser cross-linking of nucleic acids to proteins. Methodology and first applications to the phage T4 DNA replication system. J Biol Chem. 1986;261:3512. [PubMed] [Google Scholar]
- 3.Lejnine S, Durfee G, Murnane M, Kapteyn HC, Makarov VL, Langmore JP. Crosslinking of proteins to DNA in human nuclei using a 60 femtosecond 266 nm laser. Nucleic Acids Res. 1999;27:3676. doi: 10.1093/nar/27.18.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pashev IG, Dimitrov SI, Angelov D. Crosslinking proteins to nucleic acids by ultraviolet laser irradiation. Trends Biochem Sci. 1991;16:323. doi: 10.1016/0968-0004(91)90133-g. [DOI] [PubMed] [Google Scholar]
- 5.Russmann C, Stollhof J, Weiss C, Beigang R, Beato M. Two wavelength femtosecond laser induced DNA–protein crosslinking. Nucleic Acids Res. 1998;26:3967. doi: 10.1093/nar/26.17.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russmann C, Truss M, Fix A, Naumer C, Herrmann T, Schmitt J, Stollhof J, Beigang R, Beato M. Crosslinking of progesterone receptor to DNA using tuneable nanosecond, picosecond and femtosecond UV laser pulses. Nucleic Acids Res. 1997;25:2478. doi: 10.1093/nar/25.12.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angelov D, Charra M, Müller CW, Cadet J, Dimitrov S. Solution study of the NF-kB p50-DNA complex by UV laser protein-DNA cross-linking. Photochem Photobiol. 2003;77:592. doi: 10.1562/0031-8655(2003)077<0592:ssotnp>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 8.Angelov D, Yu Stefanovsky V, Dimitrov SI, Russanova VR, Keskinova E, Pashev IG. Protein-DNA crosslinking in reconstituted nucleohistone, nuclei and whole cells by picosecond UV laser irradiation. Nucleic Acids Res. 1988;16:4525. doi: 10.1093/nar/16.10.4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altucci C, Nebbioso A, Benedetti R, Esposito R, Carafa V, Conte M, Micciarelli M, Altucci L, Velotta R. Nonlinear protein – nucleic acid crosslinking induced by femtosecond UV laser pulses in living cells. Laser Phys Lett. 2012;9:234. [Google Scholar]
- 10.Kryukov P, Letokhov V, Nikogosyan D, Borodavkin A, Budowsky E, Simukova N. Multiquantum photoreactions of nucleic acid components in aqueous solution by powerful ultraviolet picosecond radiation. Chem Phys Lett. 1979;61:375. [Google Scholar]
- 11.Nikogosyan DN. Two-quantum UV photochemistry of nucleic acids: Comparison with conventional low-intensity UV photochemistry and radiation chemistry. Int J Radiat Biol. 1990;57:233. doi: 10.1080/09553009014552411. [DOI] [PubMed] [Google Scholar]
- 12.Hockensmith JW, Kubasek WL, Vorachek WR, Evertsz EM, Von Hippel PH. Laser cross-linking of protein-nucleic acid complexes. Methods Enzymol. 1991;46:211. doi: 10.1016/0076-6879(91)08015-a. [DOI] [PubMed] [Google Scholar]
- 13.Nagaich AK, Hager GL. UV laser cross-linking: a real-time assay to study dynamic protein/DNA interactions during chromatin remodeling. Sci STKE. 2004;2004:113. doi: 10.1126/stke.2562004pl13. [DOI] [PubMed] [Google Scholar]
- 14.Suchanek M, Radzikowska A, Thiele C. Photo-leucine and photo-methionine allow identification of protein-protein interactions in living cells. Nat Methods. 2005;2:261. doi: 10.1038/nmeth752. [DOI] [PubMed] [Google Scholar]
