Abstract
Salmonella enterica causes substantial morbidity and mortality in humans and animals. Infection and intestinal colonization by S. enterica require virulence factors that mediate bacterial binding and invasion of enterocytes and innate immune cells. Some S. enterica colonization factors and their alleles are host restricted, suggesting a potential role in regulation of host specificity. Recent data also suggest that colonization factors promote horizontal gene transfer of antimicrobial resistance genes by increasing the local density of Salmonella in colonized intestines. Although a profusion of genes are involved in Salmonella pathogenesis, the relative importance of their allelic variation has only been studied intensely in the type 1 fimbrial adhesin FimH. Although other Salmonella virulence factors demonstrate allelic variation, their association with specific metadata (e.g., host species, disease or carrier state, time and geographic place of isolation, antibiotic resistance profile, etc.) remains to be interrogated. To date, genome-wide association studies (GWAS) in bacteriology have been limited by the paucity of relevant metadata. In addition, due to the many variables amid metadata categories, a very large number of strains must be assessed to attain statistically significant results. However, targeted approaches in which genes of interest (e.g., virulence factors) are specifically sequenced alleviates the time-consuming and costly statistical GWAS analysis and increases statistical power, as larger numbers of strains can be screened for non-synonymous single nucleotide polymorphisms (SNPs) that are associated with available metadata. Congruence of specific allelic variants with specific metadata from strains that have a relevant clinical and epidemiological history will help to prioritize functional wet-lab and animal studies aimed at determining cause-effect relationships. Such an approach should be applicable to other pathogens that are being collected in well-curated repositories.
Keywords: Salmonella, colonization, adhesin, invasion, T3SS, SNPs
ONE TO FIVE STAR HOTELS FOR Salmonella
Salmonella enterica infections result in substantial morbidity and mortality worldwide, both in humans and livestock (Hirsh, 2004). The immunopathology induced during the infectious process is the result of both Salmonella virulence factors and host responses (de Jong et al., 2012). Salmonellosis, when not self-limiting, requires antimicrobial therapy, particularly for neonates and to treat or prevent systemic infections. As such, the increasing prevalence of multidrug resistant S. enterica raises substantial concerns regarding the efficacy of current therapy. Continuous surveillance of reported Salmonella cases (Centers for Disease and Prevention, 2013) indicates that Salmonella remains the most frequent bacterial cause of foodborne disease in the US and suggests that control programs aimed at reducing food contamination have not succeeded for Salmonella. In this regard, the persistence of S. enterica in the intestinal tract in a large number of food animals creates chronic or non-symptomatic carriers that continue to shed bacteria in feces, thereby serving as a reservoir for future spread by contaminated meat, milk, eggs, and agricultural products grown on land fertilized with Salmonella-containing manure. The Salmonella reservoir is not only maintained by transmission between animals, directly or indirectly through insect vectors (Wales et al., 2010), but also by long-term environmental contaminations. Indeed, some Salmonella strains can survive for days, weeks, or months in the environment, and can encamp in soil, water, or on plants by making protective biofilms before being re-ingested by animals or humans. Maintenance of the environmental reservoir is enhanced by Salmonella’s aggressive behavior toward competitors or predators such as fungi, ameba, and helminthes, and possibly other bacteria (Aballay et al., 2000; Wildschutte and Lawrence, 2007; Tampakakis et al., 2009; Pezoa et al., 2013). Some Salmonella can even invade and hide inside certain plants, which if edible, add another source of transmission to humans for this successful pathogen (Arthurson et al., 2011; Golberg et al., 2011).
With the latest official publications on serological grouping of Salmonella up to 2007, the number of different serovars amounts to 2625 (Grimont and Weill, 2007; Guibourdenche et al., 2010). Two species are recognized, bongori and enterica, the latter species being divided in six named or numbered subspecies (enterica or I, salamae or II, arizonae or IIIa, diarizonae or IIIb, houtenae or IV, and indica or VI; V became S. bongori; Tindall et al., 2005). Many Salmonella strains survive and multiply in a range of environments, but the most “luxurious” accommodations are warm and rich in food. For example, although most S. enterica subspecies preferentially lodge in the intestines of cold-blooded animals, S. enterica subsp. enterica (also designated subsp. I) has “upgraded” to the bowels of some warm-blooded animals, the optimal environment for bacterial multiplication and transmission. Moreover, the maintained ability of S. enterica to “camp” outside of the host in less opulent telluric biotopes further improves its chances of transmission.
While many of the approximately 1,500 characterized serovars of that subspecies seem to promiscuously colonize any mammal or bird, others show adaptation to a subset of host species, or may even be restricted to one host species. Salmonella serovar variants with distinct host range are well documented in host species-specific endemic or epidemic strains (Rabsch et al., 2002). While comparative genomic studies of Salmonella strains highlight evolutionary steps involving gene gain by horizontal gene transfer (HGT), as well as recombination, mutations and gene loss (Porwollik et al., 2002; Holt et al., 2009; den Bakker et al., 2011), determining which genetic alterations directly participate in host adaptation remains a barely started puzzle. It is clear, for example, that bovines don’t offer “room and board” to the avian-restricted S. enterica serovar Gallinarum or the human restricted S. enterica serovar Typhi. However, both the bovine-adapted S. enterica serovar Dublin as well as serovar Typhimurium, which has a broad host range, can colonize cattle whose levels of immune competence and competing flora dictate the one to five star “quality” rating of the host environment. Serovars such as serovar Dublin that have a more defined host restriction are often more virulent toward their hosts. For example, serovar Dublin causes a potentially lethal systemic infection in its bovine host. However, since not all serovar Dublin-infected hosts die, the high level of multiplication and transmission in chronically ill hosts and/or convalescent silent carriers appears to trump the occasional death of individual hosts. In contrast, the ability of broad host range serovars to subsist in any warm-blooded animal seems to compensate for their less extensive host invasion, potentially explaining why the broad host range serovar Typhimurium remains a most isolated Salmonella serovar in veterinary and human medicine.
Salmonella LIGAND ALLELES: THE DEVIL IS IN THE DETAILS
Current Salmonella genomic studies identify potential genetic evolutionary adaptations using a variety of in silico approaches that analyze genetic differences in collections of subspecies and serovars (Soyer et al., 2009). A recent study compared tree-building methods to ascertain congruent results by integrating models of gene gains and losses (Desai et al., 2013). Incremental information was built with strain-specific lists of new or missing groups of genes that might be involved in strain adaptation and virulence. However, to date, such studies have mainly investigated differences between serovars. Strains within a serovar are typically more related to each other, suggesting that genes responsible for the O and H antigens of Salmonella, which determine serovar classification, are latecomers in Salmonella’s evolutionary history. However, it is sometimes unclear whether the strains studied within a serovar are independent and not clonal, considering that many of the histories and pathotypes (origin, host species, disease and pathology) remain poorly defined. Thus, the observed lack of diversity within a serovar might be partially biased by the collection of strains studied.
Strain differences within the same serovar, particularly for those of broad host range, have long been under-evaluated, as exemplified by the increasing number of well-documented “exceptions” that illustrate distinct lineages within such serovars (Sangal et al., 2010; Lawson et al., 2011; Okoro et al., 2012; Gragg et al., 2013). However, it is quite likely that the ability of Salmonella to colonize and/or cause diseases in different hosts depends not only on the presence of a collection of specific genes, but also on the allelic variation within these genes. In surprising contrast to research in both cancer and metabolic and inheritable diseases, where mammalian single nucleotide polymorphisms (SNPs) are dissected in great detail to determine potential causality (Frazer et al., 2009), few investigators have assessed allele-mediated effects of non-synonymous SNPs on bacterial proteins that mediate or participate in host specificity and virulence (Hopkins and Threlfall, 2004). Notably, the few studies that have assessed allelic effects of virulence factors in Salmonella pathogenesis revealed altered virulence phenotypes (Wagner and Hensel, 2011; Thornbrough and Worley, 2012). The potential role of non-synonymous SNPs on host specificity is best exemplified with the orthologous fimbrial adhesin FimH of the avian-specific serovars Gallinarum and Pullorum which mediates significantly better bacterial binding to chicken leukocytes than did serovar Typhimurium FimH alleles (Guo et al., 2009). In contrast to the latter alleles, the avian-specific FimH did not mediate bacterial binding to mammalian cells. Even though the FimH of serovar Typhimurium mediates specific binding to mannosylated residues on glycoproteins, as expected of the adhesion of type 1 fimbriae in general, binding to chicken leukocytes by fimbriated bacteria expressing the avian-specific FimH was only minimally inhibited by mannose. Notably, FimH of serovar Gallinarum and Typhimurium differ by only 5–6 amino acids, but one amino acid substitution was sufficient to restore mannose-specific binding (Kisiela et al., 2005). Moreover, the mannose-inhibitable FimH of serovar Enteritidis which has the latter amino acid substitution, decreased chick colonization when studied in serovar Gallinarum (Kuzminska-Bajor et al., 2012). Together, these data demonstrated that allelic variation of the Salmonella FimH adhesin directs not only host-cell-specific recognition, but also distinctive binding to mammalian or avian receptors. Remarkably, the allele-specific binding profile parallels the host specificity of the respective FimH-expressing pathogen, highlighting its physiologic relevance.
