We recently conducted a genome-wide association study (GWAS) using data from 1,854 PrCa cases with clinically detected (not PSA screened) PrCa diagnosed at <60 years or with a family history of the disease, and 1,894 population screened controls with a prostate specific antigen (PSA) of <0.5ng/ml (Eeles et al. 2008). These were analysed for 541,129 SNPs using the Illumina Infinium platform. Putative associations were evaluated using a further 3,268 cases and 3,366 controls. After these two stages, associations at seven loci, on chromosomes 2,3,6,7,10,11,19 and X, reached genome-wide levels of significance (p=2.7×10−8 to p=8.7×10−29). The SNP rs2735839 on chromosome 19 lies between two kallikreins, PSA (KLK3) and hK2 (KLK2). It was associated with a per allele OR for PrCa of 0.83 (95%CI 0.75-0.91; ptrend in stage 2 = 0.0002; ptrend overall = 2×10−18). We also showed that rs2735839 was strongly associated with PSA level, in the direction consistent with the disease association (per allele rise in geometric mean PSA 1.12., p=6×10−8).
Ahn et al (this issue of Nature Genetics) analysed 24 tagSNPs in the kallikrein region on chromosome 19 (to include KLK1, KLK2, KLK3 and KLK15) in five studies and found that none showed a significant association with PrCa risk. They also confirm the association between several SNPs, including rs2735839, and PrCa risk. They raise the possibility that the association found with PrCa risk in our study may reflect the selection of subjects based on PSA levels rather than a causal relationship with PrCa risk.
It is clear that the selection of controls in stage 1 of our study for low PSA levels did influence the association in stage 1. This is reflected in the minor allele frequency for rs2735839, which is 21.1% in the stage 1 controls, compared with 14%-15% in the UK 1958 birth cohort and the CGEMS study (males and females). However, the controls in stage 2 were not highly selected for PSA level. The only selection was to exclude controls with PSA levels of >10, and to require a negative prostatic biopsy if the PSA was >4. The MAF in the stage 2 controls (15.2%) is similar to that in other control populations and indicates that any selection bias at this stage was minimal.
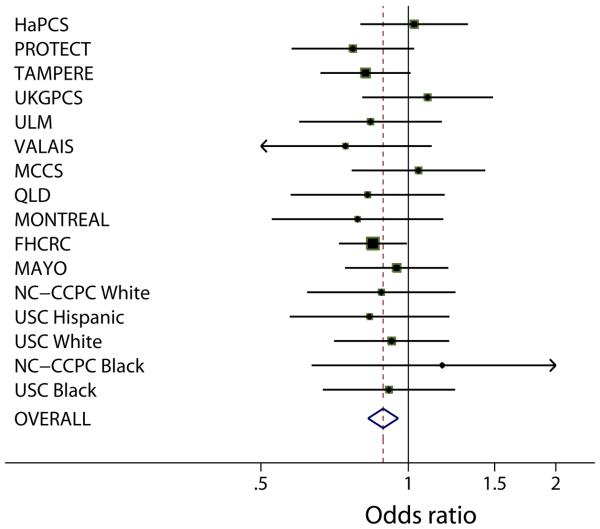
To further evaluate the evidence for this association, we have undertaken an analysis of rs2735839 (together with SNPs at the other loci identified in our GWAS) in 13 further case-control studies as part of The PRACTICAL consortium. These studies comprise 7,370 PrCa cases and 5,342 controls. The estimated per allele OR for PrCa associated with rs2735839 was 0.89 (95% CI 0.83-0.95; p=.0007), very close to our original estimate (Kote-Jarai, CEBP in press, 2008, cited with permission). There was no evidence of heterogeneity in the OR estimates among studies (see figure 1). We also note that when data from the five CGEMS studies are combined, the per allele OR is also remarkably similar (per allele OR 0.90, 0.83-0.90; P=.01), although this was not formally significant using the 4 degree freedom test given by Ahn et al (2008). If the results from our stage 2, PRACTICAL and CGEMS are combined, the overall evidence of association reaches genome-wide levels of significance (p<10−8), demonstrating that, even disregarding our stage 1 result, the association is unlikely to be due to chance. The overall effect size, while modest, is comparable to that seen for other cancer associated loci.
figure 1.
None of the control series used in PRACTICAL, nor in CGEMS, involved selection for PSA level, and for this reason and those given above, the association appears unlikely to be driven purely by control selection. Selection bias related to case ascertainment is an alternative possible explanation. We excluded from our GWAS any cases identified through PSA screening, and several of the studies included in PRACTICAL are drawn from populations where PSA screening has not been used (e.g. the study from Finland). Thus, the association is unlikely to be due to PSA screening for asymptomatic disease. PSA testing is, however, also used in the process of diagnosis of symptomatic disease. This raises the possibility of a more subtle bias, in that some cases may have raised PSA related to the genotype but not related to their disease. Whether or not this potential bias is significant could be resolved using genotyping in studies based on biopsy of whole populations not driven by the PSA level (e.g. the Prostate Cancer Prevention Trial; Thompson et al., 2007), or studies where mortality is the endpoint.
Conversely, there are plausible biological grounds for believing that the association with KLK polymorphisms may be causal. For example, polymorphisms in the promoter of KLK3 are associated with alterations in androgen receptor binding (e.g. Lai et al., 2007). Moreover, it is known that PSA level is a long-term predictor of prostate cancer risk (Lilja et al, 2007), and it is plausible that determinants of PSA level, including genetic determinants, may influence prostate cancer risk.
