Abstract
Muscle atrophy can result from inactivity or unloading on one hand or the induction of a catabolic state on the other. Muscle-specific ring finger 1 (MuRF1), a member of the tripartite motif family of E3 ubiquitin ligases, is an essential mediator of multiple conditions inducing muscle atrophy. While most studies have focused on the role of MuRF1 in protein degradation, the protein may have other roles in regulating skeletal muscle mass and metabolism. We therefore systematically evaluated the effect of MuRF1 on gene expression during denervation and dexamethasone-induced atrophy. We find that the lack of MuRF1 leads to few differences in control animals, but there were several significant differences in specific sets of genes upon denervation- and dexamethasone-induced atrophy. For example, during denervation, MuRF1 knockout mice showed delayed repression of metabolic and structural genes and blunted induction of genes associated with the neuromuscular junction. In the latter case, this pattern correlates with blunted HDAC4 and myogenin upregulation. Lack of MuRF1 caused fewer changes in the dexamethasone-induced atrophy program, but certain genes involved in fat metabolism and intracellular signaling were affected. Our results demonstrate a new role for MuRF1 in influencing gene expression in two important models of muscle atrophy.
Keywords: denervation, glucocorticoid, microarray, skeletal muscle, gene expression
two e3 ubiquitin ligase-encoding genes, muscle-specific ring finger 1 (MuRF1) and MAFbx (or atrogin-1), are upregulated in multiple skeletal muscle atrophy models, downregulated in hypertrophy, and when deleted in mice result in muscle sparing in certain atrophy-inducing conditions (4, 5, 32). The discovery of a link between these genes and muscle atrophy a decade ago led to intensive research into the roles that the encoded proteins play in atrophy and their potential downstream targets. We have focused mostly on MuRF1 since a null deletion of MuRF1, but not MAFbx spares muscle mass in both the denervation (DEN)- and dexamethasone (DEX)-induced atrophy models (4, 5). Furthermore, while MAFbx knockout (KO) mice show greater sparing of mass than MuRF1 KO mice after DEN, MAFbx KO mice show increased fiber necrosis and degeneration over time (18). Thus, MuRF1 expression is critical for induction of atrophy under multiple conditions and preventing its activity spares both muscle mass and function.
Most of the work on the function of MuRF1 has focused on its role in mediating protein degradation via the ubiquitin proteasome system (UPS). For example, MuRF1 has been proposed to target myosin heavy chain (MHC) and other thick filament proteins for polyubiquitination and degradation by the 26S proteasome (9, 10, 25). While the UPS remains a focus of current research in the field, there may be important additional roles for MuRF1 in control of muscle mass and function in response to various hormonal challenges or with changes in neuromuscular activity. For example, reductions in protein synthesis due to DEX treatment (4) or amino acid deprivation (29) are reduced in MuRF1 KO mice. Furthermore, skeletal muscle- and cardiac-specific overexpression of MuRF1 in vivo affects the expression and activity of specific metabolic enzymes (23) without obvious effects on skeletal muscle mass or cardiac muscle protein ubiquitination and contractile protein levels (62). Whether these changes are due to MuRF1 targeting proteins involved in protein synthesis or metabolism for degradation by the UPS or by another mechanism altogether is not known; regardless, the view that MuRF1 is strictly involved in degradation of structural proteins by the UPS is beginning to be re-evaluated.
In the course of studying MuRF1 function in DEN- and DEX-induced atrophy, we observed that FOXO1 expression was inhibited in DEX-treated MuRF1 KO mice (4), and MAFbx expression was prolonged in denervated muscles of MuRF1 KO mice (18). Based on these observations, we hypothesized that MuRF1 might be involved, either directly or indirectly, in changes in the expression of a larger set of specific mRNAs during atrophy. This hypothesis is supported by previous findings that MuRF1 is not only found at the M line where it was first discovered (6) but can also be detected as tagged fusion proteins in cardiac myonuclei (37) and C2C12 myotubes, (13). In addition, endogenous MuRF1 shows nuclear translocation in a model of skeletal muscle unloading (43). MuRF1 has been shown to physically interact with known transcription factors in vitro, including SRF (61), which plays an important role in cardiac and skeletal muscle structural gene expression, and GMEB-1 (29, 37), which has been implicated in glucocorticoid receptor activity. Finally, nuclear UPS activity is known to be important for transcriptional regulation, by regulated degradation of transcription factor complexes via the classical pathway (17), as well as via nonproteolytic monoubiquitination or ubiquitin chains modifying the localization and activity of multiple transcription factors (17, 45, 46). Furthermore, other members of the tripartite motif ring finger protein gene family to which MuRF1 belongs are known to act as direct transcriptional coactivators (e.g., Ref. 22).
In an ongoing effort to understand the full range of roles that MuRF1 may play in skeletal muscle structure and function, we explored the possibility that lack of MuRF1 may affect gene expression during muscle atrophy. Our data demonstrate that the expression of a suite of genes clustered into specific functional classes is altered following DEN and DEX treatment and suggests a new role for MuRF1 in mediating muscle atrophy.
MATERIALS AND METHODS
Animal treatments.
Four- to six-month-old, female MuRF1−/− (KO) and wild-type (WT) littermate mice on a C57/Bl6 background were treated for 3 or 14 days with 3 mg/kg water-soluble DEX in their drinking water (DEX, Sigma) or used as age- and sex-matched control animals for each group (DEX treatment), as previously described (4). The sciatic nerve was transected in the midthigh to produce DEN of the lower limb as described elsewhere (18). All animal experiments were carried out in accordance with an approved University of California (UC) Davis Institutional Animal Use and Care Committee protocol.
RNA isolation and microarray analysis.