Salmonella’s SWAT TEAMS FOR HOST COLONIZATION
The ability of Salmonella to colonize its host(s) relies on sets of molecules that allow it to bypass natural host defense mechanisms, such as gastric acidity and gastrointestinal proteases and defensins, as well as the aggressins of the intestinal microbiome. In addition, Salmonella utilizes surface molecules, organelles, and machineries, such as adhesins, flagella, and type 3 secretion systems (T3SS) to actively make contact with host cells and deliver molecules that initially improves host colonization, and later its transmission to new hosts. As such, allelic variation in any of these molecules could influence host specificity and pathogenicity.
Flagella direct the Salmonella toward the surface of intestinal epithelium by promoting bacterial movement across the mucus layer (Figure 1, step 1). Flagella’s near-surface swimming properties optimize the encounter of bacterial adhesins with host receptors (Humphries et al., 2001; Korhonen, 2007; Wagner and Hensel, 2011; Misselwitz et al., 2012). Flagella have highly variable alleles (which serve as the basis for H antigen serology in Salmonella). While flagellar motility is clearly required for intestinal colonization (Robertson et al., 2000), early studies suggested that flagella act as adhesins (Allen-Vercoe and Woodward, 1999) and flagella were reported to mediate bacterial binding for biofilm formation on cholesterol-coated surfaces (Crawford et al., 2010). Thus, in addition to providing antigenic variation to allow Salmonella to escape a host’s immune system, variations in Salmonella flagellin sequences may also influence the efficiency of intestinal colonization.
FIGURE 1.
Model of intestinal colonization by Salmonella. Most Salmonella remain associated to specific anatomical segments of the intestines using cyclical multiplication steps in the lumen and enterocytes, using (1) near surface swimming, (2) adhesion (several adhesins), (3) invasion with formation of SCV, (4.1) intravacuolar replication, (5) escape in the cytoplasm, pyroptosis and return to the intestinal surface and lumen. After intravacuolar replication, (4.2) Salmonella can also escape toward the lamina propria, resulting in their potential uptake by dendritic cells and macrophages. (6) Some Salmonella can be further transported to the mesenteric lymph nodes which frequently contain further spreading. Typically for host-restricted (e.g., serovar Typhi in humans) or host-adapted strains (e.g., serovar Dublin in bovines), but also for Salmonella with broad host range encountering stressed hosts or weakened host immune responses, (7) Salmonella can cause bacteremia. When disseminating Salmonella (whether or not host-adapted or restricted) can be somewhat controlled by the host, they typically settle in the liver, gallbladder (for mammals with this organ*), on calculi and in the biliary tract, where they can replicate and be released in the bile to return to the intestines (Oboegbulem and Muogbo, 1981; Cavallini et al., 1991; Wood et al., 1991; Pellegrini-Masini et al., 2004; Akoachere et al., 2009; He et al., 2010).
Salmonella encodes a large collection of adhesins, including fimbrial organelles (Yue et al., 2012a), surface associated proteins or surface-exposed domains of anchored outer membrane proteins (OMPs; Wagner and Hensel, 2011), which can attach to enterocytes (Figure 1, step 2), phagocytic cells (Figure 1, step 4.2), or gallbladder and biliary calculi (Figure 1, step 7; Reisner et al., 2003; Ledeboer et al., 2006; Dwyer et al., 2011; Gonzalez-Escobedo et al., 2011; Spurbeck et al., 2011; Wilksch et al., 2011). A subset of the adhesive OMPs are autotransporter proteins with surface-exposed adhesive N-terminal ends or passenger domain that have the potential to influence host-specificity (Kingsley et al., 2003, 2004; Morgan et al., 2004; Dorsey et al., 2005; Raghunathan et al., 2011; Wagner and Hensel, 2011; Leyton et al., 2012). Other OMPs have adhesive properties or significant sequence similarities to known adhesins, some which promote epithelial cell invasion by a zipper mechanism (Velge et al., 2012). Salmonella adhesins secreted by type 1 secretion systems (T1SS, ABC transporters) and anchored on the bacterial surface (Morgan et al., 2004; Latasa et al., 2005; Gerlach et al., 2007; Wagner and Hensel, 2011) may also participate in specific host colonization.
Salmonella optimizes transmission and intestinal colonization by invading and replicating in enterocytes, a process that is mediated by subversion of the enterocyte’s cytoskeletal machinery and signal transduction systems by secreted/injected effector proteins from the two T3SS (Stevens et al., 2009; Heffron et al., 2011). For this, adhesion-mediated attachment stabilizes bacterial docking by the Salmonella-pathogenicity island 1 (SPI-1) encoded T3SS, which then injects effector proteins into enterocytes (Misselwitz et al., 2011) to trigger bacterial uptake by macropinocytosis (Figure 1, step 3). Once inside the Salmonella-containing vacuole (SCV), Salmonella deploys a second T3SS encoded by SPI-2 to inject effector proteins that optimize conditions for bacterial survival and replication (Figure 1, step 4.1; Stevens et al., 2009; Heffron et al., 2011). Additional virulence factors subsequently allow hyper-replicating bacteria to escape the SCV, thereby promoting a cytosolic inflammatory response that results in pyroptotic cell death. Extracellular release of bacteria from dying cells potentially contributes to colonization by allowing invasion of new enterocytes (Figure 1, step 5; Knodler et al., 2010). In addition, T3SS effectors direct the transport of SCVs to the basolateral side of enterocytes, where released bacteria can be taken up directly by lamina propria phagocytes (Muller et al., 2012), including dendritic cells (DCs; Figure 1, step 4.2), a T3S-regulated process (Swart and Hensel, 2012). Finally, effectors of the two T3SS, but particularly of SPI-2 allow Salmonella to replicate intracellularly or bypass phagocyte-mediated killing in the lamina propria and to hitchhike inside DCs (Swart and Hensel, 2012) to seed mesenteric lymph nodes (Figure 1, step 6). SPI-2 effectors also interfere with MHC-presentation and activation of an immune response that would eliminate Salmonella. If not contained, the bacteria that escape the lymph nodes promote a primary bacteremia that seeds the liver (Figure 1, step 7), to initiate a circular reinfection process by taking the biliary route to return to the intestinal lumen. In addition to making biofilms on gallbladder stones, Salmonella adheres to and invades gallbladder epithelial cells (and probably biliary tract epithelial cells) in a SPI-1-dependent manner (Gonzalez-Escobedo and Gunn, 2013), creating a constant or recurring seeding source for the intestines. Some T3SS effectors might play several roles, such as the SPI-2 effector SseI/SrfH that was suggested to activate cell motility early during intestinal infection, but also to mainly inhibit cell migration later during infection, after establishment of new reservoirs in the mesenteric lymph nodes and liver (Worley et al., 2006; McLaughlin et al., 2009). Thus, effector proteins of the two T3SS promote various steps of intestinal, mesenteric, and hepatic survival and replication, some inducing anti-inflammatory responses, creating stable seeding reservoirs that contribute to host’s persistent colonization (Monack et al., 2004; Heffron et al., 2011; Gopinath et al., 2012; Ruby et al., 2012).
SNPs SNAPSHOT: Salmonella ET AL.
As discussed earlier for the type 1 fimbrial adhesin FimH, type 3 effector proteins of Salmonella harbor non-synonymous SNPs that have the capacity to modulate the infectious process. For example, the Gifsy-2 phage srfH/sseI gene encodes two protein alleles that differentially influence the motility of intestinal CD18-expressing phagocytes upon ingestion of S. enterica serovar Typhimurium (Thornbrough and Worley, 2012). Migration is stimulated by the SrfH/SseI allele with a glycine at position 103, allowing it to bind to TRIP6 to promote mobility. The same effector protein with an aspartic acid at position 103 does not interact with TRIP6, and thus does not induce phagocyte migration. Interestingly, the glycine-bearing allele is found in specific Typhimurium strains that are more invasive, cause more severe diseases or associate with outbreaks. Allelic variations among effectors of Salmonella T3SS include the SopE Cdc42 RhoGTPase (Hopkins and Threlfall, 2004; Schlumberger and Hardt, 2005). Although existing alleles were not directly compared functionally, a substitution of residue 198, which is variable in different serovars, affects the catalytic activity of the enzyme.
