Abstract
The liver is affected by many types of diseases, including metabolic disorders and acute liver failure. Orthotopic liver transplantation (OLT) is currently the only effective treatment for life-threatening liver diseases but transplantation of allogeneic hepatocytes has now become an alternative as it is less invasive than OLT and can be performed repeatedly. However, this approach is hampered by the shortage of organ donors, and the problems related to the isolation of high quality adult hepatocytes, their cryopreservation and their absence of proliferation in culture. Liver is also a key organ to assess the pharmacokinetics and toxicology of xenobiotics and for drug discovery, but appropriate cell culture systems are lacking. All these problems have highlighted the need to explore other sources of cells such as stem cells that could be isolated, expanded to yield sufficiently large populations and then induced to differentiate into functional hepatocytes. The presence of a niche of “facultative” progenitor and stem cells in the normal liver has recently been confirmed but they display no telomerase activity. The recent discovery that human induced pluripotent stem cells can be generated from somatic cells has renewed hopes for regenerative medicine and in vitro disease modelling, as these cells are easily accessible. We review here the present progresses, limits and challenges for the generation of functional hepatocytes from human pluripotent stem cells in view of their potential use in regenerative medicine and drug discovery.
Keywords: Induced pluripotent stem cells, liver diseases, hepatocyte differentiation, regenerative medicine, disease modelling.
INTRODUCTION
Regenerative medicine is one of the most exciting and rapidly advancing areas of modern biology, as it focuses on innovative approaches to repairing and replacing cells, tissues and organs. The importance of this relatively new frontier of research is reflected in the unprecedented needs of patients from around the world: there is currently about one death every 30 seconds due to organ failure, and complications and rejection remain significant problems. The cost of care for individuals that might benefit from treatment with engineered tissues or organs has been estimated at $600 billion per year in Europe.
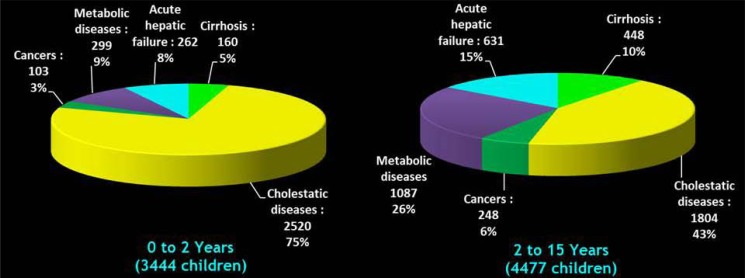
The liver is affected by many types of diseases, including inherited metabolic disorders (IMD) and acute liver failure. Orthotopic liver transplantation (OLT) is currently the only effective treatment for life-threatening liver diseases. In Europe, inherited metabolic diseases account for 26% of the indications for OLT (Fig. 1). However, the number of patients dying while on waiting lists for liver transplantation has increased in recent years due to the severe shortage of organs (11%), with this situation affecting up to 80% of the patients with acute liver failure on transplantation waiting lists (European Liver Transplant Registry). There are also
Fig. (1).
Primary indications of liver transplant in paediatric patients from European Liver Transplant Registry (1968 - 2010).
substantial risks of mortality and morbidity due to post-surgical complications (cell rejection, vascular and biliary obstruction), and immunosuppression (infections, cancer). Long-term complications include hypertension, hyperlipidemia, diabetes and obesity. In Europe, the survival rate for adult patients is 62% after 10 years, indicating that more than 30,000 transplant patients die within this period (European Liver Transplant Registry).
Cell therapies targeting the liver, with allogeneic hepatocytes for example, provide an alternative to orthotopic liver transplantation for the treatment of life-threatening metabolic disorders, with the aim of replacing a single deficient enzyme or its product. The procedure is less invasive than OLT and can be performed repeatedly.
Worldwide, the treatment of more than 30 patients in this way has been reported, mostly children with urea cycle defects, but also patients with type 1 Crigler-Najjar syndrome, FVII deficiency, type I glycogen storage disease, infantile Refsum’s disease and progressive type 2 familial intrahepatic cholestasis. In children, up to 108/kg of body weight hepatocytes (fresh or cryopreserved), matched for ABO blood group, have been transplanted, with the aim of replacing 5% of the liver cell mass [1, 2].
The safety of the procedure is well established and the results obtained are encouraging, with transient improvement in some disease phenotypes. However, these approaches are subject to several major limitations. In addition to the problems arising from the shortage of organ donors, current techniques for the isolation of adult hepatocytes yield highly variable numbers of viable, high-quality cells [3] and there is no standardized procedure for assessing the quality of cell preparations. Hepatocytes are currently isolated from unused donor livers and for donor livers that are not suitable for whole-organ transplantation, which may be steatotic [4, 5]. Moreover, hepatocytes can divide only once or twice in vitro, in the presence of Hepatocyte Growth Factor, with no further expansion possible. These cells are also difficult to cryopreserve and are highly susceptible to freeze-thaw damage [6].
Allogeneic cell transplantation is also hampered by the transient functionality of transplanted cells, partly due to immunosuppressive regimens and to a cell-mediated immune response, although other nonspecific mechanisms, such as apoptosis [7] may also contribute to cell loss.
The autologous transplantation of genetically corrected cells could be envisaged as an alternative overcoming these two limitations. However, this approach requires a lobectomy corresponding to the removal of about 20% of the liver for hepatocyte isolation, a procedure not without risk in patients with certain metabolic diseases, such as Familial Hypercholesterolemia.
Liver is a key organ in drug testing, in which it is used to assess the pharmacokinetics and toxicology of xenobiotics, but the results obtained in animal models are often misleading, due to differences in the levels and substrate specificity of liver enzymes between animals and humans. Consequently, the hepatic clearance and chemical profiles obtained for metabolites in animal models do not correctly represent what is observed in humans. Indeed, unexpected toxicity and pharmacokinetic problems account for 40 to 50 % of all failures in clinical drug development. Human cell systems, including human hepatocyte cultures, immortalized cell lines and liver microsomes, could potentially overcome these limitations, but none of the available cell systems has yet proven suitable. The expression of key liver enzymes, such as CYP450, declines rapidly after hepatocyte isolation, and cell lines, such as like HEP-G2 cells, most of which originate from tumors, have insufficiently high levels of expression for transporters and key liver enzymes (Cytochromes P450, conjugating enzymes) and do not have the correct morphology and polarization for vectorial drug transport from the plasma to the bile. A new hepatoma cell line has recently proved highly valuable as a model for studies of drug metabolism in humans. However, some Cytochromes P450 activities remain low [8].
All these limitations to direct therapeutic applications and drug discovery have highlighted the need to explore other sources of cells. Stem cells that could be isolated, expanded to yield sufficiently large clonal populations and then induced to differentiate into fully functional hepatocytes in vitro would be an ideal source of cells.
Source of Hepatocytes
Endogenous Stem Cells
Mesenchymal stem cells are cells of extra-hepatic origin and have potential therapeutic applications. However, recent reports have suggested that their role in injured livers is essentially to provide trophic support, thereby keeping endogenous hepatocytes alive and stimulating their proliferation. In culture, these cells enter a phase of replicative senescence after a limited number of population doublings [9-11].
The adult liver has a remarkable capacity for regeneration, which is achieved through proliferation of the mature cell populations making up the intact organ. However, if the regenerative capacity of mature cells is impaired by liver-damaging agents, hepatic progenitor cells are activated and expand in the liver parenchyma. Following their amplification during transit, these progenitor cells may generate new hepatocytes and biliary cells to restore liver homeostasis [12]. Hepatic progenitors constitute a heterogeneous population expressing markers of both hepatocytes and bile duct cells. In the human liver, these cells are activated by various liver diseases, including chronic viral hepatitis, and after severe hepatocellular necrosis [13], as demonstrated by morphological studies. The presence of a niche of progenitor and stem cells in the normal liver has recently been confirmed. These cells have been estimated to account for between 0.01% and 1% of liver cells in neonates and, unlike foetal liver progenitor cells, they display no telomerase activity [14-16]. The intrahepatic stem cell niches are the canals of Hering in postnatal livers and are derived from ductal plates in foetal livers [17, 18]. We have also isolated foetal liver progenitor cells and demonstrated their ability to engraft in vivo [19], but limited access to these cells and their small numbers are major drawbacks.
Pluripotent Stem Cells
Embryonic Stem Cells (ESCs)
Human embryonic stem cells (hESCs), which are derived from the inner cell masses of human blastocysts (5 days post-fertilization), were first isolated less than 15 years ago by Thomson’s group at the University of Wisconsin [20]. The embryonic origin of ESCs confers two important properties on these cells: they can grow indefinitely in vitro while maintaining pluripotency or a capacity to differentiate into any type of cell. These cells remain the gold standard for all studies in the pluripotent stem cell field, although their use has been restricted by ethical concerns, and two clinical trials have been launched by Advanced Cell Technology. These trials aim to assess the safety of hESC-derived retinal cells for treating patients with an eye disease called Stargardt's Macular Dystrophy and patients with age-related macular degeneration. Preliminary results four months after the trials suggest that the method is safe [21], [see also the article by Cramer et al. in this special issue of Current Gene Therapy].