Three separate animals were used for each time point and genotype and processed in parallel as biological triplicates. Additional animals were included for subsequent microarray validation by quantitative PCR. Total RNA from denervated, DEX-treated, or control triceps surae (TS) complexes were prepared by polytron homogenization in Trizol reagent (Invitrogen) according to the manufacturer's recommendation. Prior to labeling, RNA was quantified with a NanoDrop ND-100 spectrophotometer (NanoDrop Technologies), and RNA quality was further assessed with an Agilent 2100 Bioanalyzer (Agilent Technologies). For microarray analysis, RNA isolation and labeling were performed for control, DEX-treated, and denervated TS as described previously (18). Labeled cRNAs were hybridized to Illumina Mouse BeadChip WG-6 arrays; these arrays contain 46,632 probes representing 34,551 genes, and each probe is represented on the chip ∼90 times. Several genes had multiple probes, and in these cases we used the averaged signal over all probes. Background signals were subtracted, and we demanded at least one treatment group have an average signal twice that of background (>150 average signal DEN animals and >50 average signal DEX animals) and a detection P value < 0.01. Data was analyzed with Illumina Beadstudio v3.2.3 software, using quantile normalization. An expression difference was called significant if it changed by more than twofold from matched controls and by a two-tailed t-test with a P value < 0.05. Additional comparisons for selected genes were analyzed by one-way ANOVA with multiple comparisons and Tukey's multiple-comparison tests (GraphPad Prism). Significant, twofold differentially regulated genes were further analyzed by the DAVID algorithm (24) and clustered into Gene Ontology pathways. Microarray data was submitted to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus with the SuperSeries identification GSE44259.
Quantitative real-time PCR.
All cDNAs used for RT-PCR assays were made with a Qiagen Quantitech reverse-transcription kit with 1 μg of total TS RNA, as described previously (4, 59). Commercially available Taqman probe sets (Applied Biosystems) were used for MuRF1 (TRIM63, Mm01185221_m1), myogenin (MYOG, Mm00446195_g1), and myosin binding protein C2 (MYBPC2, Mm00525419_m1), with ribosomal protein L22 (RPL22, Mm04213302_s1) as normalizer. Unlabeled primer sets for arrestin domain containing 2 (ARRDC2; forward 5′-GGACATAAACACACCTGCCC-3′, reverse 5′-CACGTGTGCAGTACCAGGAT-3′), metallothionein 2 (MT2; forward 5′-ATAGACCATGTAGAAGCCTAGCCTTT-3′, reverse 5′-GGCTTTTATTGTCAGTTACATGCTTTATAG-3′), collagen 1a1 (COL1A1; forward 5′-AGAGCCTGAGTCAGCAGATTGAG-3′, reverse 5′-CCAGTACTCTCCGCTCTTCCA-3′), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 5′-CCAGCCTCGTCCCGTAGAC-3′, 5′-ATGGCAACAATCTCCACTTTGC-3′) were used for SYBR green-based assays. Quantitative PCR was carried out on an Applied Biosystems Prism 7900 HTA FAST machine in a 384-well format at the UC Davis Real-time PCR Research and Diagnostics Core Facility, using serially diluted cDNA as the standard curve.
Semiquantitative RT-PCR of proline-rich nuclear receptor coactivator 2 gene expression and 3′-untranslated region sequencing.
All PCR reactions were performed using cDNA made as described above using an Invitrogen PCR kit with the following cycling conditions: 10 min initial denaturation at 94°C, followed by 26 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. Primer sets were chosen to amplify regions of the gene in a relatively evenly distributed fashion across 3′-untranslated region (UTR) (set 1 upstream primer 5-CCAACCATCTTAAACCCAAAGA-3′, set 1 downstream primer 5′-AAATGAGGGGTTGCACATAATC-3′; set 2 upstream primer 5-CTTGTGCTTAAGTGCCAGAGTG-3′, set 2 downstream primer 5′-TCTTTGGGTTTAAGATGGTTGG-3′; set 3 upstream primer 5-CTTGTGCTTAAGTGCCAGAGTG-3′, set 3 downstream primer 5′-CAGGGGTGACTTAGGGTTGATA-3′; set 4 upstream primer 5-TGCTTAAGTGCCAGAGTGAATC-3′, set 4 downstream primer 5′-GGGTTGATAAAGCCAAAATGAG-3′). The specific region in the 3′-UTR of the proline-rich nuclear receptor coactivator 2 (PNRC2) gene bracketing the microarray probe was amplified by a 40-cycle reaction using the following primers: upstream primer, 5′-AAGGTTGCACAGTTCAGTTTCA-3′; downstream primer, 5′-CCAGGATCACTTTAGCAGCTT-3′, and purified with a Qiagen QIA-quick PCR purification kit following the manufacturer's instructions. Purified PCR amplicons were then ligated into a Promega pGEM T-easy vector, and subcloned plasmids were purified using a Qiagen Plasmid Mini-Prep kit. Inserts in purified plasmids were sent for sequencing by the UC Davis UCDNA sequencing facility using the common T7 primer. Sequences were compared with reference sequence available from Ensembl and aligned with the BLAST feature of the NCBI database (the sequence used by Illumina to manufacture probes). Each experimental group (WT and MuRF1 KO controls) was examined in triplicate (3 individual mice from each group), and each individual mouse was sequenced twice to verify results. All inserts from a given genotype yielded the same results.
Western blot analysis.
Western blot of total TS protein extracts was performed as described previously. Anti-histone deacetylase (HDAC)4 antisera was a kind gift of Tso-Pang Yao (Duke University). Anti-MYOG and GAPDH antibodies were purchased from Santa Cruz and Cell Signaling, respectively.
RESULTS
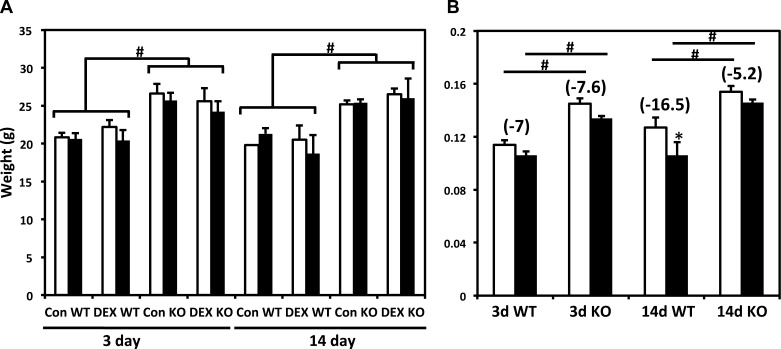
To investigate whether MuRF1 plays a role in gene expression changes during muscle atrophy, we examined DEN induced by sciatic nerve resection and chronic glucocorticoid treatment with DEX, two extensively studied atrophy models representing muscle inactivity (DEN) and catabolic states (DEX). Previously, we reported microarray results of denervated TS muscles to specifically compare the UPS-associated gene expression profiles of WT and MuRF1 KO mice (18). Interestingly, the mice showed elevated proteasome activity with reduced markers of autophagy, and protein synthesis rates were unchanged between genotypes. Here, we report an expanded genome-wide analysis and compare the gene expression profiles of DEN-induced atrophy to excess glucocorticoid-induced atrophy in TS muscle. In addition, we compared the response of WT and MuRF1 KO mice to each of these atrophy conditions. Two time points were selected for analyses: 3 days of DEN or DEX treatment, when MuRF1 expression is highly induced in both models, and slight but not significant decreases in wet weights have occurred in both WT and KO mice; and 14 days when significant wet weight loss has occurred in WT mice, and significant sparing of mass is evident in the MuRF1 KO. Figure 1 shows the muscle mass and body weights for DEX-treated animals used in this particular study. We previously demonstrated that DEX treatment induced atrophy that was blunted in MuRF1 KO mice, as was a reduction in protein synthesis, with maintenance of force output (4). The magnitude of muscle loss was much greater for denervated- vs. DEX-treated muscles; roughly 17% for 14-day DEX-treated TS (Fig. 1) and 41% for denervated WT TS that we reported elsewhere (18).