In addition to the above-mentioned virulence factors of Salmonella, this bacterium has evolved sophisticated subsistence factors or metabolic pathways to survive and multiply during host colonization (Deatherage Kaiser et al., 2013; Steeb et al., 2013). Multi-omic data by Deatherage Kaiser et al. revealed that Salmonella induces an inflammatory response that alters the intestinal microbiota, thereby increasing the availability of various metabolites, including fucose, that accumulate following depletion of commensal bacteria. Moreover, Salmonella responds to this shift in metabolites by upregulating expression of fucose utilizing proteins. Similarly, combining proteomics, microbial genetics, competitive infections, and computational approaches Steeb et al. identified at least 31 host nutrients that increased in infected tissues and demonstrated that Salmonella adjusted its catabolic pathways to utilize new host nutrients. Elegant experiments also demonstrated how Salmonella takes advantage of its induced inflammatory environment to out-compete the intestinal microbiota with specialized respiratory pathways (Winter et al., 2010; Rivera-Chavez et al., 2013).
An important caveat concerning most of the information on Salmonella virulence and infection is that it is based on in vitro studies, carried out with a restricted number of Salmonella strains, and primarily assessed in mouse models of infection, with a limited amount of data gathered in farm animals, particularly cattle and chicken. We predict that host-dependent colonization is not only modulated by specific adhesin alleles (Guo et al., 2009; Kuzminska-Bajor et al., 2012), but also by non-synonymous SNPs in additional virulence factors, such as the T3SS effectors (Schlumberger et al., 2003; Hopkins and Threlfall, 2004; Raffatellu et al., 2005; Thornbrough and Worley, 2012). It is also likely that numerous additional Salmonella protein alleles will be identified in the near future as virulence factors senso lato, including proteins of metabolic pathways that have evolved to take advantage of unique host species properties.
Because of its importance if the fields of immunology and vaccinology, allelic variation of antigenic microbial proteins has been studied for years. The vertebrate’s immune system exerts a powerful selection on such antigens, forcing an accelerated evolutionary route for the bacteria that use these virulence tools to breach immune recognition. In contrast, allelic variation in non-antigenic virulence genes has received less attention, possibly due to the uniqueness and complexity of genotype-phenotype links for each factor. However, differential interaction of allelic variants with host molecules has been reported for a variety of bacteria, supporting the global relevance of this pathoadaptive mechanism. For example, only SpeB (streptococcal pyrogenic exotoxin B) variants that harbor an integrin-binding motif (Stockbauer et al., 1999) mediate cell rounding and detachment from culture substrates. In addition, a specific amino acid polymorphism in the CagL protein of Helicobacter pylori correlates significantly with a higher risk for gastric cancer and severe corpus gastritis (Yeh et al., 2011) and OmpA allelic variants of Bacteroides vulgatus isolates from ulcerative colitis patients mediate better bacterial adhesion to a human colon cancer cell line than do those isolated from patients with colon cancer (Sato et al., 2010). Finally, allelic polymorphism between the YopJ/YopP type 3 effector proteins of the mammalian virulent Yersinia species differentially affects secretion and toxicity levels in a mouse model (Brodsky and Medzhitov, 2008). Similarly, strains of P. syringae can express three YopJ/YopP homologous proteins with various alleles that induce host-adapted hypersensitive responses in different plants. Thus, whereas allelic variation of YopJ/YopP in Yersinia modulates the type of pathology in one host, allelic variation in the corresponding proteins of the plant pathogen Pseudomonas syringae elicit host-specific spreading of infections (Ma et al., 2006).
FROM GENOMICS TO GWAS: THE IMPORTANCE OF BEING EARNEST WITH METADATA
Technological improvements have reduced the cost of next generation sequencing, allowing for increasingly larger-scale investigations based on tens to hundreds of Salmonella strains. This trove of new information has allowed extensive studies on the adaptive history of pathogens. In addition to information on gene acquisition, inactivation, or loss, comparative genomics highlights non-synonymous mutations that may alter function. Unfortunately, the number of mutations is usually too high to make firm conclusions. In contrast, genome-wide association studies (GWAS) that focus on Salmonella strains for which there is host and clinical information would help to better limit the number of genes likely involved in strain-specific phenotypes. Currently, GWAS are mainly used to identify disease-associated sequences within human genomes. Specifically, whole genomes of healthy and affected individuals are sequenced and compared for intra- and inter-genic SNPs that are associated with a specific disease to suggest a potential cause and effect relationship. Discovered associations are expected to support the development of new diagnostic, therapeutic, and preventive tools. Some studies focus on genomic regions already suspected to contribute to disease progression to identify a limited number of associated SNPs. Notably, GWAS have successfully identified several disease-specific SNPs (Stranger et al., 2011).
Analogous to studies on human genomes, GWAS can be applied to bacterial genomes. Assuming one has the needed metadata to separate groups of strains in an epidemiologically, clinically, or biologically relevant manner, GWAS should be able to characterize specific SNPs in strain variants that are associated with restriction to a particular host species or strain pathotype in-between and inside serovars. Moreover, by carrying out GWAS on strains with detailed host metadata, strains restricted to the same host could be directly compared, thereby reducing the confounding effects of differential host innate immune and adaptive defense responses on virulence. In addition to host specific associations, SNPs associated with ethnicity or breed, age, and sex are some additional factors that could be evaluated by GWAS. More complicated studies can compare additional groups or variables, however, statistical methods for GWAS can be plagued by having more variables than samples, the so called “curse of dimensionality” (Kelemen et al., 2009). Increasing the number of strains with useful metadata should improve the reliability of the conclusions, but large-scale investigations of hundreds of Salmonella genomes still remain a challenging undertaking, considering the high cost and time-consuming analysis. A palliative plan is to restrict GWAS to genes of interest, namely genes already known to play a role in virulence or multiplication (e.g., through special metabolic pathways) in at least one host species. Several technologies for targeted sequencing have been developed in recent years, and are based on DNA enrichment methods such as capture by hybridization or microfluidic PCR (Meyer et al., 2008). Incorporating sample barcoding to the latter method recently allowed us to use massive parallel sequencing for assessing allelic variation in known and predicted fimbrial adhesins of S. enterica serovar Newport (Yue et al., 2012b). Despite only targeting 8 adhesin genes from 48 independent strains, we identified 5–85 SNPs per gene, with 6–12 different alleles per gene. Phylogenic trees clearly separated each of five adhesins into two groups, which overlapped significantly. Most importantly, the two groups clearly differentiated strains of bovine and non-bovine origin. Even though these preliminary results utilized a relatively small number of isolates, the data were highly promising and clearly supported the idea that adhesin alleles contribute to host specificity.
It is safe to assume that multiple mutations in a variety of genes participate in the slow and progressive course of host adaptation. In addition to binding to intestinal host receptors, Salmonella has evolved to colonize its diverse hosts through a tailored army of virulence factors that mediate tissue invasion and subvert host immune defense mechanisms to improve short to long-term multiplication and transmission. Identification of such factors in bacterial pathogenesis studies are traditionally based on molecular Koch’s postulates that either interrogate one candidate at a time or assess libraries of random mutants of one prototypic bacterium in an animal model. However, despite the frequent insufficiency of metadata, candidate virulence genes that encode variable alleles in Salmonella strains for which genomic data are available can be detected by comparative in silico analysis. As described above, targeted sequencing of these genes, focusing particularly on the ones predicted or found experimentally to play a role in various steps of Salmonella-host interactions, can then be applied to strain collections that have relevant metadata to identify statistically significant associations. The detection of such associations will help to limit the number of confirmatory “wet lab” and animal experiments required to demonstrate the role of allelic variants in specific host colonization.