Induced Pluripotent Stem Cells (hiPSCs)
The recent discovery that human induced pluripotent stem cells (iPSC) can be generated by the forced expression of a few transcription factors in somatic cells has renewed hopes for regenerative medicine and in vitro disease modelling, as these cells are easily accessible [22, 23], [see also the article by Bayart et al. in this special issue of Current Gene Therapy]. Human iPSCs closely resemble hESCs in terms of their self-renewal capacity and ability to differentiate into cell types derived from the three primary germ layers: the ectoderm, mesoderm and endoderm. They therefore provide a unique opportunity to study patient-specific diseases, once the conditions required to induce their differentiation into the desired type of cell have been defined [24, 25]. This is particularly important for patients with liver diseases, who cannot undergo surgical biopsy for the isolation of hepatocytes for transplantation. iPSC technology should make it possible to identify the impact of mutations on the differentiation/proliferation/function of the cells. Patients’ iPSCs should also be good tools for the screening of candidate pharmacological molecules.
Two different sets of applications can be envisaged, depending on the origin of the iPSCs:
iPSC-Derived Hepatocytes from Normal Individuals
These cells can be used in the establishment of cell banks for applications in regenerative medicine. The possibility of generating iPSC-derived hepatocytes from selected adults would facilitate the construction of libraries of cell lines with known genotypes, providing patients with a close HLA/MHC match, thereby minimizing the need for immunosuppression before cell engraftment.
These cells would also be useful for modelling susceptibility to infectious diseases, providing an opportunity to elucidate the genetic basis of the mechanisms underlying cell susceptibility or resistance to viruses. In particular, iPSC-derived hepatocytes are an appropriate target for studying the interactions between the host and viruses with hepatic tropism. The infection of iPSC-derived hepatocyte-like cells with hepatitis C virus has already been reported, and permissiveness to infection has been shown to be correlated with induction of the liver-specific microRNA-122 and the modulation of cellular factors affecting HCV replication [26-28]. These cells support the entire life cycle of the HCV. The LDL receptor, genetic mutations of which have been shown to cause type IIA familial hypercholesterolemia, is one of the identified cofactors for HCV entry, and there is growing evidence to suggest that associations between HCV and LDL/VLDL are important for viral entry and release from cells [29].
In acute situations, such as stroke or acute liver injury, the need for rapid treatment would preclude the use of autologous cells, even “off-the-shelf” allogeneic iPSCs due to thawing/culture delay.
However, the differentiation of normal iPSCs into hepatocytes could be used to create bio-artificial livers for the temporary treatment of acute liver failure. Such a replacement of liver function has been accomplished by constructing extracorporeal liver assist devices from either primary porcine hepatocytes or a human hepatoma cell line. However, the use of porcine cells is restricted because the risk of zoonotic infection posed by porcine endogenous retroviruses is unknown [30]. Human hepatoma cell lines have few differentiated functions and there is thus concern that such cells might escape into the patient's bloodstream and cause tumour development. New hepatoma cell lines, such as HepaRG, have been shown to be effective in an animal model of acute liver failure, but the clinical implementation of a bio-artificial liver with a human proliferative biocomponent is still several years away [31].
iPSC-Derived Hepatocytes from Diseased Individuals
Patient-specific gene/cell therapy is an ideal way to prevent cell rejection and the need for immunosuppression in situations in which long-term hepatocyte engraftment is required, as in the genetic correction of inherited liver disorders. iPSCs should not entail the same risk of immune system rejection when used for treatment, because they are autologous cells unique to the patient concerned. It is thus assumed that these cells would yield the best possible functional outcome of transplantation: cells that function in their natural environment, without eliciting chronic immune or inflammatory reactions and without the problems associated with the use of immunosuppressive drugs [32].
The first demonstration of iPSC-based cell therapy was published by Hanna and coworkers in 2007. IPSCs were derived from a mouse model of human sickle cell anaemia. The genetic defect was corrected in the iPSCs, which were allowed to differentiate into hematopoietic cells and were then transplanted back into syngeneic mice [33]. A significant improvement was observed in the symptoms of the treated mice.
The liver is the principal organ responsible for detoxification. Thus, iPSC-derived hepatocytes from normal individuals should find a wide range of applications in drug screening. The use of iPSC models should also increase our understanding of the risk to the foetus of drugs taken by pregnant women. Furthermore, by obtaining iPSC-derived hepatocytes with various genotypes, it should be possible to identify new therapeutic compounds, as cells from patients with inherited diseases could be used for the development of high-throughput drug screens.
Pluripotent Cell Differentiation Toward Hepatocytes
In recent years, a number of laboratories have reported the hepatic specification of endoderm cells and their further differentiation into foetal hepatocyte-like cells [34-42] in diverse culture systems. However, all these approaches were based on culture media containing serum, complex matrices such as Matrigel and/or the use of mouse embryonic fibroblasts as feeder cells. All of these elements are the source of unknown factors that could obscure the molecular mechanisms controlling human liver development or render the resulting tissues incompatible with future clinical applications. Indeed, the exposure of human cells to products of animal origin may increase the risk of transmission of pathogens that do not normally infect humans, such as transmissible spongiform encephalopathies, and the immune rejection of transplanted cells. This was highlighted by Martin MJ et al. [43], who demonstrated that hESCs cultured with animal or serum products retained non-human sialic acid, which was immunogenic when these cells were transplanted into humans. Moreover, the very small number of in vivo studies of ESC-derived hepatocytes carried out was either focused on very short-term effects or reported teratomas or adenocarcinomas. Identical protocols were applied to iPSCs and the resulting differentiated cells did not display the functions of fully mature hepatocytes.
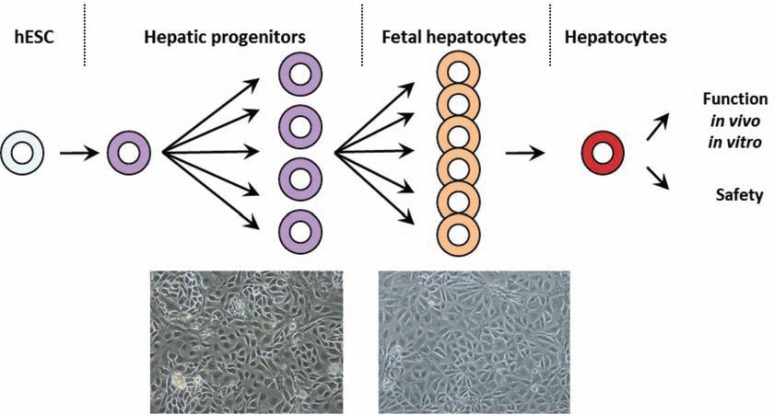
We therefore focused on the improvement of differentiation protocols on the basis of the idea that modelling the early steps of embryonic development in vitro might be the best way to produce cell types with native properties. This approach requires the identification and sequential induction of key developmental factors, potentially resembling the normal sequence of human liver development. Little is known about the molecular mechanisms regulating this process, but human liver development can be divided into four major, sequential steps: 1) Anterior endoderm differentiation, 2) Ventral foregut formation (definitive endoderm), 3) Hepatic bud specification (hepatoblasts), and 4) The differentiation of hepatic progenitors into mature foetal hepatocytes and mature hepatocytes (Fig. 2).
Fig. (2).
Sequential stages of hepatocyte differentiation for therapeutic applications.
Toward Endoderm Cells
The Activin/Nodal signalling pathway has been shown to induce endoderm differentiation during early development in fish, amphibian and mammals [44]. However, our group and others have shown that the same growth factors maintain the pluripotent status of hESCs and hiPSCs [45]. The mechanisms by which the same signalling pathway can fulfil these contradictory functions have been a major focus for the stem cells field during the past 10 years. First, the various signalling pathways controlling the early cell fate decisions of pluripotent stem cells, such as those involving Activin, FGF and BMP, were analysed. This analysis resulted in the development and validation of chemically defined culture conditions for achieving the specification of human embryonic stem cells into mesendoderm (but also into neuroectoderm and extra-embryonic tissues) [45]. Then genome wide analyses have revealed that Activin/Nodal signalling controlled divergent transcriptional networks in hESCs and endoderm cells and that the effect of Activin/Nodal signalling on pluripotency and differentiation was dictated by tissue-specific partners of Smad2/3, such as NANOG and Eomesodermin [46]. The first comprehensive molecular model connecting the transition from pluripotency to endoderm specification during mammalian development, through a hierarchy of transcription factors regulating endoderm specification, was recently described by Teo et al. [47]. We found that the pluripotency factors NANOG, OCT4, and SOX2 played an essential role in this network, by actively directing differentiation. These factors control the production of Eomesodermin (EOMES), which marks the onset of endoderm specification. In turn, EOMES interacts with SMAD2/3, to initiate the transcriptional network governing endoderm formation [47]. Finally, recent studies have shown that PI3 Kinase block the inductive effect of Activin/Nodal on endoderm differentiation by controlling the activity of WNT signalling [48]. Hence, inhibition of this pathway significantly improves endoderm differentiation of hPSCs [49].
Toward Hepatoblasts
Foetal liver development begins when the ventral foregut endoderm buds off and gives rise to the early hepatic epithelium in response to signals from both the cardiac mesoderm and the septum transversum [50, 51]. In humans, this bud forms at week 4 of gestation. During this process, hepatic progenitors arise from the maturation of an as yet unknown, multipotent stem cell. Hepatic progenitors are bipotent progenitor cells that can differentiate into either foetal hepatocytes or, if in contact with the portal mesenchyme, biliary cells. They express the markers of both lineages.