Fig. 1.
Body weights and triceps surae wet weights from control (Con) and dexamethasone (DEX)-treated animals. A: body weight measurements before (white bars) and after (black bars) treatment with (DEX) or without (Con) 3 mg/kg DEX in the drinking water for the indicated days. WT, wild-type animals; KO, muscle-specific ring finger 1 (MuRF1) knockout animals. #Bracketed groups are significantly different from each other, P < 0.05. B: right triceps surae wet weights of untreated (white bars) or 3 mg/kg DEX-treated (black bars). Numbers in parentheses indicate % change in wet weight with DEX treatment. #Significantly different from between groups, P < 0.05; *Different from controls in same group, P < 0.05. Weights in grams are shown on the y-axis.
Microarray analysis identifies the induced gene expression programs in skeletal muscle after 3 and 14 days of DEN or DEX treatment.
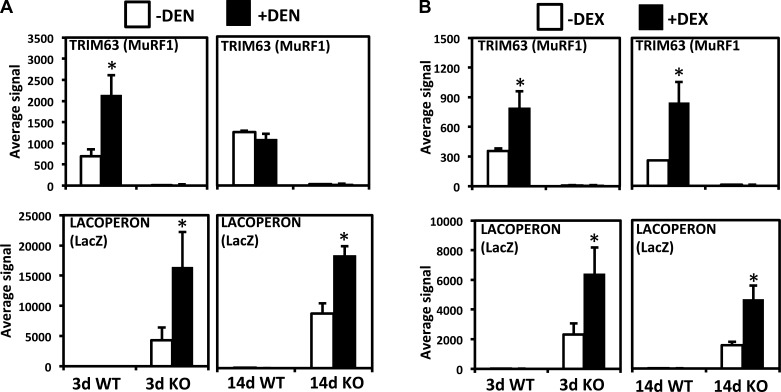
To detect differentially regulated genes by DEN or DEX treatment and between WT and MuRF1 KO mice, a genome-wide microarray analysis was performed with mouse Illumina BeadChip arrays. As a positive control, we first examined the expression of MuRF1 since it is known to be upregulated in both conditions and expression should be absent in KO mice. As shown in Fig. 2, MuRF1 expression is essentially undetectable in KO mice. In agreement with previous results, MuRF1 is induced 2.5- to 3-fold at 3 and 14 days of DEX treatment (4), whereas in denervated TS, MuRF1 expression is induced approximately fourfold at 3 days but returns toward baseline levels by 14 days (5, 49). This expression pattern was verified by quantitative PCR (Table 1). One advantage of the Illumina array platform is that it includes a probe for the prokaryotic β-galactosidase gene (lacZ, designated as LACOPERON on the array), a gene that was used to replace MuRF1 exons 1–5 in generation of the KO mouse line (5). As expected, MuRF1 and LacZ expression levels were mirror images of one another (Fig. 2), and LacZ was significantly induced in both atrophy models. However, unlike MuRF1, LacZ expression remained elevated in MuRF1 KO mice at 14 days after DEN.
Fig. 2.
MuRF1 and LacZ expression after 3 and 14 days of denervation (DEN) or DEX treatment. A: DEN experiment results. B: DEX treatment experiment results. White bars, controls; black bars, DEN or DEX treated. *Different from controls in same group, P < 0.05.
Table 1.
qPCR validation of microarray data
| Fold Change |
|||||||
|---|---|---|---|---|---|---|---|
| Treatment | Gene | Assay | Effect of Treatment | 3d WT | 3d KO | 14d WT | 14d KO |
| DEN | TRIM63 | qPCR | U | 3.88 | ND | (−)1.2 | ND |
| array | U | 3.05 | ND | (−)1.2 | ND | ||
| MYOG | qPCR | U | 44.9 | 35.5 | 21.6 | 15.1 | |
| array | U | >300* | 16.0 | 16.5 | 14.6 | ||
| MYBPC2 | qPCR | D | 2.72 | 1.38 | 21.0 | 10.1 | |
| array | D | 1.94 | 1.05 | 4.0 | 1.34 | ||
| DEX | ARRDC2 | qPCR | U | 3.40 | 2.25 | 5.02 | 0.88 |
| array | U | 2.38 | 1.34 | 5.23 | 1.54 | ||
| MT2 | qPCR | U | 7.96 | 6.91 | 2.46 | 3.19 | |
| array | U | 2.64 | 2.99 | 3.66 | 3.45 | ||
| COL1A1 | qPCR | D | 4.71 | 6.66 | 6.74 | 6.81 | |
| array | D | 7.76 | 9.39 | 11.53 | 9.11 | ||
qPCR, quantitative PCR; DEN, denervation; DEX, dexamethasone; WT, wild type; KO, knockout; U, the gene was upregulated; D, the gene was downregulated; ND, not determined.
TRIM63, MuRF1; MYOG, myogenin; MYBPC2, myosin binding protein C2; ARRDC2, arrestin domain containing 2; MT2, metallothionein 2; COL1A1, collagen 1α1.
Fold induction of myogenin is likely inaccurate since basal expression in the 3 day (d) control animals was below baseline. (−) in the 14d WT MuRF1 DEN data indicates that the gene was slightly downregulated relative to controls after being strongly upregulated at 3d.