- 15.Chin JW, Schultz PG. In vivo photocrosslinking with unnatural amino Acid mutagenesis. Chembiochem. 2002;3:1135. doi: 10.1002/1439-7633(20021104)3:11<1135::AID-CBIC1135>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 16.Forné I, Ludwigsen J, Imhof A, Becker PB, Mueller-Planitz F. Probing the conformation of the ISWI ATPase domain with genetically encoded photoreactive crosslinkers and mass spectrometry. Mol Cell Proteomics. 2012:11. doi: 10.1074/mcp.M111.012088. In press M111.012088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith DP, Anderson J, Plante J, Ashcroft AE, Radford SE, Wilson AJ, Parker MJ. Trifluoromethyldiazirine: an effective photo-induced cross-linking probe for exploring amyloid formation. Chem Commun. 2008;44:5728. doi: 10.1039/b813504e. [DOI] [PubMed] [Google Scholar]
- 18.Rotili D, Altun M, Hamed RB, Loenarz C, Thalhammer A, Hopkinson RJ, Tian YM, Ratcliffe PJ, Mai A, Kessler BM, Schofield CJ. Photoactivable peptides for identifying enzyme-substrate and protein-protein interactions. Chem Commun. 2011;47:1488. doi: 10.1039/c0cc04457a. [DOI] [PubMed] [Google Scholar]
- 19.Grosvenor AJ, Morton JD, Dyer JM. Profiling of residue-level photo-oxidative damage in peptides. Amino Acids. 2010;39:285. doi: 10.1007/s00726-009-0440-7. [DOI] [PubMed] [Google Scholar]
- 20.Davies MJ. Reactive species formed on proteins exposed to singlet oxygen. Photochem Photobiol Sci. 2004;3:17. doi: 10.1039/b307576c. [DOI] [PubMed] [Google Scholar]
- 21.Mizdrak J, Hains PG, Truscott RJW, Jamie JF, Davies MJ. Tryptophan-derived ultraviolet filter compounds covalently bound to lens proteins are photosensitizers of oxidative damage. Free Radical Biol Med. 2008;44:1108. doi: 10.1016/j.freeradbiomed.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Aquilina JA, Carver JA, Truscott RJ. Elucidation of a novel polypeptide cross-link involving 3-hydroxykynurenine. Biochemistry. 1999;38:11455. doi: 10.1021/bi990458h. [DOI] [PubMed] [Google Scholar]
- 23.Aquilina JA, Carver JA, Truscott RJ. Polypeptide modification and cross-linking by oxidized 3-hydroxykynurenine. Biochemistry. 2000;39:16176. doi: 10.1021/bi001230t. [DOI] [PubMed] [Google Scholar]
- 24.Hawkins CL, Davies MJ. Generation and propagation of radical reactions on proteins. Biochim Biophys Acta. 2001;1504:196. doi: 10.1016/s0005-2728(00)00252-8. [DOI] [PubMed] [Google Scholar]
- 25.Igarashi N, Onoue S, Tsuda Y. Photoreactivity of amino acids: tryptophan-induced photochemical events via reactive oxygen species generation. Anal Sci. 2007;23:943. doi: 10.2116/analsci.23.943. [DOI] [PubMed] [Google Scholar]
- 26.Fonseca C, Domingues MR, Simões C, Amado F, Domingues P. Reactivity of Tyr-Leu and Leu-Tyr dipeptides: identification of oxidation products by liquid chromatography-tandem mass spectrometry. J Mass Spectrom. 2009;44:681. doi: 10.1002/jms.1543. [DOI] [PubMed] [Google Scholar]
- 27.Domingues MR, Domingues P, Reis A, Fonseca C, Amado FM, Ferrer-Correia AJ. Identification of oxidation products and free radicals of tryptophan by mass spectrometry. J Am Soc Mass Spectrom. 2003;14:406. doi: 10.1016/S1044-0305(03)00127-2. [DOI] [PubMed] [Google Scholar]
- 28.Stadtman ER, Levine RL. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids. 2003;25:207. doi: 10.1007/s00726-003-0011-2. [DOI] [PubMed] [Google Scholar]
- 29.Maleknia SD, Downard K. Radical approaches to probe protein structure, folding, and interactions by mass spectrometry. Mass Spectrom Rev. 2001;20:388. doi: 10.1002/mas.10013. [DOI] [PubMed] [Google Scholar]
- 30.Middleton CT, de La Harpe K, Su C, Law YK, Crespo-Hernández CE, Kohler B. DNA excited-state dynamics: from single bases to the double helix. Annu Rev Phys Chem. 2009;60:217. doi: 10.1146/annurev.physchem.59.032607.093719. [DOI] [PubMed] [Google Scholar]