Progenitor cells from rodent foetal liver have been purified by flow cytometry-based positive selection [52]. Clonal populations of murine self-renewing stem cells can differentiate into cells of various lineages, such as those found in the liver, pancreas, intestine and stomach. After transplantation, they repopulate hepatocyte populations and bile duct, pancreas (pancreatic ductal cells) and intestinal-epithelium tissues. However, these stem cells are rare, even in mouse foetal liver, and their abundance appears to decline further with aging. Such foetal progenitors have been isolated from human foetal liver at late (18 to 22 weeks) [53, 54] and early [19] stages of development. We have also shown that progenitors/hepatocytes derived from human foetal livers have specific molecular properties different from those of adult hepatocytes, enabling them to engraft and migrate within the recipient liver parenchyma more efficiently than adult hepatocytes after transplantation [19, 55, 56].
Following the generation of endoderm cells from pluripotent cells, we generated hepatoblasts, which displayed phenotypic markers of foetal cells, such as A1AT, AFP, CK19.
Upon further differentiation, foetal hepatocytes and more mature cells were generated from both ESCs and iPSCs, and these cells displayed hepatic functions, such as albumin production and secretion, urea excretion, CYP3A7 then CYP3A4 expression [57].
Toward Fully Differentiated Hepatocytes
Despite recent advances, the differentiation of human ESCs and iPSCs is highly variable, cell line-dependent and generates immature cells different from those found in mature organs in vivo. This is also true for other cell types (e.g. for neurons [58]).
It is essential to maintain stable differentiated hepatic cell function in culture, particularly if hiPSC-derived hepatocytes are to be used for disease modelling, the assessment of drug toxicity or even as a bio-artificial liver.
Differences in differentiation capacity have been reported between ESC and iPSC lines, necessitating the testing of defined protocols on different lines. Yamanaka and co-workers recently analysed differences in hepatic differentiation between 28 hiPSC lines [59]. The cell lines originated from various somatic cells (peripheral blood cells, dermal fibroblasts) and were generated with retroviruses, Sendai virus or episomal plasmids. Differences in hepatic differentiation were largely attributed to differences between donors rather than to differences in the type of cell originally used, although iPSC clones derived from peripheral blood lymphocytes (PBL) consistently differentiated more efficiently than those derived from dermal fibroblasts.
In the liver, heterotypic cell interactions between parenchymal cells and their non-parenchymal neighbours result in the regulation of differentiation and tissue proliferation in a three-dimensional (3D) microenvironment.
In vitro, hepatocyte viability and liver-specific function are stabilized for several weeks by coculture with other cell types, by culture with extracellular matrix components or by using 3D bioreactor [60]. In other conditions, the cells rapidly dedifferentiate and display poor drug inducibility. ECM and coculture with non-parenchymal cells have been shown to modulate ESC differentiation [38, 61].
Paracrine signals produced by the different subpopulations of liver-derived mesenchymal cells, purified with immunoselection technologies, have been identified and shown to induce the differentiation of human hepatic stem cells into fully mature and functional parenchymal cells [62]. Thus, one goal of in vitro cell systems is to reconstitute a cell microenvironment suitable for cell differentiation, and 3D systems are being developed for this purpose. A polyurethane matrix has recently been shown to promote drug-inducible hepatocyte metabolism and function [63].
The most important functions required from a mature hepatocyte are the polarized expression of gap and adherent junction proteins, such as Connexin 32, Zona occludens protein 1 and Integrins, all of which are involved in cell engraftment, and of membrane transporters, such as multidrug resistance proteins and bile acid transporters. For effective detoxification, the cells must produce large amounts of drug-metabolizing enzymes, cytochromes P450, such as CYP 3A4, UDP-glucuronosyltransferase and glutathione S-transferases. The cytochromes P450 of the liver are particularly important because they are involved in the maintenance of lipid homeostasis (cholesterol, vitamin D, oxysterol and bile acid metabolism) and in the detoxification of endogenous compounds (e.g. bile acids) and xenochemicals (drugs).
In Vivo Assay
Specific human cells differentiated in vitro must be transplanted into rodent models to demonstrate their functionality. However, the following problems must be taken into account: the rejection of human cells, and the low level of engraftment of transplanted cells, which do not proliferate under normal conditions in rodent livers [64]. IPSC-derived hepatocytes can be transplanted into immunodeficient mice or into immunosuppressed mice as disease models. Alternatively, as the repopulation of the liver by donor hepatocytes has been demonstrated in animal models in which transplanted hepatocytes display a selective growth advantage over endogenous hepatocytes, such models could be used. For example, in some models, the survival and/or proliferation of native hepatocytes is impaired by a genetic or inherited inability to regenerate, as in fumarylacetoacetate hydrolase (FAH)-deficient mice and urokinase (alb-uPA) transgenic mice, [65, 66]. These two types of mouse model have been crossed with immunodeficient mice with a different genetic background.
To date, very few studies have assessed the functionality of ESC/iPSC-derived hepatocyte-like cells in vivo. HESC-derived hepatocyte-like cells were recently injected into the spleens of acutely injured NOD/SCID(IL-2Rγ)-null mice and the persistence and function of these cells were demonstrated for up to three months after transplantation. However, all the recipient mice developed large spleen and liver tumours, suggesting that there may have been undifferentiated cells in the cell population infused [67]. We transplanted ESC-derived GFP-hepatoblasts and iPSC-derived hepatocyte-like cells into uPAxRag2gammac-/- mice. Three months later, some clusters of GFP-expressing cells and a small percentage of iPSC-derived hepatocytes were found to have engrafted within the host parenchyma [57, 68]. Nevertheless, hepatocytes generated from pluripotent or multipotent stem cells currently repopulate transplanted livers less efficiently than human adult hepatocytes (up to 80%) [69], but they can be used to develop in vivo models of human cell infection in mice with chimeric livers [70].
Modelling Human Diseases
Disease modelling using iPSCs was achieved for a variety of genetic diseases, [71], [see also the article by Sille et al. in this special issue of Current Gene Therapy]. The first model was achieved for spinal muscular atrophy, a leading inherited genetic disease that often leads to death in infancy. These iPSCs were differentiated into motor neurons, which displayed selective deficits with respect to motor neurons generated from an individual not affected by spinal muscular atrophy [72, 73].
About 50 reprogramming-based disease models have since been published and have been the subject of recent review articles [74-76]. Most of the targeted diseases are neurodegenerative disorders, because of the difficulty obtaining neurons from patients and the lack of appropriate disease models. They include Huntington’s disease, Alzheimer’s disease and Parkinson’s disease [77, 78]. Other diseases have been targeted, including type 1 diabetes, which results from the autoimmune destruction of pancreatic beta cells [24], muscular dystrophy and liver diseases. The recent successful rescue of diseased iPSCs by drug screening highlights the potential of this approach to identify candidate drugs for potential treatment. Motor neurons have been generated from the iPSCs of patients with familial amyotrophic lateral sclerosis. ALS is a late-onset, fatal disorder in which the motor neurons degenerate, due to mutations of the gene encoding Tar DNA-binding protein-43. In tests of a number of chemical compounds, a histone acetyltransferase inhibitor, anacardic acid, was found to rescue the abnormal ALS motor neuron phenotype. In another report, neural crest precursors derived from iPSCs were generated from individuals with familial dysautonomia (FD), a fatal genetic disorder affecting neural crest lineages. Of the 6,912 small molecules tested, eight were found to rescue the expression of IKBKAP, the gene responsible for FD. One of the small molecules also rescued the disease-specific loss of autonomic neuronal marker expression [79].
Modelling Inherited Liver Metabolic Diseases
Inherited liver disorders can be classified into two categories:
Genetic defects affecting a specific hepatic function with extra hepatic symptoms, such as Crigler Najjar (CN), Familial Hypercholesterolemia (FH) clotting factor deficiencies. Hepatocytes are normal and can proliferate.
Diseases in which hepatocytes are injured due to the accumulation of a toxic product, such as alpha1 anti-trypsin (A1AT), or copper in Wilson’s disease, and cannot proliferate.
We have focused on inherited metabolic disorders caused by genetic mutations in key proteins. We first performed a study in which iPSCs were generated from patients with several disorders, including A1AT and FH, and differentiated into hepatocyte-like cells. These cells were shown to display the disease phenotypes [68]. These two diseases, together with haemophilia B (HB), have been chosen as paradigms of hepatic deficiency of a cell-surface receptor (FH), and secreted (HB and A1AT) proteins. The loss of hepatocyte protein activity results in various downstream manifestations specific to the associated metabolic pathway: extracellular lipid uptake mediated by the deficient receptor, impaired secretion and toxic accumulation of the mutated protein and impaired enzyme-dependent coagulation cascade.