Comparison of the gene expression program induced by the two atrophy-inducing conditions revealed that a much larger number of genes was induced by DEN than DEX treatment (Table 2), consistent with its greater effect on loss of muscle mass over the 14-day period. Further, the fold changes induced by DEN were generally more robust than those induced by DEX-treated muscle. A large number of genes were strongly induced from a very low or undetectable level such as MYOG, embryonic acetylcholine receptor (AchR) subunits (gamma and delta; CHRNG and CHRND), and muscle-specific tyrosine kinase (MUSK), making a “fold induction” determination for these genes only an estimate. Downregulation by DEN was also robust and included FGF signaling pathway components as well as contractile protein and metabolic enzyme genes (Table 3). The somewhat more modest DEX-induced changes reflect a more modulatory effect on muscle gene expression, although strong effects were observed on repression of extracellular matrix components and immune system-related genes (Table 4). As expected, several genes scored as significantly regulated by DEX are known to be glucocorticoid responsive in skeletal muscle or in other tissues and, in some cases, are known to be direct response genes with well characterized glucocorticoid response elements. These genes include lipocalin 2 (LCN2) (21), FK506 binding protein 5 (FKBP5) (55), and metallothioneins 1 and 2 (MT1, MT2) (26).
Table 2.
Number of significant, differentially up- and downregulated genes after DEN or DEX treatment in WT or MuRF1 KO mice
| 3d WT | 3d KO | 14d WT | 14d KO | |
|---|---|---|---|---|
| DEN | ||||
| Upregulated | 603 | 414 | 753 | 588 |
| Downregulated | 632 | 482 | 502 | 572 |
| DEX | ||||
| Upregulated | 112 | 89 | 91 | 87 |
| Downregulated | 110 | 102 | 65 | 95 |
Summary of Illumina microarray analysis of skeletal muscle genes differentially expressed 2-fold or greater, comparing paired sets of non-DEN or untreated controls vs. DEN or DEX-treated animals, at 3d or 14d post-DEN or of treatment, and in WT siblings vs. MuRF1 KO mice. Differential expression was called significant by a 2-tailed t-test if P < 0.05.
Table 3.
Gene expression changes in 3d and 14d DEN triceps surae
| Gene Symbol | Definition | Fold Induction/Repression |
|---|---|---|
| A. 3d DEN upregulated | ||
| Highest fold change | ||
| ORM2 | orosomucoid 2 | 3,046.6 |
| CHRNG | cholinergic receptor, nicotinic, gamma polypeptide | 2,955.6 |
| SLPI | secretory leukocyte peptidase inhibitor | 1,465.1 |
| KRT8 | keratin 8 | 899.1 |
| AGMAT | agmatine ureohydrolase (agmatinase) | 441.7 |
| MYOG | myogenin | 362.7 |
| MUSK | muscle, skeletal, receptor tyrosine kinase, transcript 5 | 361.2 |
| XEDAR EDA-A2R | X-linked ectodysplasin A2 receptor | 345.3 |
| A630038E17RIK | unknown | 339.6 |
| 2310043L19RIK | unknown | 320.7 |
| Others of interest | ||
| TRIM63 | MuRF1 | 3.0 |
| FBXO32 | MAFbx, atrogin-1 | 2.2 |
| HDAC4 | histone deacetylase 4 | 3.0 |
| ANKRD1 | ankyrin repeat domain containing 1 (Carp) | 72.3 |
| TNFRSF12A | TWEAK receptor (Fn14) | 19.4 |
| B. 14d DEN upregulated | ||
| Highest fold change | ||
| SLPI | secretory leukocyte peptidase inhibitor | 595.0 |
| KRT8 | keratin 8 | 447.2 |
| CHRNG | cholinergic receptor, nicotinic, gamma polypeptide | 407.6 |
| MUSK | muscle, skeletal, receptor tyrosine kinase, transcript 5 | 326.1 |
| MEIG1 | meiosis expressed gene 1 | 258.2 |
| KRT18 | keratin 18 | 174.2 |
| 4930432N10RIK | unknown | 173.0 |
| 9830123M21RIK | Ig-like and fibronectin type III domain containing 1 | 122.9 |
| PIGH | phosphatidylinositol glycan anchor biosynthesis, class H | 96.8 |
| RUNX1 | runt-related transcription factor 1 | 86.6 |
| Others of interest | ||
| MYOG | myogenin | 16.5 |
| TRIM63 | MuRF1 | NS |
| FBXO32 | MAFbx, atrogin-1 | NS |
| HDAC4 | histone deacetylase 4 | 8.9 |
| ANKRD1 | ankyrin repeat domain containing 1 (Carp) | 36.4 |
| TNFRSF12A | TWEAK receptor (Fn14) | 5.8 |
| C. 3d DEN downregulated | ||
| Highest fold change | ||
| FGF6 | fibroblast growth factor 6 | 158.8 |
| 4933401F05RIK | protease, serine, 55 | 69.4 |
| 2310015K22RIK | unknown | 42.3 |
| 2310010M20RIK | unknown | 41.7 |
| MAP2K6 | mitogen-activated protein kinase kinase 6 | 40.0 |
| LRRC38 | PREDICTED: leucine-rich repeat-containing 38 | 39.3 |
| FGFBP1 | fibroblast growth factor binding protein 1 | 36.3 |
| 2310042D19RIK | unknown | 31.6 |
| 2310007J06RIK | unknown | 29.7 |
| 2310065F04RIK | unknown | 27.3 |
| Others of interest | ||
| MYH1 | myosin, heavy polypeptide 1, skeletal muscle, adult | 2.7 |
| MYH4 | myosin, heavy polypeptide 4, skeletal muscle | 1.5 |
| MYL2 | myosin, light chain 2, regulatory, cardiac, slow | 3.4 |
| MYL3 | myosin, light chain 3, alkali; ventricular, skeletal, slow | 2.9 |
| MYLK2 | myosin light chain kinase 2 | 6.0 |
| ENO3 | enolase 3, beta muscle | 3.0 |
| FBP2 | fructose bisphosphatase 2 | 4.2 |
| D. 14d DEN downregulated | ||
| Highest fold change | ||
| FBP2 | fructose bisphosphatase 2 | 12.2 |
| 2310042D19RIK | unknown | 11.9 |
| PTP4A3 | protein tyrosine phosphatase 4a3 | 11.2 |
| CES3 | carboxylesterase 3 | 11.0 |
| 2310007J06RIK | unknown | 10.0 |
| CMBL | carboxymethylenebutenolidase-like | 9.9 |
| HHATL | hedgehog acyltransferase-like | 8.6 |
| WFDC1 | WAP four-disulfide core domain 1 | 8.6 |
| D0H4S114 | unknown | 7.9 |
| GNGBP1 | gametogenetin binding protein 1 | 7.9 |
| Others of interest | ||
| MYH1 | myosin, heavy polypeptide 1, skeletal muscle, adult | NS |
| MYH4 | myosin, heavy polypeptide 4, skeletal muscle | 2.2 |
| MYL2 | myosin, light chain 2, regulatory, cardiac, slow | 1.8 |
| MYL3 | myosin, light chain 3, alkali; ventricular, skeletal, slow | 2.3 |
| MYLK2 | myosin light chain kinase 2 | 7.8 |
| ENO3 | enolase 3, beta muscle | 2.6 |
The top 10 most highly, significantly upregulated genes (A and B) and downregulated genes (C and D) after 3d (A and C) and 14d (B and D) in denervated (DEN) triceps surae muscle compared with control muscles. Other genes of interest related to muscle atrophy are also listed. Fold induction/repression column shows fold induction in A and B and fold repression in C and D. NS, not significant.