HOARDING ANTIMICROBIAL RESISTANCE BY STICKING AROUND
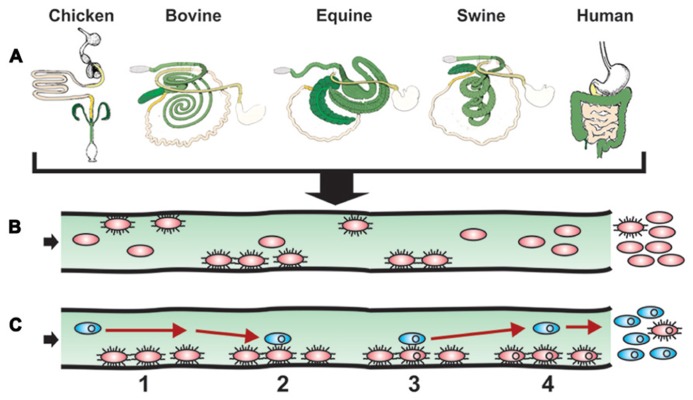
Although Salmonella-induced gastroenteritis is typically self-limiting, antibiotherapy is generally indicated for salmonellosis in patients or animals of young age or at risk for septicemia. As many farm animals are asymptomatic carriers of Salmonella, farm animals given antibiotics either routinely as food additives or therapeutically likely contribute to the alarming increase in multiple drug-resistant (MDR) Salmonella (McDermott, 2006). The corresponding uncontrolled access to and self-prescription of oral antibiotics in the human population of developing countries and the frequently inappropriate use of antibiotics in developed countries amplify the problem. Antibiotic resistance genes in enteric bacteria, including Salmonella, are frequently located on mobile DNA elements such as transposons, integrons, integrative conjugative, or mobilizable elements (including conjugative transposons) and plasmids (Vo et al., 2007; Welch et al., 2007; Su et al., 2008; Ajiboye et al., 2009; Fricke et al., 2009; Lindsey et al., 2009; Call et al., 2010; Douard et al., 2010; Johnson et al., 2010; Wozniak and Waldor, 2010). HGT of these mobile DNA elements occurs in the wet and warm environment of the intestines (Nijsten et al., 1995; Lester et al., 2004; Rowe-Magnus and Mazel, 2006; Schjorring et al., 2008; Trobos et al., 2009; Faure et al., 2010), where antibiotics have been speculated to select for successful MDR gene dissemination (Beaber et al., 2004; Hastings et al., 2004). Host-specific bacterial adhesion and local invasion of the intestinal mucosa (Figure 2A) result in a local inflammatory response that can profit HGT events (Stecher et al., 2012). Moreover cyclical colonization of the intestines (Figure 1) that leads to intestinal persistence (Figure 2B), likely creates optimal conditions and timespans for HGT events (Figure 2C), thereby increasing the antibiotic resistance gene pool (further stabilized by clonal expansion and selection if antibiotics are administered). Consistent with this notion, specific colonization of the ileum can increase the rate of intra-intestinal conjugation (Garcia-Quintanilla et al., 2008). Notably, HGT is increased at high bacterial density such as in biofilms (Licht et al., 1999; Roberts et al., 2001), the generation of which is influenced by adhesins (Reisner et al., 2003; Ledeboer et al., 2006; Dwyer et al., 2011; Spurbeck et al., 2011; Wilksch et al., 2011). In addition, HGT of antibiotic resistance genes can increase expression of some adhesins (Sahly et al., 2008), suggesting a positive feedback mechanism between intestinal colonization and HGT. Interestingly, the enhanced colonization or enteritis by Salmonella in swine or calves, respectively (Bearson et al., 2010; Pullinger et al., 2010; Verbrugghe et al., 2012) that occurs in response to host stress (transport, feed withdrawal) is associated with enhanced intestinal CF expression and increased conjugative transfer of antibiotic-resistance plasmids (Peterson et al., 2011), further supporting a link between adhesins and HGT. As the repertoire and alleles of Salmonella colonization factors influence cell binding and invasion of specific hosts, one predicts that they will have a direct impact on both intestinal colonization efficiencies and the stability and rate of conjugation.
FIGURE 2.
Model of intestinal surface colonization and HGT. (A) The success story of Salmonella evolution and adaptation to a multitude of environments and hosts is illustrated by showing how diverse the intestinal tract anatomies of just a few warm blooded animals are when comparing chicken, bovine, equine swine, and human. In addition to the distinct anatomies, histological, immunological, and microbiota particularities further participate to create unique compartments in which varieties of Salmonella developed ways to live and multiply, with a simple escape route for future settlements. (B) S. enterica expressing specific allelic adhesins for the cognate host intestinal receptors and cellular targets initiate successful colonization. Other strains lacking such ligands will not be retained as well in the host. A resulting gastroenteritis and/or persistence will result in the multiplication and possible long-term excretion of such Salmonella strains (not shown). (C) Intestinal colonization mediated by specific allelic adhesins of Salmonella (red ovals) optimizes contact (event numbers and time span) with a constant flow of new bacteria (blue ovals), some carrying antimicrobial resistance genes on conjugative or mobilizable elements (small circles in ovals), resulting in increased HGT efficiency and antibiotic-resistant S. enterica colonizing carrier hosts. With time, excretion of antibiotic-resistant S. enterica will be excreted in greater numbers than antibiotic-susceptible S. enterica (not shown).
We recently tested this prediction by investigating the association of fimbrial adhesin alleles with antimicrobial resistance for eight adhesin genes from 48 strains of S. enterica serovar Newport (Yue et al., 2012b). Phylogenic trees clearly separated each of five adhesins into two separate groups. The allelic adhesin groups correlated significantly with strains that did or did not show antimicrobial resistance. A corresponding association was found with the presence or absence of mobile DNA elements carrying antibiotic resistance genes such as plasmids or integrons. These findings further supported the notion that some Salmonella adhesin alleles might participate in the persistent colonization of specific hosts and optimize the opportunity for bacterial encounters and HGT in intestines, resulting in the accumulation of antimicrobial resistance genes in such strains. Consistent with this, a new study just determined that the resistance genes of S. enterica serovar Typhimurium DT104 were largely maintained within animal and human populations separately (Mather et al., 2013), potentially suggesting limitations in successful transmission between host species. This novel concept deserves further investigations to confirm its biological relevance by in vitro and in vivo experiments.
A PROMISING FUTURE, “NOW THIS IS NOT THE END, THIS IS NOT EVEN THE BEGINNING OF THE END, BUT IT IS PERHAPS THE END OF THE BEGINNING” (Churchill, 1943)
Future work aimed at assessing the relative importance of allelic variation in pathogenic bacteria will clearly benefit from the investigation of strains that have a complete clinical and epidemiological history. Whereas full sequencing will best serve GWAS when relevant strain metadata are available, targeted sequencing of genes of interest might be more productive by (1) permitting analysis of more strains at the same cost, (2) improving statistical power, and (3) simplifying the bioinformatics and statistical analysis. This latter approach best suits pathogens for which there are already long lists of predicted or known proteins that act as virulence factors in vitro or in animal models. Notably, the recent use of genomics and metabolomics have also renewed interest in bacterial metabolism by highlighting the importance of specific metabolic pathways to the intestinal, the extra-intestinal and particularly the intra-cellular lifestyle of Salmonella (Dandekar et al., 2012). Studies that characterized non-synonymous SNPs in metabolic enzyme genes (Allard et al., 2013), or identified distinct invasive Salmonella subpopulations characterized by allelic enzyme profiles (Saeed et al., 2006) strongly suggest that allelic proteins involved in Salmonella metabolism deserve further investigation. Similarly, allelic and phenotypic properties can be identified in regulatory proteins that participate directly or indirectly in virulence (Chen et al., 2012). Finally, while SNPs in open reading frames clearly controls the expression of allelic proteins, SNPs in cis-regulatory regions that act as operators may elicit differential recognition by downstream virulence effectors. For example, SNPs in the operator region of the srfN gene of Salmonella created a binding site for the SPI-2 regulator SsrB and a corresponding fitness gain in an oral murine model of infection (Osborne et al., 2009).
To summarize, the identification of biological consequences for allelic variants have reached the critical threshold required to stimulate more systematic investigations on well-curated strain collections, which can now exploit recently developed tools and technologies, such as GWAS and targeted sequencing.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Leslie King for critically reading the manuscript and Deborah Argento for the graphic design. This work was supported by NIH/NIAID grant AI098041, USDA grant 2013-67015-21285 and by a University of Pennsylvania Research Foundation grant.