1- Type IIa Familial Hypercholesterolemia (FH)
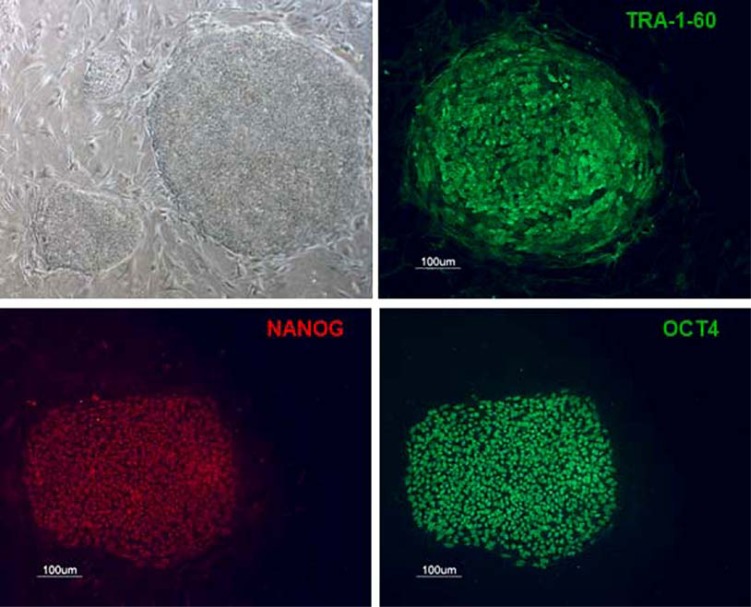
Cardiovascular diseases constitute a major health problem in Europe. Familial hypercholesterolemia is one such disease, for which there is no curative treatment. FH is a genetic disorder caused by mutations of the gene encoding the low-density lipoprotein receptor (LDLR) that results in an abnormal level of circulating cholesterol conjugated to LDL particles (LDLc). In normal individuals, LDLc can be taken up and degraded only by hepatocytes, via the LDLR. Heterozygous patients (prevalence 1:500) are treated with a combination of drugs, including statins, but the efficacy of treatment is variable and these patients frequently present premature cardiovascular disease from the age of 40 years [80, 81]. Homozygous patients (prevalence 1:106) have severe cardiovascular disease from childhood. LDL apheresis is the most effective treatment currently available for lowering serum cholesterol levels in these patients, but is highly aggressive. Nevertheless, even treated patients die from cardiovascular disease at about 50 years of age [82, 83]. Given the difficulty in obtaining liver biopsy samples from patients with FH for the isolation of primary hepatocytes, there is currently no in vitro cellular model of this disease. Five patients with homozygous familial hypercholesterolemia received autologous hepatocytes transduced ex vivo with a retroviral vector carrying the human LDL receptor gene [84], in the first ex vivo cell/gene therapy trial, reported more than 15 years ago. This treatment transiently decreased LDL levels in some of the patients and the trial showed this approach to be safe. However, this approach was not subsequently repeated due to the complexity of the procedure which involved the removal of the left lobe of the patient for perfusion, the culture and retroviral transduction of the hepatocytes and their infusion into the patient. Furthermore, future developments of this approach were limited by the number of hepatocytes isolated and the rate of transduction of non-dividing cells with the retroviral vector. To bypass these limitations and to develop a relevant in vitro model for drug screening, we have generated new iPSC lines from patients homozygous for the mutation causing FH. These resulting cells displayed the morphological features and markers of pluripotent stem cells (Fig. 3) and once differentiated into hepatocytes internalized the LDLR ligand poorly, suggesting that they reproduce the diseased phenotype.
Fig. (3).
Induced pluripotent stem cells derived from fibroblasts of a patient homozygous for Familial Hypercholesterolemia and expression of pluripotency markers.
2- Haemophilia B (HB)
HB is caused by mutations of the gene encoding the clotting factor IX (FIX) located on the X chromosome. HB is a haemorrhagic disease (incidence: 1:30,000 males). Hepatocytes secrete large amounts of FIX, in an inactive form, into the bloodstream. The severity of the disease is inversely correlated with residual FIX activity, and as little as 5% more FIX activity is sufficient to transform a severe life-threatening disease into mild haemophilia with a much better quality of life. Substitutive treatment with recombinant FIX or with FIX purified from donor plasma is available, but costly, due to the limited supply. However, this treatment is not devoid of side effects, including the production of neutralising anti-FIX antibodies. Several trials of human gene therapy, as an alternative treatment, have been conducted although the stable production of therapeutic coagulation protein has not yet been achieved. This nevertheless remains an area of intense investigation [85, 86]. A successful gene therapy trial has recently been reported in which a serotype-8-pseudotyped, self-complementary adenovirus-associated virus (AAV) was used [87]. The infusion of a single dose of human factor IX vector resulted in the AAV-mediated production of FIX in amounts corresponding to 2 to 11% normal levels. This was sufficient to improve the bleeding phenotype in the six participants, with only a few side effects. So far, this disease has not been modelled using hiPSCs and thus remains a major focus for the development of new drug screening platform. Accordingly, it is has been demonstrated that ESC- and iPSC-derived hepatocytes express FIX [88, 89].
3- Alpha-1-Anti-Trypsin
α1 antitrypsin (A1AT) deficiency (A1ATD) is one of the most common IMDs (1:2000 North Caucasian) which is associated with severe clinical manifestations. The A1AT protein, encoded by the SERPINA1 gene, is normally secreted from hepatocytes as a glycoprotein of 52 kDa serum. A1AT acts as an inhibitor of neutrophil elastase in the lungs. Disease principally arises from homozygous inheritance of the Z allele, which consists of point mutation in the last exon (Glu342Lys, Z) of the SERPINA1 gene. The ZZ genotype results in a well-characterized defect in A1AT folding, and predilection for formation of protein polymers by the loop-sheet insertion mechanism [90]. Subsequent retention of Z A1AT within hepatocytes causes protein overload that is manifest as periodic acid Schiff (PAS)-positive inclusions associated with neonatal hepatitis, cirrhosis, and hepatocellular carcinoma. Moreover, as plasma levels of A1AT in ZZ homozygotes fall to only 10–15 % of normal levels, the lungs are left exposed to enzymatic damage by neutrophil elastase and thus predisposed to early onset of panlobular emphysema [91]. Curative treatment for this disorder is currently lacking, leaving researchers pursuing different therapeutic strategies including gene therapy. To date two Phase I clinical trials have been undertaken to investigate the efficacy of gene augmentation from ectopic sites such as skeletal muscle as a means to treat A1AT lung disease [92]. Such strategies are not however able to address the liver-based complications resulting from intra-hepatocytic polymer accumulation. Instead transplantation of genetically corrected hepatocytes into the livers of affected patients may facilitate a complete correction of all aspects of the disease. Such an approach already carries a precedence within clinical practice since at least two patients treated for α1-antitrypsin deficiency [93]. These treatments have unfortunately yielded limited long-term clinical success due to the limited number and poor quality of donor hepatic cells used, as well as the persistent requirement of immunosuppressant drugs to prevent rejection of allogeneic cells. More recently, our group have shown that hiPSC could be used to model α1-antitrypsin deficiency in vitro and that the resulting culture system could be applicable for large-scale drug screening.
Gene Correction
Personalised gene/cell therapy of genetic disorders using hiPSCs will avoid cell rejection and the need for immunosuppression. However, such therapeutic approach will require new technology to correct the genetic anomalies inducing the disease. Gene therapy of hepatocytes derived from hiPSCs represents a first alternative to achieve this major goal. Indeed, since the trial performed on FH patients, significant progress has been made with therapeutic vectors and, in particular, lentiviral vectors have been designed and protocols developed for the efficient transduction of human hepatocytes (>80%, versus 20-25% with retroviral vectors) [94, 95]. This work made it possible to correct hepatocytes isolated from a patient with type 1 Crigler-Najjar syndrome [96]. This strategy based on the genetic modification of autologous cells from the patient could ultimately be applied to many other metabolic diseases affecting the liver, including Crigler-Najjar syndrome, A1ATD, urea cycle disorders and other coagulation disorders. However, studies have shown that the use of integrative vectors such as retroviruses could present risks including vector immunogenicity, hepatic toxicity and induction of harmful genetic mutation [97]. Furthermore, the continuous expression of non-integrative vectors such as AAV is often difficult to achieve. Moreover, constant proliferation of hepatocytes might limit the efficiency of episomal viral systems at long term in the liver [98]. Most importantly, IMD often involve gene whose expression needs to be tightly controlled and/or regulated through complex physiological mechanisms that cannot be replicated by ubiquitously active promoters commonly found in gene therapy vectors. The spread of vector and thus gene expression is also difficult to control and despite the availability of tissue-specific promoters, the potential effects of systemic integration represent other pitfall of such therapies. While interesting, gene therapy for IMDs will require further development for personalised therapy of liver diseases.
Genome editing represents an interesting alternative and several publications have reported efficient genetic mutation correction in hiPSCs [99-101]. Homologous Recombination (HR) represents the most commonly used approach to target specific sites of the mammalian genome. However, this approach has so far proved prohibitively time and resource consuming in hESCs and in hiPSCs. It also requires use of the potentially toxic Cre recombinase enzyme and leaves behind residual “loxP” sequences [102]. Such limitations render HR inappropriate for genetic correction of hiPSC lines that would ultimately result in derivatives that were to be re-introduced back into the human body. Advent of a new technology based on Zinc Finger Nucleases (ZFNs) and transcription activator–like effector nucleases (TALENs) has been shown to greatly increase the efficiency of gene targeting in human pluripotent stem cells [103, 104]. Recently, ZFNs in combination with the PiggyBAC transposon system (Z-Pig technology) has been used to correct the point mutation in the serpin 1a responsible for A1ATD [105]. This approach allowed the correction of both abnormal alleles simultaneously and the production of fully corrected hiPSC lines in less than 4 months. Furthermore, the PiggyBAC transposon does not require the Cre/LoxP technology and thus enable gene correction without leaving any artificial sequence in the genome. Importantly, a major drawback of ZFNs could be their lack of specificity (i.e. they could target a broad number of cryptic sites in the genome) which could result in broad number of genetic abnormalities [106]. However, whole genome sequencing showed that gene correction using ZFNs did not induce additional genetic changes in hiPSCs [105]. Taken together, these data demonstrated the efficiency of the Z-pig to edit point mutation in the mammalian genome which represents an important step toward the use of hiPSCs for cell based therapy in the context of monogenic disorders.