Table 4.
Gene expression changes in 3d and 14d DEX-treated triceps surae
| Gene Symbol | Definition | Fold Induction/Repression |
|---|---|---|
| A. 3d DEX-treated upregulated | ||
| Highest fold change | ||
| LCN2 | lipocalin 2 | 120.6 |
| ODF3L2 | outer dense fiber of sperm tails 3-like 2 | 71.0 |
| GCK | glucokinase | 19.2 |
| SV2B | synaptic vesicle glycoprotein 2 b | 11.3 |
| FKBP5 | FK506 binding protein 5 | 11.0 |
| CXCL13 | chemokine (C-X-C motif) ligand 13 | 9.5 |
| OPN1MW | opsin 1 (cone pigments), medium-wave-sensitive | 8.6 |
| KLK1B26 | kallikrein 1-related petidase b26 | 7.7 |
| CDKN1A | cyclin-dependent kinase inhibitor 1A (p21) | 7.7 |
| MT1 | metallothionein I | 6.6 |
| Others of interest | ||
| TRIM63 | MuRF1 | 2.2 |
| FBXO32 | MAFbx, atrogin-1 | 2.8 |
| MT2 | metallothionein II | 2.7 |
| EIF4EBP1 | eif4E binding protein 1 | 2.7 |
| ANKRD1 | ankyrin repeat domain containing 1 (Carp) | 2.1 |
| TNFRSF12A | TWEAK receptor (Fn14) | 5.8 |
| B. 14d DEX-treated upregulated | ||
| Highest fold change | ||
| ODF3L2 | outer dense fiber of sperm tails 3-like 2 | 53.1 |
| SV2B | synaptic vesicle glycoprotein 2 b | 21.9 |
| CXCL13 | chemokine (C-X-C motif) ligand 13 | 20.0 |
| GSTA3 | glutathione S transferase 3 | 14.4 |
| LCN2 | lipocalin 2 | 11.5 |
| GLUL | glutamine synthase | 10.1 |
| GCK | glucokinase | 9.6 |
| KLK1B26 | kallikrein 1-related petidase b26 | 9.0 |
| CYP2F2 | cytochrome P450, 2f2 | 7.7 |
| HP | haptoglobin | 7.7 |
| Others of interest | ||
| TRIM63 | MuRF1 | 3.3 |
| FBXO32 | MAFbx, atrogin-1 | 3.5 |
| FKBP5 | FK506 binding protein 5 | 6.6 |
| CDKN1A | cyclin-dependent kinase inhibitor 1A (p21) | 7.0 |
| MT1 | metallothionein I | 4.8 |
| MT2 | metallothionein II | 3.7 |
| ANKRD1 | ankyrin repeat domain containing 1 (Carp) | NS |
| TNFRSF12A | TWEAK receptor (Fn14) | NS |
| C. 3d DEX-treated downregulated | ||
| Highest fold change | ||
| FGFBP1 | FGF binding protein 1 | 62.0 |
| ACTC1 | alpha cardiac actin | 27.2 |
| DBP | D site-binding protein | 12.4 |
| RSPH1 | radial spoke head 1 homolog | 11.9 |
| B230312C02RIK | unknown | 9.8 |
| D0H4S114 | unknown | 9.4 |
| COL1A2 | collagen 1A2 | 9.1 |
| GAMT | guanidinoacetate methyltransferase | 8.2 |
| DUPD1 | dual-specificity phosphatase and pro isomerase domain 1 | 7.2 |
| KERA | keratocan | 7.1 |
| Others of interest | ||
| COL15A1 | collagen 15A1 | 3.4 |
| COL5A1 | collagen 5A1 | 2.4 |
| COL6A1 | collagen 6A1 | 2.7 |
| D. 14d DEX-treated downregulated | ||
| Highest fold change | ||
| DBP | D site-binding protein | 13.0 |
| COL1A2 | collagen 1A2 | 12.6 |
| RSPH1 | radial spoke head 1 homolog | 9.1 |
| COL15A1 | collagen 15A1 | 8.8 |
| ACTC1 | alpha cardiac actin | 6.6 |
| KDELR3 | KDEL ER protein retention receptor 3 | 6.2 |
| D0H4S114 | unknown | 4.4 |
| H2-AA | histocompatibility 2, class II antigen A, alpha | 4.3 |
| TNMD | tenomodulin | 4.1 |
| CD74 | CD74 antigen | 4.1 |
| Others of interest | ||
| COL5A1 | collagen 5A1 | 2.4 |
| COL6A1 | collagen 6A1 | 2.9 |
The top 10 most highly, significantly upregulated genes (A and B) and downregulated genes (C and D) after 3d (A and C) and 14 days (B and D) in DEX-treated triceps surae muscle compared with control muscles. Other genes of interest related to muscle atrophy are also listed. Fold induction/repression column shows fold induction in A and B and fold repression in C and D.