REFERENCES
- Aballay A., Yorgey P., Ausubel F. M. (2000). Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr. Biol. 10 1539–1542 10.1016/S0960-9822(00)00830-7 [DOI] [PubMed] [Google Scholar]
- Ajiboye R. M., Solberg O. D., Lee B. M., Raphael E., Debroy C., Riley L. W. (2009). Global spread of mobile antimicrobial drug resistance determinants in human and animal Escherichia coli and Salmonella strains causing community-acquired infections. Clin. Infect. Dis. 49 365–371 10.1086/600301 [DOI] [PubMed] [Google Scholar]
- Akoachere J. F., Tanih N. F., Ndip L. M., Ndip R. N. (2009). Phenotypic characterization of Salmonella typhimurium isolates from food-animals and abattoir drains in Buea, Cameroon. J. Health Popul. Nutr. 27 612–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard M. W., Luo Y., Strain E., Pettengill J., Timme R., Wang C., et al. (2013). On the evolutionary history, population genetics and diversity among isolates of Salmonella enteritidis PFGE pattern JEGX01.0004. PLoS ONE 8:e55254 10.1371/journal.pone.0055254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen-Vercoe E., Woodward M. J. (1999). The role of flagella, but not fimbriae, in the adherence of Salmonella enterica serotype Enteritidis to chick gut explant. J. Med. Microbiol. 48 771–780 10.1099/00222615-48-8-771 [DOI] [PubMed] [Google Scholar]
- Arthurson V., Sessitsch A., Jaderlund L. (2011). Persistence and spread of Salmonella enterica serovar Weltevreden in soil and on spinach plants. FEMS Microbiol. Lett. 314 67–74 10.1111/j.1574-6968.2010.02140.x [DOI] [PubMed] [Google Scholar]
- Beaber J. W., Hochhut B., Waldor M. K. (2004). SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427 72–74 10.1038/nature02241 [DOI] [PubMed] [Google Scholar]
- Bearson B. L., Bearson S. M., Lee I. S., Brunelle B. W. (2010). The Salmonella enterica serovar Typhimurium QseB response regulator negatively regulates bacterial motility and swine colonization in the absence of the QseC sensor kinase. Microb. Pathog. 48 214–219 10.1016/j.micpath.2010.03.005 [DOI] [PubMed] [Google Scholar]
- Brodsky I. E., Medzhitov R. (2008). Reduced secretion of YopJ by Yersinia limits in vivo cell death but enhances bacterial virulence. PLoS Pathog. 4:e1000067 10.1371/journal.ppat.1000067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Call D. R., Singer R. S., Meng D., Broschat S. L., Orfe L. H., Anderson J. M., et al. (2010). blaCMY-2-positive IncA/C plasmids from Escherichia coli and Salmonella enterica are a distinct component of a larger lineage of plasmids. Antimicrob. Agents Chemother. 54 590–596 10.1128/AAC.00055-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallini A., Messa C., Mangini V., Linsalata M., Guerra V., Misciagna G., et al. (1991). Prevalence of pigment gallstones in sheep. Am. J. Vet. Res. 522043–2045 [PubMed] [Google Scholar]
- Centers for Disease and Prevention. (2013). Incidence and trends of infection with pathogens transmitted commonly through food – foodborne diseases active surveillance network, 10 U.S. sites, 1996–2012. MMWR Morb. Mortal. Wkly. Rep. 62 283–287 [PMC free article] [PubMed] [Google Scholar]
- Chen H. D., Jewett M. W., Groisman E. A. (2012). An allele of an ancestral transcription factor dependent on a horizontally acquired gene product. PLoS Genet. 8:e1003060 10.1371/journal.pgen.1003060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill W. S. (1943). “The end of the beginning. A speech at the Lord Mayor’s Day Luncheon at the Mansion House, London, 9 November 1942,” in The End of the Beginning War Speeches 1st Edn ed. Eade C. (Boston: Little, Brown and company; ) 264–269 [Google Scholar]
- Crawford R. W., Reeve K. E., Gunn J. S. (2010). Flagellated but not hyperfimbriated Salmonella enterica serovar Typhimurium attaches to and forms biofilms on cholesterol-coated surfaces. J. Bacteriol. 192 2981–2990 10.1128/JB.01620-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar T., Astrid F., Jasmin P., Hensel M. (2012). Salmonella enterica: a surprisingly well-adapted intracellular lifestyle. Front. Microbiol. 3:164. 10.3389/fmicb.2012.00164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong H. K., Parry C. M., Van Der Poll T., Wiersinga W. J. (2012). Host-pathogen interaction in invasive Salmonellosis. PLoS Pathog. 8:e1002933 10.1371/journal.ppat.1002933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatherage Kaiser B. L., Li J., Sanford J. A., Kim Y. M., Kronewitter S. R., Jones M. B., et al. (2013). A multi-omic view of host-pathogen-commensal interplay in Salmonella-mediated intestinal infection. PLoS ONE 8:e67155 10.1371/journal.pone.0067155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Bakker H. C., Moreno Switt A. I., Govoni G., Cummings C. A., Ranieri M. L., Degoricija L., et al. (2011). Genome sequencing reveals diversification of virulence factor content and possible host adaptation in distinct subpopulations of Salmonella enterica. BMC Genomics 12:425 10.1186/1471-2164-12-425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai P. T., Porwollik S., Long F., Cheng P., Wollam A., Bhonagiri-Palsikar V., et al. (2013). Evolutionary genomics of Salmonella enterica subspecies. MBio 4 e00579–e00612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsey C. W., Laarakker M. C., Humphries A. D., Weening E. H., Baumler A. J. (2005). Salmonella enterica serotype Typhimurium MisL is an intestinal colonization factor that binds fibronectin. Mol. Microbiol. 57 196–211 10.1111/j.1365-2958.2005.04666.x [DOI] [PubMed] [Google Scholar]
- Douard G., Praud K., Cloeckaert A., Doublet B. (2010). The Salmonella genomic island 1 is specifically mobilized in trans by the IncA/C multidrug resistance plasmid family. PLoS ONE 5:e15302 10.1371/journal.pone.0015302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer B. E., Newton K. L., Kisiela D., Sokurenko E. V., Clegg S. (2011). Single nucleotide polypmorphisms of fimH associated with adherence and biofilm formation by serovars of Salmonella enterica. Microbiology 157 3162–3171 10.1099/mic.0.051425-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure S., Perrin-Guyomard A., Delmas J. M., Chatre P., Laurentie M. (2010). Transfer of plasmid-mediated CTX-M-9 from Salmonella enterica serotype Virchow to Enterobacteriaceae in human flora-associated rats treated with cefixime. Antimicrob. Agents Chemother. 54 164–169 10.1128/AAC.00310-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K. A., Murray S. S., Schork N. J., Topol E. J. (2009). Human genetic variation and its contribution to complex traits. Nat. Rev. Genet. 10 241–251 10.1038/nrg2554 [DOI] [PubMed] [Google Scholar]
- Fricke W. F., Mcdermott P. F., Mammel M. K., Zhao S., Johnson T. J., Rasko D. A., et al. (2009). Antimicrobial resistance-conferring plasmids with similarity to virulence plasmids from avian pathogenic Escherichia coli strains in Salmonella enterica serovar Kentucky isolates from poultry. Appl. Environ. Microbiol. 75 5963–5971 10.1128/AEM.00786-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Quintanilla M., Ramos-Morales F., Casadesus J. (2008). Conjugal transfer of the Salmonella enterica virulence plasmid in the mouse intestine. J. Bacteriol. 190 1922–1927 10.1128/JB.01626-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach R. G., Jackel D., Geymeier N., Hensel M. (2007). Salmonella pathogenicity island 4-mediated adhesion is coregulated with invasion genes in Salmonella enterica. Infect. Immun. 75 4697–4709 10.1128/IAI.00228-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golberg D., Kroupitski Y., Belausov E., Pinto R., Sela S. (2011). Salmonella typhimurium internalization is variable in leafy vegetables and fresh herbs. Int. J. Food Microbiol. 145 250–257 10.1016/j.ijfoodmicro.2010.12.031 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Escobedo G., Gunn J. S. (2013). Gallbladder epithelium as a niche for chronic Salmonella carriage. Infect. Immun. 81 2920–2930 10.1128/IAI.00258-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Escobedo G., Marshall J. M., Gunn J. S. (2011). Chronic and acute infection of the gall bladder by Salmonella typhi: understanding the carrier state. Nat. Rev. Microbiol. 9 9–14 10.1038/nrmicro2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath S., Carden S., Monack D. (2012). Shedding light on Salmonella carriers. Trends Microbiol. 20 320–327 10.1016/j.tim.2012.04.004 [DOI] [PubMed] [Google Scholar]