Importance of Preclinical Studies in Large-Animal Models for Regenerative Medicine
Before autologous or allogeneic cell therapy can be applied to human patients, a thorough preclinical assessment of iPSCs in suitable large-animal models is required, to ensure that the proposed treatment with iPSC-derived cells is both safe and effective. It is now well established that non-human primates constitute the best model for such trials, because they are the animals most closely related to humans and have a similar physiology, particularly as concerns the liver [107] and the central nervous system. The long-term follow-up of transplanted cells, which is impossible in rodents, can be carried out in monkeys. The first iPSCs were derived from the rhesus macaque, as reported by Liu et al. in 2008 [108]. These pluripotent cells gave rise to the three lineages. iPSCs have also been obtained from pigtailed and cynomolgus macaques, marmosets and drills [108-110].
Blin et al. reported the isolation and characterization of a group of early mesodermal cardiovascular progenitor cells, induced by BMP2 and expressing the cell surface protein stage-specific embryonic antigen 1 (SSEA-1). BMP2-induced SSEA-1+ cells were purified from iPSCs and their differentiation into cardiomyocytes, endothelial cells and smooth muscle cells was induced by treatment with defined cytokines and signalling molecules. Purified SSEA+ progenitor cells from rhesus monkey ES cells were engrafted into non-human primate hearts, in which they differentiated into cardiac cells and reconstituted 20% of the scar tissue without forming teratomas. Primates receiving grafts of an un-purified population of cardiac-committed cells, including SSEA-1- cells, developed teratomas in the scar tissue [111].
One major hurdle highlighted by hepatocyte transplantation is the low efficiency of donor hepatocyte engraftment into the host parenchyma. Many studies on various rodent models have demonstrated that, once infused into the liver, hepatocytes remain entrapped in the portal spaces and sinusoids (up to 70%), causing portal hypertension and ischemia–reperfusion injury. Most of these cells are then cleared by the innate immune system, including Kupffer cells and granulocytes. This may account for the lack of long-term clinical improvement in patients undergoing transplantation [112].
In an effort to improve cell engraftment in a model transposable to clinical practice, we have developed an original approach to autologous hepatocyte transplantation into macaque liver. We have shown that transient partial portal embolization improves cell engraftment, resulting in the engraftment of 7% of the transplanted hepatocytes, versus only 0.5 to 1% in non embolised animals. Before transplantation, isolated hepatocytes were transduced with a lentiviral vector in which the GFP gene was placed under the control of the liver-specific promoter of the human Apolipoprotein A-II (APOA-II) gene. This led to the detection of GFP in vivo 12 weeks after transplantation, suggesting that the APOA-II promoter was functional in the long term in the liver and could be used for gene correction in diseased iPSC-derived hepatocytes [113-115].
These studies pave the way for experimentation with macaque-derived iPSCs and the transplantation of iPSC-derived hepatocytes into the autologous macaque liver.
Challenges
Models appearing to reproduce the human disease phenotype for monogenic diseases have now been developed, including some for liver disorders, but most of these models are based on only a few patients. The use of larger numbers of donors will be required to model faithfully the importance of genetic variability on individual variability.
Genome wide comparisons have revealed some divergence between hiPSCs and hESCs which remain the gold standard for clinical applications. Whole-genome profiles of DNA methylation at a single-base resolution in five human iPSC lines have been reported, together with the methylomes of ESCs, somatic cells, differentiated iPSCs and ESCs. iPSCs display significant variability of reprogramming, including that of somatic memory, and aberrant reprogramming of DNA methylation and differences in CG methylation and histone modifications. Finally, the differentiation of iPSCs into trophoblastic cells revealed that errors in the reprogramming of CG methylation are transmitted at a high frequency, providing an iPSC reprogramming signature that is maintained after differentiation. This epigenetic variation can be accounted for by environmentally induced and stochastic epigenetic changes, which accumulate over time in the epigenome of the individual [116-119]. In addition, the biological consequences of these differences remain to be understood if one wants to use hiPSC for long term engraftment in patients. Finally, it has to be borne in mind that disease-specific ESC lines from embryos obtained following preimplantation genetic diagnosis are probably more appropriate than iPSC lines for certain studies. An example is provided by fragile X syndrome (FX), in which the FMR1 gene is inappropriately silenced during development. FX-iPSCs do not express the FMR1 gene due to a failure to reactivate the mutant locus during reprogramming [120]. Thus FX-iPSCs may give rise to FMR-deficient neurons [121], but they cannot be used for studies of the mechanisms by which pathological gene silencing occurs during development.
Genetic instability remains a major issue for pluripotent stem cells. Indeed, hiPSCs as any others primary cells, accumulate somatic mutations during extensive in vitro culture [122, 123]. Importantly, recent reports also suggest that genetic anomalies observed in hiPSCs could reflect the genetic diversity of the somatic cells reprogrammed [124] and thus underlines the importance to understand the biological relevance of these mutations for therapeutic applications.
The pluripotent status of hiPSCs imposes on them an embryonic identity. While this characteristic is advantageous for basic developmental biology studies, it also represents a major drawback for the production of fully functional cells. Indeed, it implies that the method of differentiation must recapitulate in vitro the entire development to produce adult cells, which is extremely complex if not impossible with conventional cell culture systems. Hence, most of the cell types generated from hiPSCs have a foetal identity and have limited functional characteristics.
Direct Reprogramming As An Alternative
Direct reprogramming strategies — the direct conversion of one cell type to another, developmentally non permissive, lineage-specific cell type — involve the use of transcription factors or other genes or miRNAs, selected on the basis of their key roles in cell fate specification in embryonic development. Several cell types have already been successfully generated from fibroblasts, including neurons, cardiomyocytes, blood progenitors, macrophages and hepatocytes (For review [125-128]).
Huang et al. demonstrated the direct induction of functional hepatocyte-like (iHep) cells from mouse tail-tip fibroblasts by the transduction of Gata4, Hnf1α and Foxa3, and the inactivation of p19Arf. iHep cells had a typical epithelial morphology, expressed hepatic genes and acquired hepatocyte functions. In particular, transplanted iHep cells were able to repopulate the livers of fumarylacetoacetate hydrolase-deficient (Fah−/−) mice sufficiently to restore liver function and to rescue 40% of recipient mice from death [129].
Another group identified three specific combinations of two transcription factors — Hnf4α plus Foxa1, Foxa2 or Foxa3 — that converted mouse embryonic and adult fibroblasts into cells closely resembling hepatocytes in vitro. They suggest that HNF4a and Foxa3 could be defined as the minimal set of genes required for the generation of iHep, although Foxa3 cannot compensate for the lack of both Foxa1 and Foxa2 in mouse liver development. In repopulation experiments in the fah-/- model of liver failure, the survival of 40% of transplanted animals was increased [130]. However, in both studies, major differences in gene expression were observed between iHep cells and primary hepatocytes, resulting in the partial rescue of transplanted animals. It remains to be determined whether the different combinations of factors induce the generation of hepatocytes in humans.
Melton and co-workers found that the combination of a defined set of transcription factors (Ngn3, Pdx1 and MafA) was sufficient to convert pancreatic exocrine cells into functional insulin-secreting beta-like cells in vivo, with a high efficiency. This technique may have allowed the cells to develop in their niches, and to respond to pancreatic signalling [131].
This approach can be used for the rapid production of models of human "diseases in a dish", without the need for pluripotent cell production, thus avoiding the potential problems associated with the time-consuming and labour-intensive generation of hiPSC lines. This strategy may ultimately be more attractive for cell-based therapy and would presumably be less tumorigenic, provided that integration-free gene delivery methods are used. However, a number of the questions raised by Vierbuchen T [127] remain unanswered, including:
How do transcription factors downregulate the transcriptional program of the initial cell?
How do transcription factors find their binding sites in a cell type with a pattern of chromatin modification different from that normally encountered?
Why is direct reprogramming more difficult with human cells than with mouse cells?
CONCLUSION
Human iPSCs have had an immediate impact, through the generation of new human cell–based disease and pharmacological models of enormous potential benefit for the development of novel disease treatments. Significant challenges and obstacles to the use of iPSC technology in personalized therapies, including safety issues (in vivo phenotypic stability and absence of teratomas/carcinomas) remain, but it is hoped that these obstacles can be overcome, making it possible for iPSCs to have a genuine and substantial impact on the lives of patients.
ACKNOWLEDGEMENTS
Our work was supported by the European Commission’s Seventh Framework Programme (ND, CS, AW, LV, ADK) (grant agreement N° 223317 (LIV-ES), and N°278152 (INNOVALIV), by the ANR (RFCS Liv-iPS), by Agence de la Biomédecine (ND, CS, AW, ADK), by Fondation pour la Recherche Médicale (ND, ADK, AW) by the Région Ile de France DIM STEM POLE (ND), by the Association Française contre les Myopathies (CS), by the Medical Research Council senior non-clinical fellowships (LV) and by the Cambridge Hospitals National Institute for Health Research Biomedical Research Center (LV).
CONFLICT OF INTEREST
Authors declare having no potential competing financial interest.
PATIENT CONSENT
Declared none.