Acknowledgements
This work was supported by Cancer Research UK Grant C5047/A3354. DFE is a Principal Research Fellow of Cancer Research UK. John Hopper is an Australia Fellow of the NHMRC. We would also like to thank the following for funding support: The Institute of Cancer Research and The Everyman Campaign, The Prostate Cancer Research Foundation, Prostate Research Campaign UK, The National Cancer Research Network UK, The National Cancer Research Institute (NCRI) UK, grants from the National Health and Medical Research Council, Australia (209057, 251533, 450104), VicHealth, The Cancer Council Victoria, The Whitten Foundation, and Tattersall's. The ProtecT study is ongoing and is funded by the Health Technology Assessment Programme (projects 96/20/06, 96/20/99). The ProtecT trial and its linked ProMPT and CAP (Comparison Arm for ProtecT) studies are supported by Department of Health, England; Cancer Research UK grant number C522/A8649, Medical Research Council of England grant number G0500966, ID 75466 and The NCRI, UK. The epidemiological data for ProtecT were generated though funding from the Southwest National Health Service Research and Development. The views and opinions expressed therein are those of the authors and do not necessarily reflect those of the Department of Health of England.
The PRACTICAL Consortium
UK Genetic Prostate Cancer Study.
The Institute of Cancer Research & The Royal Marsden NHS Foundation Trust, Sutton UK.
Rosalind A. Eeles
Michelle Guy
Zsofia Kote-Jarai
Steve Edwards
Audrey Ardern-Jones
Rosemary Wilkinson
Amanda Hall
Rosemary Wilkinson
Lynne O'Brien
Daniel Leongamornlert
Malgorzata Tymrakiewitz
Sameer Jhavar
David P. Dearnaley
Alan Horwich
Robert A. Huddart
Vincent S. Khoo
Christopher C. Parker
Christopher J. Woodhouse
Alan Thompson
Tim Christmas
Chris Ogden
Cyril Fisher
Charles Jamieson
Colin S. Cooper
The UK Genetic Prostate Cancer Study Collaborators
The British Association of Urological Surgeons' Section of Oncology
Cancer Research UK Genetic Epidemiology Group, Cambridge, UK:
Douglas Easton
Ali Amin Al Olama
Jonathan Morrison
The ProtecT Study, UK:
David Neal
Jenny Donovan
Freddie Hamdy
Angela Cox
Sarah Lewis
Paul M. Brown
Gemma Marsden
The UK ProtecT Study Collaborators
University of Nottingham, UK:
Kenneth Muir
Artitaya Lophatananon
Chulabhorn Cancer Research Centre, Thailand:
Jo-fen Liu
The Melbourne Studies, Australia:
Graham Giles
John Hopper
Gianluca Severi
Melissa Southey
Dallas English
John Pedersen
Fred Hutchinson Cancer Research Center, Seattle, Washington, USA:
Janet L. Stanford
Claudia A. Salinas
Joseph S. Koopmeiners
National Human Genome Research Institute, National Institutes of Health, Bethesda MD, USA:
Elaine A. Ostrander
Danielle M. Karyadi
Bo Johanneson
University of Southern California Keck School of Medicine, Los Angeles CA, USA:
Sue A. Ingles
Mariana C. Stern
Roman Corral
Northern California Cancer Center, Fremont, California, USA and Stanford University School of Medicine, Stanford, California, USA:
Esther M. John
Amit D. Joshi
The Montreal Study:
William D. Foulkes
Nancy Hamel
Kimberley Kotar
Ulm, Germany:
Walter Vogel
Christiane Maier
Rainer Kuefer
Manuel Luedeke
Harald Surowy
Bärbel Weber
Kathleen Herkommer
Thomas Paiss
Tampere, Finland:
Johanna Schleutker
Tiina Wahlfors
Henna Mattila
Sanna Siltanen
Sanna Pakkanen
Jarkko Isotalo
Teuvo L. Tammela
Mika Matikainen
The Hannover Prostate Cancer Study:
Thilo Dörk
Peter Schürmann
Andreas Meyer
Stefan Machtens
Jörn Hagemanns
Peter Hillemanns
Michael Bremer
Johann H. Karstens
Prostate Cancer Study in Valais, Switzerland:
Pierre O. Chappuis
Pierre Hutter
Cédric Biedermann
Hans-Ulrich Peter
Nicolas Defabiani
Sabine Bieri
Christophe Girardet
Isabelle Konzelmann
Michéle Stalder
Marie-Mathilde Meier
Queensland, Australia:
Mary-Anne Kedda
Kimberly Hinze
Amanda Spurdle
Judith Clements
Beth Newman
Suzanne Steginga
Tracy O'Mara
John Yaxley
David Nicol
Megan Ferguson
David Nicol
R.A. (Frank) Gardiner
Joanne Aitken
Mayo Clinic:
Daniel Schaid
Stephen Thibodeau
Liang Wang
Julie Cunningham
Shannon K. McDonnell
References
- Eeles RA, et al. Nature Genetics. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- Kote-Jarai Z, et al. Multiple novel prostate cancer predisposition loci confirmed by an international study: The PRACTICAL Consortium. Cancer Epidemiology Biomarkers and Prevention. 2008 doi: 10.1158/1055-9965.EPI-08-0317. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J, et al. Carcinogenesis. 2007;28(5):1032–9. doi: 10.1093/carcin/bgl236. [DOI] [PubMed] [Google Scholar]
- Lilja H, et al. J Clin Oncol. 2007;25(4):431–6. doi: 10.1200/JCO.2006.06.9351. [DOI] [PubMed] [Google Scholar]
- Thompson IM, et al. J Clin Oncol. 2007;25(21):3076–81. doi: 10.1200/JCO.2006.07.6836. [DOI] [PubMed] [Google Scholar]