Overall the program of gene expression between the two conditions was fairly divergent; however, the array was able to identify a commonly regulated set of genes, besides MuRF1 and MAFbx, that included FOXO1, EIF4EBP1, ANKRD1 (mCARP), CDKN1A (p21), and TNFRSF12A (TWEAK), genes that have been previously linked to muscle atrophy in various conditions (16, 34, 38, 40, 47, 49, 51). Furthermore, several proteasome subunits were at least modestly upregulated in both atrophy models. Interestingly, while both conditions showed a modest (DEX) to strong (DEN) suppression of genes involved in glycolysis and oxidative phosphorylation, one of the most highly yet transiently repressed genes in both atrophy conditions was FGFBP1, which is interesting since FGF signaling has been implicated in motor neuron regeneration after injury (60). Overall, the microarray was successful in detecting genes known to be up- and downregulated in inactivity and catabolic state-induced atrophy, thus enabling us to take the next step in determining whether deletion of MuRF1 resulted in differential gene expression in one or both of the atrophy models.
Few genes are differentially expressed in WT vs. MuRF1 KO muscle under resting conditions.
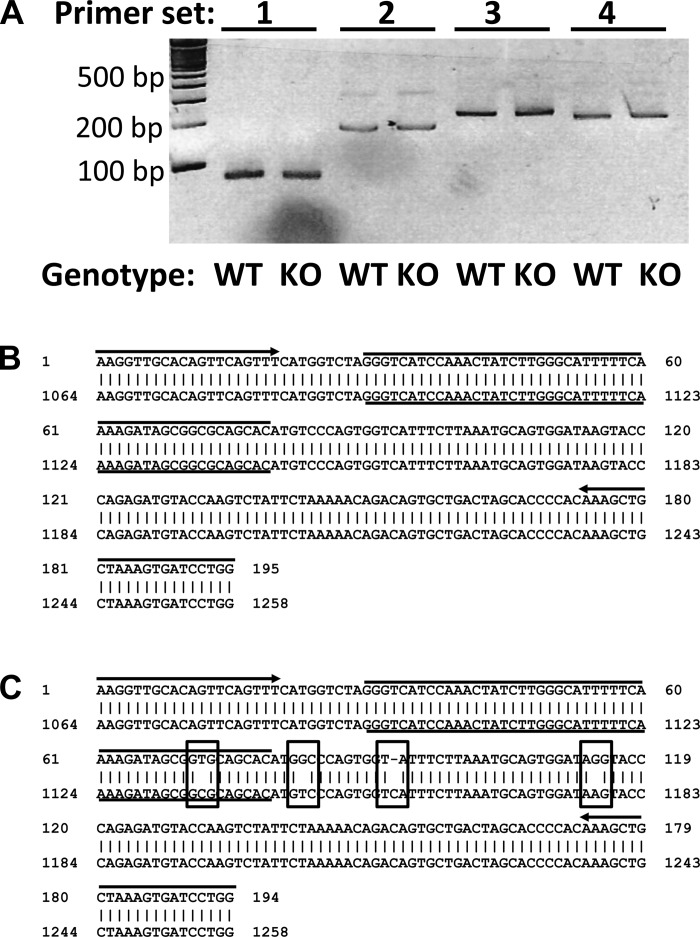
To examine a potential role for MuRF1 in gene expression, we first examined the number of differentially regulated genes in mice under untreated, resting conditions. The analysis was limited to genes that averaged at least twofold difference between WT and MuRF1 KO mice across all experimental groups (Table 5). As noted above, MuRF1 was highly enriched in all WT groups. Also as noted, LacZ expression was enriched across the board in all MuRF1 KO animals, as was the neomycin resistance gene (NEOR), a selectable marker and part of the targeting vector used to generate MuRF1 KO embryonic stem cells. Interestingly, of the relatively small number of differentially regulated genes, the vast majority localized near the MuRF1 gene on the long arm of chromosome 4, either in the same or adjacent cytoband (4qD or 4qE). Semiquantitative PCR, using a set of probes across the coding and noncoding region of the predicted mRNA, was used to verify one gene in particular, PNRC2 that showed one of the highest apparent signal differences between genotypes (Fig. 3). No clear differences were observed between WT and MuRF1 KO mice after a subsaturating number of amplification cycles, in stark contrast to the 20- to 30-fold difference reported by the microarray. PCR amplification and sequencing of the probe binding region found in the 3′-UTR of PNRC2 from two WT and two MuRF1 KO mice revealed a single base pair polymorphism in the KO-derived cDNA that is not seen in WT-derived cDNA. Thus, one reason that genes in the vicinity of MuRF1 show apparent altered expression may be the presence of polymorphisms in genes that co-segregate with MuRF1 during the several rounds of backcrosses, since the embryonic stem cells used to create the MuRF1 KO are of a different strain (CJ7) than the background (C57/Bl6), as was the BAC targeting vector (129 Sv/J) (5). Currently, we do not know the basis for the higher chromosome 4 localized gene signals in MuRF1 KO mice (Table 5), nor have we validated the very few genes that show consistent differential signals on the array and are not localized to chromosome 4. To avoid potential artifacts of chromosomal location, regardless of the source, our subsequent screening was focused on genes that 1) did not show significant baseline expression between genotypes and 2) are not located near the MuRF1 locus on chromosome 4.
Table 5.