ABBREVIATIONS
- hESCs
= Human Embryonic Stem Cells
- hiPSCs
= Human Induced Pluripotent Stem Cells
- FH
= Familial Hypercholesterolemia
- HB
= Haemophilia B
- FIX
= Clotting Factor IX
- CN
= Crigler-Najjar
- IMD
= Inherited Metabolic Disease
- A1AT
= Alpha1-Antitrypsin
- A1ATD
= Alpha1-Antitrypsin Disease
- AFP
= Alpha Foetoprotein
- CK19
= Cytokeratin 19
- CYP3A7, CYP3A4
= Cytochromes P450 3A7, 3A4
- LDLR
= LDL Receptor
REFERENCES
- 1.Dhawan A, Puppi J, Hughes RD, Mitry RR. Human hepatocyte transplantation: current experience and future challenges. Nat Rev Gastroenterol Hepatol. 2010;7:288–98. doi: 10.1038/nrgastro.2010.44. [DOI] [PubMed] [Google Scholar]
- 2.Ribes-Koninckx C, Ibars EP, Agrasot MA. Clinical outcome of hepatocyte transplantation in four pediatric patients with inherited metabolic diseases. Cell Transplant. 2012;21:2267–82. doi: 10.3727/096368912X637505. [DOI] [PubMed] [Google Scholar]
- 3.Kawahara T, Toso C, Douglas DN. Factors affecting hepatocyte isolation. engraftent.and replication in an in vivo model. Liver Transpl. 2010; 16:974–82. doi: 10.1002/lt.22099. [DOI] [PubMed] [Google Scholar]
- 4.Hughes RD, Mitry RR, Dhawan A. Current status of hepatocyte transplantation. Transplantation. 2012;93:342–7. doi: 10.1097/TP.0b013e31823b72d6. [DOI] [PubMed] [Google Scholar]
- 5.Puppi J, Strom SC, Hughes RD. Improving the techniques for human hepatocyte transplantation: report from a consensus meeting in London. Cell Transplant. 2012;21:1–10. doi: 10.3727/096368911X566208. [DOI] [PubMed] [Google Scholar]
- 6.Terry C, Dhawan A, Mitry RR. Optimization of the cryopreservation and thawing protocol for human hepatocytes for use in cell transplantation. Liver Transpl. 2010;16:229–37. doi: 10.1002/lt.21983. [DOI] [PubMed] [Google Scholar]
- 7.Najimi M, Smets F, Sokal E. Hepatocyte apoptosis. Methods Mol Biol. 2009;481:59–74. doi: 10.1007/978-1-59745-201-4_6. [DOI] [PubMed] [Google Scholar]
- 8.Antherieu S, Chesne C, Li R. Optimization of the HepaRG cell model for drug metabolism and toxicity studies. Toxicol In Vitro. 2012;26:1278–85. doi: 10.1016/j.tiv.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Popp FC, Slowik P, Eggenhofer E. No contribution of multipotent mesenchymal stromal cells to liver regeneration in a rat model of prolonged hepatic injury. Stem Cells. 2007;25:639–45. doi: 10.1634/stemcells.2006-0515. [DOI] [PubMed] [Google Scholar]
- 10.Kuo TK, Hung SP, Chuang CH. Stem cell therapy for liver disease: parameters governing the success of using bone marrow mesenchymal stem cells. Gastroenterology. 2008;134:2111–21. doi: 10.1053/j.gastro.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai PC, Fu TW, Chen YM. The therapeutic potential of human umbilical mesenchymal stem cells from Wharton's jelly in the treatment of rat liver fibrosis. Liver Transpl. 2009;15:484–95. doi: 10.1002/lt.21715. [DOI] [PubMed] [Google Scholar]
- 12.Fausto N, Campbell JS. The role of hepatocytes and oval cells in liver regeneration and repopulation. Mech Dev. 2003;120:117–30. doi: 10.1016/s0925-4773(02)00338-6. [DOI] [PubMed] [Google Scholar]
- 13.Roskams TA, Libbrecht L, Desmet VJ. Progenitor cells in diseased human liver. Semin Liver Dis. 2003;23:385–96. doi: 10.1055/s-2004-815564. [DOI] [PubMed] [Google Scholar]
- 14.Schmelzer E, Zhang L, Bruce A. Human hepatic stem cells from fetal and postnatal donors. J Exp Med. 2007;204:1973–87. doi: 10.1084/jem.20061603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang C, Guo X, Jiang G. CpG island methylator phenotype association with upregulated telomerase activity in hepatocellular carcinoma. Int J Cancer. 2008;123:998–1004. doi: 10.1002/ijc.23650. [DOI] [PubMed] [Google Scholar]
- 16.Schmelzer E, Reid LM. Human telomerase activity. telomerase and telomeric template expression in hepatic stem cells and in livers from fetal and postnatal donors. Eur J Gastroenterol Hepatol. 2009;21:1191–8. doi: 10.1097/MEG.0b013e32832973fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turner R, Lozoya O, Wang Y. Human hepatic stem cell and maturational liver lineage biology. Hepatology. 2011;53:1035–45. doi: 10.1002/hep.24157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemaigre F. [Lineage fate decisions in normal and regenerating liver]. Med Sci (Paris) 2012;28:958–62. doi: 10.1051/medsci/20122811014. [DOI] [PubMed] [Google Scholar]
- 19.Mahieu-Caputo D, Allain JE, Branger J. Repopulation of athymic mouse liver by cryopreserved early human fetal hepatoblasts. Hum Gene Ther. 2004;15:1219–28. doi: 10.1089/hum.2004.15.1219. [DOI] [PubMed] [Google Scholar]
- 20.Thomson JA, Itskovitz-Eldor J, Shapiro SS. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz SD, Hubschman JP, Heilwell G. Embryonic stem cell trials for macular degeneration: a preliminary report. Lancet. 2012;379:713–20. doi: 10.1016/S0140-6736(12)60028-2. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi K, Tanabe K, Ohnuki M. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 23.Park IH, Zhao R, West JA. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–6. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 24.Maehr R, Chen S, Snitow M. Generation of pluripotent stem cells from patients with type 1 diabetes. Proc Natl Acad Sci U S A. 2009;106:15768–73. doi: 10.1073/pnas.0906894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soldner F, Hockemeyer D, Beard C. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–77. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida T, Takayama K, Kondoh M. Use of human hepatocyte-like cells derived from induced pluripotent stem cells as a model for hepatocytes in hepatitis C virus infection. Biochem Biophys Res Commun. 2011;416:119–24. doi: 10.1016/j.bbrc.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Roelandt P, Obeid S, Paeshuyse J. Human pluripotent stem cell-derived hepatocytes support complete replication of hepatitis C virus. J Hepatol. 2012;57:246–51. doi: 10.1016/j.jhep.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz RE, Trehan K, Andrus L. Modeling hepatitis C virus infection using human induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2012;109:2544–8. doi: 10.1073/pnas.1121400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tao W, Xu C, Ding Q. A single point mutation in E2 enhances hepatitis C virus infectivity and alters lipoprotein association of viral particles. Virology. 2009;395:67–76. doi: 10.1016/j.virol.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Martin U, Winkler ME, Id M. Productive infection of primary human endothelial cells by pig endogenous retrovirus (PERV). Xenotransplantation. 2000;7:138–42. doi: 10.1034/j.1399-3089.2000.00052.x. [DOI] [PubMed] [Google Scholar]
- 31.Nibourg GA, Chamuleau RA, van Gulik TM, Hoekstra R. Proliferative human cell sources applied as biocomponent in bioartificial livers: a review. Expert Opin Biol Ther. 2012;12:905–21. doi: 10.1517/14712598.2012.685714. [DOI] [PubMed] [Google Scholar]
- 32.Zhang S, Chen S, Li W. Rescue of ATP7B function in hepatocyte-like cells from Wilson's disease induced pluripotent stem cells using gene therapy or the chaperone drug curcumin. Hum Mol Genet. 2011;20:3176–87. doi: 10.1093/hmg/ddr223. [DOI] [PubMed] [Google Scholar]
- 33.Hanna J, Wernig M, Markoulaki S. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–3. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 34.Lavon N, Yanuka O, Benvenisty N. Differentiation and isolation of hepatic-like cells from human embryonic stem cells. Differentiation. 2004;72:230–8. doi: 10.1111/j.1432-0436.2004.07205002.x. [DOI] [PubMed] [Google Scholar]
- 35.Schwartz RE, Linehan JL, Painschab MS. Defined conditions for development of functional hepatic cells from human embryonic stem cells. Stem Cells Dev. 2005;14:643–55. doi: 10.1089/scd.2005.14.643. [DOI] [PubMed] [Google Scholar]
- 36.Duan Y, Catana A, Meng Y. Differentiation and enrichment of hepatocyte-like cells from human embryonic stem cells in vitro and in vivo. Stem Cells. 2007;25:3058–68. doi: 10.1634/stemcells.2007-0291. [DOI] [PubMed] [Google Scholar]
- 37.Cai J, Zhao Y, Liu Y. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology. 2007;45:1229–39. doi: 10.1002/hep.21582. [DOI] [PubMed] [Google Scholar]
- 38.Soto-Gutierrez A, Navarro-Alvarez N, Zhao D. Differentiation of mouse embryonic stem cells to hepatocyte-like cells by co-culture with human liver nonparenchymal cell lines. Nat Protoc. 2007;2:347–56. doi: 10.1038/nprot.2007.18. [DOI] [PubMed] [Google Scholar]
- 39.Hay DC, Fletcher J, Payne C. Highly efficient differentiation of hESCs to functional hepatic endoderm requires ActivinA and Wnt3a signaling. Proc Natl Acad Sci U S A. 2008;105:12301–6. doi: 10.1073/pnas.0806522105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Basma H, Soto-Gutierrez A, Yannam GR. Differentiation and transplantation of human embryonic stem cell-derived hepatocytes. Gastroenterology. 2009;136:990–9. doi: 10.1053/j.gastro.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brolen G, Sivertsson L, Bjorquist P. Hepatocyte-like cells derived from human embryonic stem cells specifically via definitive endoderm and a progenitor stage. J Biotechnol. 2010;145:284–94. doi: 10.1016/j.jbiotec.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 42.Si-Tayeb K, Noto FK, Nagaoka M. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin MJ, Muotri A, Gage F, Varki A. Human embryonic stem cells express an immunogenic nonhuman sialic acid. Nat Med. 2005;11:228–32. doi: 10.1038/nm1181. [DOI] [PubMed] [Google Scholar]