- Gragg S. E., Loneragan G. H., Nightingale K. K., Brichta-Harhay D. M., Ruiz H., Elder J. R., et al. (2013). Substantial within-animal diversity of Salmonella isolates from lymph nodes, feces, and hides of cattle at slaughter. Appl. Environ. Microbiol. 79 4744–4750 10.1128/AEM.01020-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimont P. A. D., Weill F.-X. (eds). (2007). “Antigenic formulae of the Salmonella Serovars,” in WHO Collaborating Center for Reference and Research on Salmonella, 9th Edn (Paris: Institut Pasteur) 1–167 [Google Scholar]
- Guibourdenche M., Roggentin P., Mikoleit M., Fields P. I., Bockemuhl J., Grimont P. A., et al. (2010). Supplement 2003–2007 (No. 47) to the White-Kauffmann-Le minor scheme. Res. Microbiol. 161 26–29 10.1016/j.resmic.2009.10.002 [DOI] [PubMed] [Google Scholar]
- Guo A., Cao S., Tu L., Chen P., Zhang C., Jia A., et al. (2009). FimH alleles direct preferential binding of Salmonella to distinct mammalian cells or to avian cells. Microbiology 155 1623–1633 10.1099/mic.0.026286-0 [DOI] [PubMed] [Google Scholar]
- Hastings P. J., Rosenberg S. M., Slack A. (2004). Antibiotic-induced lateral transfer of antibiotic resistance. Trends Microbiol. 12 401–404 10.1016/j.tim.2004.07.003 [DOI] [PubMed] [Google Scholar]
- He G. Z., Tian W. Y., Qian N., Cheng A. C., Deng S. X. (2010). Quantitative studies of the distribution pattern for Salmonella enteritidis in the internal organs of chicken after oral challenge by a real-time PCR. Vet. Res. Commun. 34 669–676 10.1007/s11259-010-9438-6 [DOI] [PubMed] [Google Scholar]
- Heffron F., Niemann G., Yoon H., Kidwai A., Brown R. N. E., Mcdermott J. D., et al. (2011). “Salmonella-secreted virulence factors,” in Salmonella: From Genome to Function ed. Porwollik S. (Norflok: Caister Academic Press; ) 187–223 [Google Scholar]
- Hirsh D. C. (2004). “Enterobacteriaceae: Salmonella,” in Veterinary Microbiology eds Hirsh D. C., Maclachlan N. J., Walker R. L. (Ames, IA: Blackwell Publishing; ) 69–74 [Google Scholar]
- Holt K. E., Thomson N. R., Wain J., Langridge G. C., Hasan R., Bhutta Z. A., et al. (2009). Pseudogene accumulation in the evolutionary histories of Salmonella enterica serovars Paratyphi A and Typhi. BMC Genomics 10:36 10.1186/1471-2164-10-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins K. L., Threlfall E. J. (2004). Frequency and polymorphism of sopE in isolates of Salmonella enterica belonging to the ten most prevalent serotypes in England and Wales. J. Med. Microbiol. 53 539–543 10.1099/jmm.0.05510-0 [DOI] [PubMed] [Google Scholar]
- Humphries A. D., Townsend S. M., Kingsley R. A., Nicholson T. L., Tsolis R. M., Baumler A. J. (2001). Role of fimbriae as antigens and intestinal colonization factors of Salmonella serovars. FEMS Microbiol. Lett. 201 121–125 10.1111/j.1574-6968.2001.tb10744.x [DOI] [PubMed] [Google Scholar]
- Johnson T. J., Thorsness J. L., Anderson C. P., Lynne A. M., Foley S. L., Han J., et al. (2010). Horizontal gene transfer of a ColV plasmid has resulted in a dominant avian clonal type of Salmonella enterica serovar Kentucky. PLoS ONE 5:e15524 10.1371/journal.pone.0015524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen A., Vasilakos A. V., Liang Y. (2009). Computational intelligence in bioinformatics: SNP/haplotype data in genetic association study for common diseases. IEEE Trans. Inf. Technol. Biomed. 13 841–847 10.1109/TITB.2009.2024144 [DOI] [PubMed] [Google Scholar]
- Kingsley R. A., Abi Ghanem D., Puebla-Osorio N., Keestra A. M., Berghman L., Baumler A. J. (2004). Fibronectin binding to the Salmonella enterica serotype Typhimurium ShdA autotransporter protein is inhibited by a monoclonal antibody recognizing the A3 repeat. J. Bacteriol. 186 4931–4939 10.1128/JB.186.15.4931-4939.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley R. A., Humphries A. D., Weening E. H., De Zoete M. R., Winter S., Papaconstantinopoulou A., et al. (2003). Molecular and phenotypic analysis of the CS54 island of Salmonella enterica serotype Typhimurium: identification of intestinal colonization and persistence determinants. Infect. Immun. 71 629–640 10.1128/IAI.71.2.629-640.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisiela D., Sapeta A., Kuczkowski M., Stefaniak T., Wieliczko A., Ugorski M. (2005). Characterization of FimH adhesins expressed by Salmonella enterica serovar Gallinarum biovars Gallinarum and Pullorum: reconstitution of mannose-binding properties by single amino acid substitution. Infect. Immun. 73 6187–6190 10.1128/IAI.73.9.6187-6190.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler L. A., Vallance B. A., Celli J., Winfree S., Hansen B., Montero M., et al. (2010). Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc. Natl. Acad. Sci. U.S.A. 107 17733–17738 10.1073/pnas.1006098107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korhonen T. K. (2007). “Adhesins of Salmonella and their putative roles in infection,” in Salmonella: Molecular biology and pathogenesis eds Rhen M., Maskell D., Mastroeni P., Threlfall J. (Norfolk: Horizon Bioscience; ) 53–66 [Google Scholar]
- Kuzminska-Bajor M., Kuczkowski M., Grzymajlo K., Wojciech L., Sabat M., Kisiela D., et al. (2012). Decreased colonization of chicks by Salmonella enterica serovar Gallinarum expressing mannose-sensitive FimH adhesin from Salmonella enterica serovar Enteritidis. Vet. Microbiol. 158 205–210 10.1016/j.vetmic.2012.01.029 [DOI] [PubMed] [Google Scholar]
- Latasa C., Roux A., Toledo-Arana A., Ghigo J. M., Gamazo C., Penades J. R., et al. (2005). BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol. Microbiol. 58 1322–1339 10.1111/j.1365-2958.2005.04907.x [DOI] [PubMed] [Google Scholar]
- Lawson B., Hughes L. A., Peters T., De Pinna E., John S. K., Macgregor S. K., et al. (2011). Pulsed-field gel electrophoresis supports the presence of host-adapted Salmonella enterica subsp. enterica serovar Typhimurium strains in the British garden bird population. Appl. Environ. Microbiol. 77 8139–8144 10.1128/AEM.00131-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledeboer N. A., Frye J. G., McClelland M., Jones B. D. (2006). Salmonella enterica serovar Typhimurium requires the Lpf, Pef, and Tafi fimbriae for biofilm formation on HEp-2 tissue culture cells and chicken intestinal epithelium. Infect. Immun. 74 3156–3169 10.1128/IAI.01428-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester C. H., Frimodt-Moller N., Hammerum A. M. (2004). Conjugal transfer of aminoglycoside and macrolide resistance between Enterococcus faecium isolates in the intestine of streptomycin-treated mice. FEMS Microbiol. Lett. 235 385–391 10.1111/j.1574-6968.2004.tb09614.x [DOI] [PubMed] [Google Scholar]
- Leyton D. L., Rossiter A. E., Henderson I. R. (2012). From self sufficiency to dependence: mechanisms and factors important for autotransporter biogenesis. Nat. Rev. Microbiol. 10 213–225 10.1038/nrmicro2733 [DOI] [PubMed] [Google Scholar]
- Licht T. R., Christensen B. B., Krogfelt K. A., Molin S. (1999). Plasmid transfer in the animal intestine and other dynamic bacterial populations: the role of community structure and environment. Microbiology 145 2615–2622 [DOI] [PubMed] [Google Scholar]
- Lindsey R. L., Fedorka-Cray P. J., Frye J. G., Meinersmann R. J. (2009). Inc A/C plasmids are prevalent in multidrug-resistant Salmonella enterica isolates. Appl. Environ. Microbiol. 75 1908–1915 10.1128/AEM.02228-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Dong F. F., Stavrinides J., Guttman D. S. (2006). Type III effector diversification via both pathoadaptation and horizontal transfer in response to a coevolutionary arms race. PLoS Genet. 2:e209 10.1371/journal.pgen.0020209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather A. E., Reid S. W., Maskell D. J., Parkhill J., Fookes M. C., Harris S. R., et al. (2013). Distinguishable epidemics of multidrug-resistant Salmonella typhimurium DT104 in different hosts. Science 341 1514–1517 10.1126/science.1240578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott P. F. (2006). “Antimicrobial resistance in nontyphoidal Salmonellae,” in Antimicrobial Resistance in Bacteria of Animal Origin ed. Aarestrup F. M. (Washington, DC: ASM Press; ) 293–314 [Google Scholar]