- 31.Jiménez CR, Li KW, Dreisewerd K, Mansvelder HD, Brussaard AB, Reinhold BB, Van der Schors RC, Karas M, Hillenkamp F, Burbach JP, Costello CE, Geraerts WP. Pattern changes of pituitary peptides in rat after salt-loading as detected by means of direct, semiquantitative mass spectrometric profiling. Proc Natl Acad Sci USA. 1997;94:9481. doi: 10.1073/pnas.94.17.9481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fecko CJ, Munson KM, Saunders A, Sun G, Begley TP, Lis JT, Webb WW. Comparison of femtosecond laser and continuous wave UV sources for protein-nucleic acid crosslinking. Photochem Photobiol. 2007;83:1394. doi: 10.1111/j.1751-1097.2007.00179.x. [DOI] [PubMed] [Google Scholar]
- 33.Stadtman ER. Oxidation of free amino acids and amino acid residues in proteins by radiolysis and by metal-catalyzed reactions. Annu Rev Biochem. 1993;62:797. doi: 10.1146/annurev.bi.62.070193.004053. [DOI] [PubMed] [Google Scholar]
- 34.Davies MJ, Hawkins CL. EPR spin trapping of protein radicals. Free Radical Biol Med. 2004;36:1072. doi: 10.1016/j.freeradbiomed.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 35.Augusto O, Vaz SM. EPR spin-trapping of protein radicals to investigate biological oxidative mechanisms. Amino Acids. 2007;32:535. doi: 10.1007/s00726-006-0429-4. [DOI] [PubMed] [Google Scholar]
- 36.Chen YR, Chen CL, Zhang L, Green-Church KB, Zweier JL. Superoxide generation from mitochondrial NADH dehydrogenase induces self-inactivation with specific protein radical formation. J Biol Chem. 2005;280:37339. doi: 10.1074/jbc.M503936200. [DOI] [PubMed] [Google Scholar]
- 37.Deterding LJ, Bhattacharjee S, Ramirez DC, Mason RP, Tomer KB. Identification of the myoglobin tyrosyl radical by immuno-spin trapping and its dimerization. Anal Chem. 2007;79:6236. doi: 10.1016/j.freeradbiomed.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 38.Deterding LJ, Ramirez DC, Dubin JR, Mason RP, Tomer KB. Identification of free radicals on hemoglobin from its self-peroxidation using mass spectrometry and immuno-spin trapping: observation of a histidinyl radical. J Biol Chem. 2004;279:11600. doi: 10.1074/jbc.M310704200. [DOI] [PubMed] [Google Scholar]
- 39.Fenwick CW, AM English Trapping and LC–MS Identification of Protein Radicals Formed in the Horse Heart Metmyoglobin–H2O2 Reaction. J Am Chem Soc. 1996;118:12236. [Google Scholar]
- 40.Harris MN, Burchiel SW, Winyard PG, Engen JR, Mobarak CD, Timmins GS. Determining the site of spin trapping of the equine myoglobin radical by combined use of EPR, electrophoretic purification, and mass spectrometry. Chem Res Toxicol. 2002;15:1589. doi: 10.1021/tx025594t. [DOI] [PubMed] [Google Scholar]
- 41.Lardinois OM, Tomer KB, Mason RP, Deterding LJ. Identification of protein radicals formed in the human neuroglobin-H2O2 reaction using immuno-spin trapping and mass spectrometry. Biochemistry. 2008;47:10440. doi: 10.1021/bi800771k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Detweiler CD, Lardinois OM, Deterding LJ, De Montellano PRO, Tomer KB, Mason RP. Identification of the myoglobin tyrosyl radical by immuno-spin trapping and its dimerization. Free Radical Biol Med. 2005;38:969. doi: 10.1016/j.freeradbiomed.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 43.Robinson CV, Sali A, Baumeister W. The molecular sociology of the cell. Nature. 2007;450:973. doi: 10.1038/nature06523. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.