- 44.Wu MY, Hill CS. Tgf-beta superfamily signaling in embryonic development and homeostasis. Dev Cell. 2009;16:329–43. doi: 10.1016/j.devcel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 45.Vallier L, Touboul T, Brown S. Signaling pathways controlling pluripotency and early cell fate decisions of human induced pluripotent stem cells. Stem Cells. 2009;27:2655–66. doi: 10.1002/stem.199. [DOI] [PubMed] [Google Scholar]
- 46.Brown S, Teo A, Pauklin S. Activin/Nodal signaling controls divergent transcriptional networks in human embryonic stem cells and in endoderm progenitors. Stem Cells. 2011;29:1176–85. doi: 10.1002/stem.666. [DOI] [PubMed] [Google Scholar]
- 47.Teo AK, Arnold SJ, Trotter MW. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 2011;25:238–50. doi: 10.1101/gad.607311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh AM, Reynolds D, Cliff T. Signaling network crosstalk in human pluripotent cells: a Smad2/3-regulated switch that controls the balance between self-renewal and differentiation. Cell Stem Cell. 2012;10:312–26. doi: 10.1016/j.stem.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McLean AB, D'Amour KA, Jones KL. Activin a efficiently specifies definitive endoderm from human embryonic stem cells only when phosphatidylinositol 3-kinase signaling is suppressed. Stem Cells. 2007;25:29–38. doi: 10.1634/stemcells.2006-0219. [DOI] [PubMed] [Google Scholar]
- 50.Lemaigre F, Zaret KS. Liver development update: new embryo models. cell lineage conrol.and morphogenesis. Curr Opin Genet Dev. 2004; 14:582–90. doi: 10.1016/j.gde.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 51.Si-Tayeb K, Lemaigre FP, Duncan SA. Organogenesis and development of the liver. Dev Cell. 2010;18:175–89. doi: 10.1016/j.devcel.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki A, Zheng YW, Kaneko S. Clonal identification and characterization of self-renewing pluripotent stem cells in the developing liver. J Cell Biol. 2002;156:173–84. doi: 10.1083/jcb.200108066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dan YY, Riehle KJ, Lazaro C. Isolation of multipotent progenitor cells from human fetal liver capable of differentiating into liver and mesenchymal lineages. Proc Natl Acad Sci U S A. 2006;103:9912–7. doi: 10.1073/pnas.0603824103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malhi H, Irani AN, Gagandeep S, Gupta S. Isolation of human progenitor liver epithelial cells with extensive replication capacity and differentiation into mature hepatocytes. J Cell Sci. 2002;115:2679–88. doi: 10.1242/jcs.115.13.2679. [DOI] [PubMed] [Google Scholar]
- 55.Allain JE, Dagher I, Mahieu-Caputo D. Immortalization of a primate bipotent epithelial liver stem cell. Proc Natl Acad Sci U S A. 2002;99:3639–44. doi: 10.1073/pnas.062038599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Delgado JP, Vanneaux V, Branger J. The role of HGF on invasive properties and repopulation potential of human fetal hepatic progenitor cells. Exp Cell Res. 2009;315:3396–405. doi: 10.1016/j.yexcr.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 57.Touboul T, Hannan NR, Corbineau S. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology. 2010;51:1754–65. doi: 10.1002/hep.23506. [DOI] [PubMed] [Google Scholar]
- 58.Hu BY, Weick JP, Yu J. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci U S A. 2010;107:4335–40. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kajiwara M, Aoi T, Okita K. Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2012;109:12538–43. doi: 10.1073/pnas.1209979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nat Biotechnol. 2008;26:120–6. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 61.Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nat Methods. 2005;2:119–25. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 62.Wang Y, Yao HL, Cui CB. Paracrine signals from mesenchymal cell populations govern the expansion and differentiation of human hepatic stem cells to adult liver fates. Hepatology. 2010;52:1443–54. doi: 10.1002/hep.23829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hay DC, Pernagallo S, Diaz-Mochon JJ. Unbiased screening of polymer libraries to define novel substrates for functional hepatocytes with inducible drug metabolism. Stem Cell Res. 2011;6:92–102. doi: 10.1016/j.scr.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 64.Weber A, Groyer-Picard MT, Franco D, Dagher I. Hepatocyte transplantation in animal models. Liver Transpl. 2009;15:7–14. doi: 10.1002/lt.21670. [DOI] [PubMed] [Google Scholar]
- 65.Rhim JA, Sandgren EP, Palmiter RD, Brinster RL. Complete reconstitution of mouse liver with xenogeneic hepatocytes. Proc Natl Acad Sci U S A. 1995;92:4942–6. doi: 10.1073/pnas.92.11.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meuleman P, Libbrecht L, De Vos R. Morphological and biochemical characterization of a human liver in a uPA-SCID mouse chimera. Hepatology. 2005;41:847–56. doi: 10.1002/hep.20657. [DOI] [PubMed] [Google Scholar]
- 67.Payne CM, Samuel K, Pryde A. Persistence of functional hepatocyte-like cells in immune-compromised mice. Liver Int. 2011;31:254–62. doi: 10.1111/j.1478-3231.2010.02414.x. [DOI] [PubMed] [Google Scholar]
- 68.Rashid ST, Corbineau S, Hannan N. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest. 2010;120:3127–36. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Azuma H, Paulk N, Ranade A. Robust expansion of human hepatocytes in Fah-/-/Rag2-/-/Il2rg-/- mice. Nat Biotechnol. 2007;25:903–10. doi: 10.1038/nbt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vaughan AM, Mikolajczak SA, Wilson EM. Complete Plasmodium falciparum liver-stage development in liver-chimeric mice. J Clin Invest. 2012;122:3618–28. doi: 10.1172/JCI62684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park IH, Arora N, Huo H. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–86. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dimos JT, Rodolfa KT, Niakan KK. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–21. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 73.Ebert AD, Yu J, Rose FFJr. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–80. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grskovic M, Javaherian A, Strulovici B, Daley GQ. Induced pluripotent stem cells--opportunities for disease modelling and drug discovery. Nat Rev Drug Discov. 2011;10:915–29. doi: 10.1038/nrd3577. [DOI] [PubMed] [Google Scholar]
- 75.Tiscornia G, Vivas EL, Izpisua Belmonte JC. Diseases in a dish: modeling human genetic disorders using induced pluripotent cells. Nat Med. 2011;17:1570–6. doi: 10.1038/nm.2504. [DOI] [PubMed] [Google Scholar]
- 76.Trounson A, Shepard KA, DeWitt ND. Human disease modeling with induced pluripotent stem cells. Curr Opin Genet Dev. 2012;22:509–16. doi: 10.1016/j.gde.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 77.Perrier A, Peschanski M. How can human pluripotent stem cells help decipher and cure Huntington's disease?. Cell Stem Cell. 2012;11:153–61. doi: 10.1016/j.stem.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 78.Seibler P, Graziotto J, Jeong H. Mitochondrial Parkin recruitment is impaired in neurons derived from mutant PINK1 induced pluripotent stem cells. J Neurosci. 2011;31:5970–6. doi: 10.1523/JNEUROSCI.4441-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Egawa N, Kitaoka S, Tsukita K. Drug screening for ALS using patient-specific induced pluripotent stem cells. Sci Transl Med. 2012;4:145ra104. doi: 10.1126/scitranslmed.3004052. [DOI] [PubMed] [Google Scholar]
- 80.Brown MS, Goldstein JL. Biomedicine.Lowering LDL--not only how low.but how long?. Science. 2006;311:1721–3. doi: 10.1126/science.1125884. [DOI] [PubMed] [Google Scholar]
- 81.Bruckert E. New lipid-modifying therapies. Expert Opin Investig Drugs. 2003;12:325–35. doi: 10.1517/13543784.12.3.325. [DOI] [PubMed] [Google Scholar]
- 82.Hudgins LC, Kleinman B, Scheuer A. Long-term safety and efficacy of low-density lipoprotein apheresis in childhood for homozygous familial hypercholesterolemia. Am J Cardiol. 2008;102:1199–204. doi: 10.1016/j.amjcard.2008.06.049. [DOI] [PubMed] [Google Scholar]
- 83.Huijgen R, Kindt I, Verhoeven SB. Two years after molecular diagnosis of familial hypercholesterolemia: majority on cholesterol-lowering treatment but a minority reaches treatment goal. PLoS One. 2010;5:e9220. doi: 10.1371/journal.pone.0009220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Grossman M, Rader DJ, Muller DW. A pilot study of ex vivo gene therapy for homozygous familial hypercholesterolaemia. Nat Med. 1995;1:1148–54. doi: 10.1038/nm1195-1148. [DOI] [PubMed] [Google Scholar]
- 85.Petrus I, Chuah M, VandenDriessche T. Gene therapy strategies for hemophilia: benefits versus risks. J Gene Med. 2010;12:797–809. doi: 10.1002/jgm.1500. [DOI] [PubMed] [Google Scholar]