- McLaughlin L. M., Govoni G. R., Gerke C., Gopinath S., Peng K., Laidlaw G., et al. (2009). The Salmonella SPI2 effector SseI mediates long-term systemic infection by modulating host cell migration. PLoS Pathog. 5:e1000671 10.1371/journal.ppat.1000671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M., Stenzel U., Hofreiter M. (2008). Parallel tagged sequencing on the 454 platform. Nat. Protoc. 3 267–278 10.1038/nprot.2007.520 [DOI] [PubMed] [Google Scholar]
- Misselwitz B., Barrett N., Kreibich S., Vonaesch P., Andritschke D., Rout S., et al. (2012). Near surface swimming of Salmonella typhimurium explains target-site selection and cooperative invasion. PLoS Pathog. 8:e1002810 10.1371/journal.ppat.1002810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misselwitz B., Kreibich S. K., Rout S., Stecher B., Periaswamy B., Hardt W. D. (2011). Salmonella enterica serovar Typhimurium binds to HeLa cells via Fim-mediated reversible adhesion and irreversible type three secretion system 1-mediated docking. Infect. Immun. 79 330–341 10.1128/IAI.00581-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monack D. M., Bouley D. M., Falkow S. (2004). Salmonella typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNgamma neutralization. J. Exp. Med. 199 231–241 10.1084/jem.20031319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan E., Campbell J. D., Rowe S. C., Bispham J., Stevens M. P., Bowen A. J., et al. (2004). Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 54 994–1010 10.1111/j.1365-2958.2004.04323.x [DOI] [PubMed] [Google Scholar]
- Muller A. J., Kaiser P., Dittmar K. E., Weber T. C., Haueter S., Endt K., et al. (2012). Salmonella gut invasion involves TTSS-2-dependent epithelial traversal, basolateral exit, and uptake by epithelium-sampling lamina propria phagocytes. Cell. Host Microbe 11 19–32 10.1016/j.chom.2011.11.013 [DOI] [PubMed] [Google Scholar]
- Nijsten R., London N., Van Den Bogaard A., Stobberingh E. (1995). In-vivo transfer of resistance plasmids in rat, human or pig-derived intestinal flora using a rat model. J. Antimicrob. Chemother. 36 975–985 10.1093/jac/36.6.975 [DOI] [PubMed] [Google Scholar]
- Oboegbulem S. I., Muogbo E. N. (1981). A survey of Salmonellae in trade cattle slaughtered at Nsukka abattoir. Int. J. Zoonoses 8 107–110 [PubMed] [Google Scholar]
- Okoro C. K., Kingsley R. A., Connor T. R., Harris S. R., Parry C. M., Al-Mashhadani M. N., et al. (2012). Intracontinental spread of human invasive Salmonella typhimurium pathovariants in sub-Saharan Africa. Nat. Genet. 44 1215–1221 10.1038/ng.2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne S. E., Walthers D., Tomljenovic A. M., Mulder D. T., Silphaduang U., Duong N., et al. (2009). Pathogenic adaptation of intracellular bacteria by rewiring a cis-regulatory input function. Proc. Natl. Acad. Sci. U.S.A. 106 3982–3987 10.1073/pnas.0811669106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini-Masini A., Dolente B. A., Habecker P. L., Jesty S. A. (2004). Myonecrosis and cutaneous infarction associated with Salmonella serovar Infantum infection in a horse. J. Am. Vet. Med. Assoc. 225 722–725 10.2460/javma.2004.225.722 [DOI] [PubMed] [Google Scholar]
- Peterson G., Kumar A., Gart E., Narayanan S. (2011). Catecholamines increase conjugative gene transfer between enteric bacteria. Microb. Pathog. 51 1–8 10.1016/j.micpath.2011.03.002 [DOI] [PubMed] [Google Scholar]
- Pezoa D., Yang H. J., Blondel C. J., Santiviago C. A., Andrews-Polymenis H. L., Contreras I. (2013). The type VI secretion system encoded in SPI-6 plays a role in gastrointestinal colonization and systemic spread of Salmonella enterica serovar Typhimurium in the chicken. PLoS ONE 8:e63917 10.1371/journal.pone.0063917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porwollik S., Wong R. M., Mcclelland M. (2002). Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl. Acad. Sci. U.S.A. 99 8956–8961 10.1073/pnas.122153699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullinger G. D., Carnell S. C., Sharaff F. F., Van Diemen P. M., Dziva F., Morgan E., et al. (2010). Norepinephrine augments Salmonella enterica-induced enteritis in a manner associated with increased net replication but independent of the putative adrenergic sensor kinases QseC and QseE. Infect. Immun. 78 372–380 10.1128/IAI.01203-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabsch W., Andrews H. L., Kingsley R. A., Prager R., Tschape H., Adams L. G., et al. (2002). Salmonella enterica serotype Typhimurium and its host-adapted variants. Infect. Immun. 70 2249–2255 10.1128/IAI.70.5.2249-2255.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M., Sun Y. H., Wilson R. P., Tran Q. T., Chessa D., Andrews-Polymenis H. L., et al. (2005). Host restriction of Salmonella enterica serotype Typhi is not caused by functional alteration of SipA, SopB, or SopD. Infect. Immun. 73 7817–7826 10.1128/IAI.73.12.7817-7826.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan D., Wells T. J., Morris F. C., Shaw R. K., Bobat S., Peters S. E., et al. (2011). SadA, a trimeric autotransporter from Salmonella enterica serovar Typhimurium, can promote biofilm formation and provides limited protection against infection. Infect. Immun. 79 4342–4352 10.1128/IAI.05592-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisner A., Haagensen J. A., Schembri M. A., Zechner E. L., Molin S. (2003). Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48 933–946 10.1046/j.1365-2958.2003.03490.x [DOI] [PubMed] [Google Scholar]
- Rivera-Chavez F., Winter S. E., Lopez C. A., Xavier M. N., Winter M. G., Nuccio S. P., et al. (2013). Salmonella uses energy taxis to benefit from intestinal inflammation. PLoS Pathog. 9:e1003267 10.1371/journal.ppat.1003267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A. P., Mullany P., Wilson M. (2001). Gene transfer in bacterial biofilms. Methods Enzymol. 336 60–65 10.1016/S0076-6879(01)36578-3 [DOI] [PubMed] [Google Scholar]
- Robertson J. M., Grant G., Allen-Vercoe E., Woodward M. J., Pusztai A., Flint H. J. (2000). Adhesion of Salmonella enterica var Enteritidis strains lacking fimbriae and flagella to rat ileal explants cultured at the air interface or submerged in tissue culture medium. J. Med. Microbiol. 49 691–696 [DOI] [PubMed] [Google Scholar]
- Rowe-Magnus D., Mazel D. (2006). “The evolution of antibiotic resistance,” in Evolution of Bacterial Pathogens eds Seifert H. S., Dirita V. J. (Washington, DC: ASM Press; ) 221–244 [Google Scholar]
- Ruby T., McLaughlin L., Gopinath S., Monack D. (2012). Salmonella’s long-term relationship with its host. FEMS Microbiol. Rev. 36 600–615 10.1111/j.1574-6976.2012.00332.x [DOI] [PubMed] [Google Scholar]
- Saeed A. M., Walk S. T., Arshad M., Whittam T. S. (2006). Clonal structure and variation in virulence of Salmonella enteritidis isolated from mice, chickens, and humans. J. AOAC Int. 89 504–511 [PubMed] [Google Scholar]
- Sahly H., Navon-Venezia S., Roesler L., Hay A., Carmeli Y., Podschun R., et al. (2008). Extended-spectrum beta-lactamase production is associated with an increase in cell invasion and expression of fimbrial adhesins in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 52 3029–3034 10.1128/AAC.00010-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangal V., Harbottle H., Mazzoni C. J., Helmuth R., Guerra B., Didelot X., et al. (2010). Evolution and population structure of Salmonella enterica serovar Newport. J. Bacteriol. 192 6465–6476 10.1128/JB.00969-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Kumita W., Ode T., Ichinose S., Ando A., Fujiyama Y., et al. (2010). OmpA variants affecting the adherence of ulcerative colitis-derived Bacteroides vulgatus. J. Med. Dent. Sci. 57 55–64 [PubMed] [Google Scholar]