Differentially expressed genes in control WT and MuRF1 KO animals
| Chromsome | Gene Symbol | 3d DEN Expt | 14d DEN Expt | 3d DEX Expt | 14d DEX Expt | Av. Difference | Gene ID | Definition |
|---|---|---|---|---|---|---|---|---|
| Control average signals higher in MuRF1 KO mice | ||||||||
| 4qD3 | TRIM63 | 175.1 | 34.9 | 42.7 | 19.1 | 68.0 | 433766 | tripartite motif-containing 63; MuRF1 |
| 4qD3 | PNRC2 | 38.9 | 28.1 | 17.7 | 23.8 | 27.1 | 52830 | proline-rich nuclear receptor coactivator 2 |
| 4qE1 | HSPB7 | 10.6 | 2.4 | 19.1 | 16.9 | 12.2 | 29818 | heat shock protein family, member 7 (cardiovascular) |
| 4qD3 | PADI2 | 11.0 | 5.3 | 7.6 | 9.4 | 8.3 | 18600 | peptidyl arginine deiminase, type II |
| 4qE2 | 2810408P10RIK | 3.9 | 2.0 | 9.8 | 6.3 | 5.5 | 242747 | unknown |
| 4qD3 | NECAP2 | 5.9 | 1.9 | 5.9 | 7.8 | 5.4 | 66147 | NECAP endocytosis associated 2 |
| 2qC1.3 | XIRP2 | 3.5 | 1.5 | 6.3 | 9.1 | 5.1 | 241431 | xin actin-binding repeat containing 2, transcript 1 |
| 4qD2.3 | GPN2 | 2.8 | 2.7 | 3.1 | 3.2 | 2.9 | 100210 | GPN-loop GTPase 2 |
| 4qD2.2 | BSDC1 | 3.3 | 1.9 | 3.5 | 2.8 | 2.9 | 100383 | BSD domain containing 1 |
| 11qE2 | 2610035D17RIK | 2.7 | 1.5 | 1.8 | 3.0 | 2.2 | 72386 | unknown |
| 8qB3.3 | GATAD2A | 1.4 | 0.6 | 3.7 | 2.5 | 2.1 | 234366 | GATA zinc finger domain containing 2A |
| 11 D | KRT1-10 | 1.1 | 1.4 | 3.1 | 2.3 | 2.0 | 16661 | keratin 10 |
| Control average signals higher in WT mice | ||||||||
| (4qD3) | LACOPERON | 4290.0 | 346.4 | 482.6 | 299.4 | 1354.6 | LacZ; β galactosidase gene | |
| (4qD3) | NEOR | 95.8 | 81.8 | 344.2 | 14.4 | 134.0 | neomycin resistance gene | |
| X | ERDR1 | 1.5 | 0.9 | 6.5 | 15.6 | 6.1 | 170942 | erythroid differentiation regulator 1 |
| 4qD2.1 | HYI | 5.5 | 1.8 | 5.5 | 6.9 | 4.9 | 68180 | hydroxypyruvate isomerase homolog (E. coli) |
| 1 C4 | WDFY1 | 5.0 | 2.1 | 2.8 | 2.0 | 3.0 | 69368 | WD repeat and FYVE domain containing 1 |
| 4qD2.2 | CAP1 | 3.8 | 1.2 | 2.9 | 3.1 | 2.8 | 12331 | CAP, adenylate cyclase-associated protein 1 (yeast) |
| 4qE2 | ANGPTL7 | 3.6 | 2.4 | 1.1 | 2.7 | 2.5 | 654812 | angiopoietin-like 7 |
| 13qD2.3 | PAIP1 | 1.3 | 0.9 | 3.6 | 3.7 | 2.4 | 218693 | polyadenylate binding protein-interacting protein 1, transcript 1 |
| X | FMR1 | 1.9 | 1.9 | 3.2 | 2.3 | 2.3 | 14265 | fragile X mental retardation syndrome 1 |
Genes significantly differentially expressed on average >2-fold across all control groups used in this study are listed. Genes whose average array signals were higher in MuRF1 KO mice are shown at top; genes whose average signals were higher in WT mice are shown at bottom. Boldfaced genes are localized near MuRF1 on chromosome 4 (cytoband for each gene is listed). Chromosomal location for the LACOPERON and NEOR genes are denoted as 4qD3 (in parentheses) since these genes were knocked into the MuRF1 locus in generation of the KO mouse line.
Fig. 3.
Apparent differential expression of proline-rich nuclear receptor coactivator 2 (PNRC2) by Illumina microarray is likely due to an artifact generated by polymorphisms in the region of the gene encoding the 3′-untranslated region (UTR) of the mRNA. A: gel of semiquantitative PCR to detect PNRC2 expression in WT and KO triceps surae, using primer sets designed to correspond to different regions of the 3′-UTR of the PNRC2 mRNA. B: representative of an alignment of sequenced amplicon from PNRC2 cDNA, from 1 of 3 WT animals vs. the GenBank-deposited Mus musculus PNRC2 gene. Samples from all 3 animals and 2 separate PCR reactions per animal yielded identical sequences to the example shown here. C: representative of an alignment of sequenced amplicon from PNRC2 cDNA, from 1 of 3 MuRF1 KO animals vs. the GenBank-deposited M. musculus PNRC2 gene. Samples from all 3 animals and 2 separate PCR reactions per animal yielded identical sequences to the example shown here. Boxed areas show 4 single base differences between the PNRC2 amplicon generated from MuRF1 KO animals and the GenBank sequence based on the C57/Bl6 genome. Arrows, up- and downstream PCR primer sites; over and underlined region, Illumina microarray PNRC2 probe site.
MuRF1 KO mice show significantly altered expression patterns after DEN.
In contrast to the small number of genes with average signal differences in controls, a much larger number showed more than twofold differences in array signal after DEN, even when we excluded those genes near the MuRF1 locus (Table 6). These inferred expression differences mostly fall into three categories of gene expression patterns: category I, genes that show blunted or delayed repression in MuRF1 KO compared with WT at 3 days but are similarly repressed by 14 days; category II, genes that are upregulated similarly in both genotypes at 3 days but return to baseline more quickly in WT mice than in MuRF1 KO mice; and category III, genes that have a significantly greater induction in WT than MuRF1 KO mice at either 3 or 14 days. There were relatively few genes whose signals were repressed to a greater degree in MuRF1 KO than WT mice (category IV) or were induced to a much greater extent (or even exclusively) at 3 days or 3 and 14 days in MuRF1 KO mice (category V).
Table 6.