- 86.Wu YM, Kao CY, Huang YJ. Genetic modification of donor hepatocytes improves therapeutic efficacy for hemophilia B in mice. Cell Transplant. 2010;19:1169–80. doi: 10.3727/096368910X503398. [DOI] [PubMed] [Google Scholar]
- 87.Nathwani AC, Tuddenham EG, Rangarajan S. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N Engl J Med. 2011;365:2357–65. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kasuda S, Tatsumi K, Sakurai Y. Expression of coagulation factors from murine induced pluripotent stem cell-derived liver cells. Blood Coagul Fibrinolysis. 2011;22:271–9. doi: 10.1097/MBC.0b013e328344c63b. [DOI] [PubMed] [Google Scholar]
- 89.Cao J, Shang CZ, Lu LH. Differentiation of embryonic stem cells into hepatocytes that coexpress coagulation factors VIII and IX. Acta Pharmacol Sin. 2010;31:1478–86. doi: 10.1038/aps.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lomas DA, Evans DL, Finch JT, Carrell RW. The mechanism of Z alpha 1-antitrypsin accumulation in the liver. Nature. 1992;357:605–7. doi: 10.1038/357605a0. [DOI] [PubMed] [Google Scholar]
- 91.Gooptu B, Ekeowa UI, Lomas DA. Mechanisms of emphysema in alpha1-antitrypsin deficiency: molecular and cellular insights. Eur Respir J. 2009;34:475–88. doi: 10.1183/09031936.00096508. [DOI] [PubMed] [Google Scholar]
- 92.Griesenbach U, Geddes DM, Alton EW. Advances in cystic fibrosis gene therapy. Curr Opin Pulm Med. 2004;10:542–6. doi: 10.1097/01.mcp.0000142102.91202.04. [DOI] [PubMed] [Google Scholar]
- 93.Fisher RA, Strom SC. Human hepatocyte transplantation: worldwide results. Transplantation. 2006;82:441–9. doi: 10.1097/01.tp.0000231689.44266.ac. [DOI] [PubMed] [Google Scholar]
- 94.Nguyen TH, Birraux J, Wildhaber B. Ex vivo lentivirus transduction and immediate transplantation of uncultured hepatocytes for treating hyperbilirubinemic Gunn rat. Transplantation. 2006;82:794–803. doi: 10.1097/01.tp.0000234675.56598.35. [DOI] [PubMed] [Google Scholar]
- 95.Nguyen TH, Mainot S, Lainas P. Ex vivo liver-directed gene therapy for the treatment of metabolic diseases: advances in hepatocyte transplantation and retroviral vectors. Curr Gene Ther. 2009;9:136–49. doi: 10.2174/156652309787909481. [DOI] [PubMed] [Google Scholar]
- 96.Birraux J, Menzel O, Wildhaber B. A step toward liver gene therapy: efficient correction of the genetic defect of hepatocytes isolated from a patient with Crigler-Najjar syndrome type 1 with lentiviral vectors. Transplantation. 2009;87:1006–12. doi: 10.1097/TP.0b013e31819ca245. [DOI] [PubMed] [Google Scholar]
- 97.Hacein-Bey-Abina S, Garrigue A, Wang GP. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–42. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Connelly S. Adenoviral vectors for liver-directed gene therapy. Curr Opin Mol Ther. 1999;1:565–72. [PubMed] [Google Scholar]
- 99.Howden SE, Gore A, Li Z. Genetic correction and analysis of induced pluripotent stem cells from a patient with gyrate atrophy. Proc Natl Acad Sci U S A. 2011;108:6537–42. doi: 10.1073/pnas.1103388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li H, Haurigot V, Doyon Y. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475:217–21. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu GH, Suzuki K, Qu J. Targeted gene correction of laminopathy-associated LMNA mutations in patient-specific iPSCs. Cell Stem Cell. 2011;8:688–94. doi: 10.1016/j.stem.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vallier L, Alexander M, Pedersen R. Conditional gene expression in human embryonic stem cells. Stem Cells. 2007;25:1490–7. doi: 10.1634/stemcells.2006-0825. [DOI] [PubMed] [Google Scholar]
- 103.Urnov FD, Rebar EJ, Holmes MC. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–46. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 104.Hockemeyer D, Wang H, Kiani S. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–4. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yusa K, Rashid ST, Strick-Marchand H. Targeted gene correction of alpha1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478:391–4. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gupta A, Meng X, Zhu LJ. Zinc finger protein-dependent and -independent contributions to the in vivo off-target activity of zinc finger nucleases. Nucleic Acids Res. 2011;39:381–92. doi: 10.1093/nar/gkq787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vons C, Beaudoin S, Helmy N. First description of the surgical anatomy of the cynomolgus monkey liver. Am J Primatol. 2009;71:400–8. doi: 10.1002/ajp.20667. [DOI] [PubMed] [Google Scholar]
- 108.Liu H, Zhu F, Yong J. Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell. 2008;3:587–90. doi: 10.1016/j.stem.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 109.Okahara-Narita J, Umeda R, Nakamura S. Induction of pluripotent stem cells from fetal and adult cynomolgus monkey fibroblasts using four human transcription factors. Primates. 2012;53:205–13. doi: 10.1007/s10329-011-0283-1. [DOI] [PubMed] [Google Scholar]
- 110.Wu Y, Mishra A, Qiu Z. Nonhuman primate induced pluripotent stem cells in regenerative medicine. Stem Cells Int. 2012;2012:767195. doi: 10.1155/2012/767195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Blin G, Nury D, Stefanovic S. A purified population of multipotent cardiovascular progenitors derived from primate pluripotent stem cells engrafts in postmyocardial infarcted nonhuman primates. J Clin Invest. 2010;120:1125–39. doi: 10.1172/JCI40120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Joseph B, Malhi H, Bhargava KK. Kupffer cells participate in early clearance of syngeneic hepatocytes transplanted in the rat liver. Gastroenterology. 2002;123:1677–85. doi: 10.1053/gast.2002.36592. [DOI] [PubMed] [Google Scholar]
- 113.Dagher I, Boudechiche L, Branger J. Efficient hepatocyte engraftment in a nonhuman primate model after partial portal vein embolization. Transplantation. 2006;82:1067–73. doi: 10.1097/01.tp.0000236103.99456.8f. [DOI] [PubMed] [Google Scholar]
- 114.Lainas P, Boudechiche L, Osorio A. Liver regeneration and recanalization time course following reversible portal vein embolization. J Hepatol. 2008;49:354–62. doi: 10.1016/j.jhep.2008.01.034. [DOI] [PubMed] [Google Scholar]
- 115.Dagher I, Nguyen TH, Groyer-Picard MT. Efficient hepatocyte engraftment and long-term transgene expression after reversible portal embolization in nonhuman primates. Hepatology. 2009;49:950–9. doi: 10.1002/hep.22739. [DOI] [PubMed] [Google Scholar]
- 116.Lister R, Pelizzola M, Kida YS. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mekhoubad S, Bock C, de Boer AS. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell. 2012;10:595–609. doi: 10.1016/j.stem.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ohi Y, Qin H, Hong C. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol. 2011;13:541–9. doi: 10.1038/ncb2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ruiz S, Diep D, Gore A. Identification of a specific reprogramming-associated epigenetic signature in human induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2012;109:16196–201. doi: 10.1073/pnas.1202352109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Urbach A, Bar-Nur O, Daley GQ, Benvenisty N. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell. 2010;6:407–11. doi: 10.1016/j.stem.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sheridan SD, Theriault KM, Reis SA. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS One. 2011;6:e26203. doi: 10.1371/journal.pone.0026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gore A, Li Z, Fung HL. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–7. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ji J, Ng SH, Sharma V. Elevated coding mutation rate during the reprogramming of human somatic cells into induced pluripotent stem cells. Stem Cells. 2012;30:435–40. doi: 10.1002/stem.1011. [DOI] [PubMed] [Google Scholar]
- 124.Abyzov A, Mariani J, Palejev D. Somatic copy number mosaicism in human skin revealed by induced pluripotent stem cells. Nature. 2012;492:438–42. doi: 10.1038/nature11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nie B, Wang H, Laurent T, Ding S. Cellular reprogramming: a small molecule perspective. Curr Opin Cell Biol. 2012;24:784–92. doi: 10.1016/j.ceb.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pournasr B, Khaloughi K, Salekdeh GH. Concise review: alchemy of biology: generating desired cell types from abundant and accessible cells. Stem Cells. 2011;29:1933–41. doi: 10.1002/stem.760. [DOI] [PubMed] [Google Scholar]
- 127.Vierbuchen T, Wernig M. Direct lineage conversions: unnatural but useful?. Nat Biotechnol. 2011;29:892–907. doi: 10.1038/nbt.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–76. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 129.Huang P, He Z, Ji S. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–9. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- 130.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–3. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 131.Zhou Q, Brown J, Kanarek A. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–32. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]