- Schjorring S., Struve C., Krogfelt K. A. (2008). Transfer of antimicrobial resistance plasmids from Klebsiella pneumoniae to Escherichia coli in the mouse intestine. J. Antimicrob. Chemother. 62 1086–1093 10.1093/jac/dkn323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlumberger M. C., Friebel A., Buchwald G., Scheffzek K., Wittinghofer A., Hardt W. D. (2003). Amino acids of the bacterial toxin SopE involved in G nucleotide exchange on Cdc42. J. Biol. Chem. 278 27149–27159 10.1074/jbc.M302475200 [DOI] [PubMed] [Google Scholar]
- Schlumberger M. C., Hardt W. D. (2005). Triggered phagocytosis by Salmonella: bacterial molecular mimicry of RhoGTPase activation/deactivation. Curr. Top. Microbiol. Immunol. 291 29–42 10.1007/3-540-27511-8_3 [DOI] [PubMed] [Google Scholar]
- Soyer Y., Orsi R. H., Rodriguez-Rivera L. D., Sun Q., Wiedmann M. (2009). Genome wide evolutionary analyses reveal serotype specific patterns of positive selection in selected Salmonella serotypes. BMC Evol. Biol. 9:264 10.1186/1471-2148-9-264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurbeck R. R., Stapleton A. E., Johnson J. R., Walk S. T., Hooton T. M., Mobley H. L. (2011). Fimbrial profiles predict virulence of uropathogenic E. coli strains: contribution of Ygi and Yad fimbriae. Infect. Immun. 79 4753–4763 10.1128/IAI.05621-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B., Denzler R., Maier L., Bernet F., Sanders M. J., Pickard D. J., et al. (2012). Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc. Natl. Acad. Sci. U.S.A. 109 1269–1274 10.1073/pnas.1113246109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeb B., Claudi B., Burton N. A., Tienz P., Schmidt A., Farhan H., et al. (2013). Parallel exploitation of diverse host nutrients enhances Salmonella virulence. PLoS Pathog. 9:e1003301 10.1371/journal.ppat.1003301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens M. P., Humphrey T. J., Maskell D. J. (2009). Molecular insights into farm animal and zoonotic Salmonella infections. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364 2709–2723 10.1098/rstb.2009.0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockbauer K. E., Magoun L., Liu M., Burns E. H., Jr., Gubba S., Renish S., et al. (1999). A natural variant of the cysteine protease virulence factor of group A Streptococcus with an arginine-glycine-aspartic acid (RGD) motif preferentially binds human integrins alphavbeta3 and alphaIIbbeta3. Proc. Natl. Acad. Sci. U.S.A. 96 242–247 10.1073/pnas.96.1.242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger B. E., Stahl E. A., Raj T. (2011). Progress and promise of genome-wide association studies for human complex trait genetics. Genetics 187 367–383 10.1534/genetics.110.120907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L. H., Chu C., Cloeckaert A., Chiu C. H. (2008). An epidemic of plasmids? Dissemination of extended-spectrum cephalosporinases among Salmonella and other Enterobacteriaceae. FEMS Immunol. Med. Microbiol. 52 155–168 10.1111/j.1574-695X.2007.00360.x [DOI] [PubMed] [Google Scholar]
- Swart A. L., Hensel M. (2012). Interactions of Salmonella enterica with dendritic cells. Virulence 3 660–667 10.4161/viru.22761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tampakakis E., Peleg A. Y., Mylonakis E. (2009). Interaction of Candida albicans with an intestinal pathogen, Salmonella enterica serovar Typhimurium. Eukaryot. Cell 8 732–737 10.1128/EC.00016-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornbrough J. M., Worley M. J. (2012). A naturally occurring single nucleotide polymorphism in the Salmonella SPI-2 type III effector srfH/sseI controls early extraintestinal dissemination. PLoS ONE 7:e45245 10.1371/journal.pone.0045245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tindall B. J., Grimont P. A., Garrity G. M., Euzeby J. P. (2005). Nomenclature and taxonomy of the genus Salmonella. Int. J. Syst. Evol. Microbiol. 55 521–524 10.1099/ijs.0.63580-0 [DOI] [PubMed] [Google Scholar]
- Trobos M., Lester C. H., Olsen J. E., Frimodt-Moller N., Hammerum A. M. (2009). Natural transfer of sulphonamide and ampicillin resistance between Escherichia coli residing in the human intestine. J. Antimicrob. Chemother. 63 80–86 10.1093/jac/dkn437 [DOI] [PubMed] [Google Scholar]
- Velge P., Wiedemann A., Rosselin M., Abed N., Boumart Z., Chausse A. M., et al. (2012). Multiplicity of Salmonella entry mechanisms, a new paradigm for Salmonella pathogenesis. Microbiologyopen 1 243–258 10.1002/mbo3.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbrugghe E., Boyen F., Gaastra W., Bekhuis L., Leyman B., Van Parys A., et al. (2012). The complex interplay between stress and bacterial infections in animals. Vet. Microbiol. 155 115–127 10.1016/j.vetmic.2011.09.012 [DOI] [PubMed] [Google Scholar]
- Vo A. T., Van Duijkeren E., Fluit A. C., Gaastra W. (2007). A novel Salmonella genomic island 1 and rare integron types in Salmonella typhimurium isolates from horses in The Netherlands. J. Antimicrob. Chemother. 59 594–599 10.1093/jac/dkl531 [DOI] [PubMed] [Google Scholar]
- Wagner C., Hensel M. (2011). Adhesive mechanisms of Salmonella enterica. Adv. Exp. Med. Biol. 715 17–34 10.1007/978-94-007-0940-9_2 [DOI] [PubMed] [Google Scholar]
- Wales A. D., Carrique-Mas J. J., Rankin M., Bell B., Thind B. B., Davies R. H. (2010). Review of the carriage of zoonotic bacteria by arthropods, with special reference to Salmonella in mites, flies and litter beetles. Zoonoses Public Health 57 299–314 10.1111/j.1863-2378.2008.01222.x. [DOI] [PubMed] [Google Scholar]
- Welch T. J., Fricke W. F., Mcdermott P. F., White D. G., Rosso M. L., Rasko D. A., et al. (2007). Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS ONE 2:e309 10.1371/journal.pone.0000309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildschutte H., Lawrence J. G. (2007). Differential Salmonella survival against communities of intestinal amoebae. Microbiology 153 1781–1789 10.1099/mic.0.2006/003616-0 [DOI] [PubMed] [Google Scholar]
- Wilksch J. J., Yang J., Clements A., Gabbe J. L., Short K. R., Cao H., et al. (2011). MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS pathogens 7:e1002204 10.1371/journal.ppat.1002204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter S. E., Thiennimitr P., Winter M. G., Butler B. P., Huseby D. L., Crawford R. W., et al. (2010). Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467 426–429 10.1038/nature09415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood R. L., Rose R., Coe N. E., Ferris K. E. (1991). Experimental establishment of persistent infection in swine with a zoonotic strain of Salmonella newport. Am. J. Vet. Res. 52 813–819 [PubMed] [Google Scholar]
- Worley M. J., Nieman G. S., Geddes K., Heffron F. (2006). Salmonella typhimurium disseminates within its host by manipulating the motility of infected cells. Proc. Natl. Acad. Sci. U.S.A. 103 17915–17920 10.1073/pnas.0604054103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak R. A., Waldor M. K. (2010). Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8 552–563 10.1038/nrmicro2382 [DOI] [PubMed] [Google Scholar]
- Yeh Y. C., Chang W. L., Yang H. B., Cheng H. C., Wu J. J., Sheu B. S. (2011). H. pylori cagL amino acid sequence polymorphism Y58E59 induces a corpus shift of gastric integrin alpha5beta1 related with gastric carcinogenesis. Mol. Carcinog. 50 751–759 10.1002/mc.20753 [DOI] [PubMed] [Google Scholar]
- Yue M., Rankin S. C., Blanchet R. T., Nulton J. D., Edwards R. A., Schifferli D. M. (2012a). Diversification of the Salmonella fimbriae: a model of macro- and microevolution. PLoS ONE 7:e38596 10.1371/journal.pone.0038596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue M., Schmieder R., Edwards R. A., Rankin S. C., Schifferli D. M. (2012b). Microfluidic PCR combined with pyrosequencing for identification of allelic variants with phenotypic associations among targeted Salmonella genes. Appl. Environ. Microbiol. 78 7480–7482 [DOI] [PMC free article] [PubMed] [Google Scholar]