Most differentially expressed genes between WT vs. MuRF1 KO mice after DEN
| Category | Fold Difference | Effect of DEN | Gene Symbol | Definition | Gene ID |
|---|---|---|---|---|---|
| A. 3d DEN genes expressed at higher levels in MuRF1 KO mice | |||||
| I | 4.09 | D | COPS8 | COP9 (constitutive photomorphogenic), subunit 8) | 108679 |
| I | 3.30 | D | Gm4980 | predicted gene 4980 | 245190 |
| I | 3.27 | D | DDIT4L | DNA-damage-inducible transcript 4-like | 73284 |
| I | 2.93 | D | NEU2 | neuraminidase 2 | 23956 |
| I | 2.80 | D | LOC667597 | unknown | 667597 |
| I | 2.45 | D | CHAC1 | ChaC, cation transport regulator-like 1 (E. coli) | 69065 |
| I | 2.41 | D | THAP4 | THAP domain containing 4 | 67026 |
| I | 2.39 | D | PHKG1 | phosphorylase kinase gamma 1 | 18682 |
| V | 2.18 | U | TMEM100 | transmembrane protein 100 | 67888 |
| I | 2.11 | D | MYH1 | myosin, heavy polypeptide 1, skeletal muscle, adult | 17879 |
| I | 2.07 | D | GPD1 | glycerol-3-phosphate dehydrogenase 1 (soluble) | 14555 |
| I | 2.05 | D | MYLK2 | myosin, light polypeptide kinase 2, skeletal muscle | 228785 |
| B. 3d DEN Genes expressed at higher levels in WT mice | |||||
| III | 3.25 | U | DUPD1 | Dual-specificity phosphatase and proisomerase domain 1 | 435391 |
| III | 2.52 | U | SNHG6 | small nucleolar RNA host gene (nonprotein coding) 6 | 73824 |
| III | 2.44 | U | A630038E17RIK | unknown | 219065 |
| III | 2.38 | U | ORM1 | orosomucoid 1 | 18405 |
| III | 2.36 | U | EGR1 | early growth response 1 | 13653 |
| III | 2.09 | U | GM6462 | predicted gene 6462 | 638892 |
| III | 2.03 | U | GSTA2 | glutathione S-transferase, alpha 2 (Yc2) | 14858 |
| III | 2.02 | U | SIDT1 | SID1 transmembrane family, member 1 | 320007 |
| III | 2.01 | U | GSTA4 | glutathione S-transferase, alpha 4 | 14860 |
| C. 14d DEN Genes expressed at higher levels in MuRF1 KO mice | |||||
| V | 7.39 | U | 2310015A16RIK | unknown | 69561 |
| V | 4.82 | U | CCDC110 | coiled-coil domain containing 110 | 212392 |
| II | 3.40 | U | MT2 | metallothionein 2 | 17750 |
| II | 3.36 | U | CCL8 | chemokine (C-C motif) ligand 8 | 20307 |
| I | 3.23 | D | 9630033F20RIK | unknown | 319801 |
| V | 3.16 | U | LONRF1 | LON peptidase N-terminal domain and ring finger 1 | 244421 |
| II | 3.16 | U | HP | haptoglobin | 15439 |
| I | 3.14 | D | EG245190 | predicted gene 4980 | 100503386 |
| I | 2.96 | D | GNG2 | guanine nucleotide binding protein (G protein), gamma 2 | 14702 |
| V | 2.83 | U | CSRP1 | cysteine and glycine-rich protein 1 | 13007 |
| II | 2.83 | U | CCL7 | chemokine (C-C motif) ligand 7 | 20306 |
| I | 2.77 | D | FCOR | FOXO corepressor | 100503924 |
| II | 2.72 | U | TSC22D3 | TSC22 domain family, member 3, transcript variant 2 | 14605 |
| II | 2.70 | U | MT1 | metallothionein 1 | 17748 |
| I | 2.69 | D | KLHL38 | kelch-like 38 (Drosophila) | 268807 |
| V | 2.68 | U | SSH2 | slingshot homolog 2 (Drosophila) | 237860 |
| I | 2.67 | D | NOS1 | nitric oxide synthase 1, neuronal | 18125 |
| V | 2.63 | U | CAR1 | carbonic anhydrase 1, transcript variant 1. | 12346 |
| II | 2.56 | U | FCER1G | Fc receptor, IgE, high affinity I, gamma polypeptide | 14127 |
| I | 2.54 | D | ZFP467 | zinc finger protein 467, transcript variant 3 | 68910 |
| II | 2.49 | U | SRFBP1 | serum response factor binding protein 1 | 67222 |
| II | 2.40 | U | PLEKHO2 | pleckstrin homology domain containing, family O2 | 102595 |
| II | 2.40 | U | WISP2 | WNT1 inducible signaling pathway protein 2 | 22403 |
| II | 2.34 | U | A430104N18RIK | unknown | 78591 |
| II | 2.33 | D | ADH1 | alcohol dehydrogenase 1 (class I) | 11522 |
| II | 2.32 | U | FBXO32 | F-box protein 32, (MAFbx, atrogin-1) | 67731 |
| II | 2.29 | U | GLUL | glutamate-ammonia ligase (glutamine synthetase) | 14645 |
| I | 2.26 | D | CLEC3B | C-type lectin domain family 3 | 21922 |
| II | 2.22 | U | TRAM2 | translocating chain-associating membrane protein 2 | 170829 |
| II | 2.22 | U | SERPINA3N | serine (or cysteine) peptidase inhibitor, clade A 3N | 20716 |
| V | 2.21 | U | 2900073C17RIK | unknown | 73020 |
| II | 2.20 | U | GPR146 | G protein-coupled receptor 146, transcript variant 1 | 80290 |
| I | 2.19 | D | TUBA8 | tubulin, alpha 8 | 53857 |
| V | 2.14 | U | H2-DMA | histocompatibility 2, class II, locus DMa | 14998 |
| I | 2.14 | D | SEC61B | Sec61 beta subunit | 66212 |
| II | 2.12 | U | BCL2A1B | B-cell leukemia/lymphoma 2 related protein A1b | 12045 |
| I | 2.12 | D | ZMYND17 | PREDICTED: zinc finger, MYND domain containing 17 | 74843 |
| I | 2.09 | D | LRG1 | leucine-rich alpha-2-glycoprotein 1 | 76905 |
| II | 2.06 | U | CCL9 | chemokine (C-C motif) ligand 9 | 20308 |
| II | 2.06 | U | MGL1 | macrophage galactose N-acetyl-galactosamine lectin 1 | 17312 |
| V | 2.04 | U | OLIG1 | oligodendrocyte transcription factor 1 | 50914 |
| II | 2.03 | U | H2-AA | histocompatibility 2, class II antigen A, alpha | 14960 |
| I | 2.02 | D | CIDEC | cell death-inducing DFFA-like effector c | 14311 |
| D. 14d DEN Genes expressed at higher levels in WT mice | |||||
| III | 5.74 | U | NRCAM | neuron-glia-CAM-related cell adhesion molecule | 319504 |
| III | 4.08 | U | AIF1L | allograft inflammatory factor 1-like | 108897 |
| III | 3.85 | U | GSTA1 | glutathione S transferase A1 | 14857 |
| III | 3.55 | U | FAM83D | family with sequence similarity 83, member D | 71878 |
| III | 3.51 | U | KRT8 | keratin 8 | 16691 |
| III | 3.02 | U | CHRND | cholinergic receptor, nicotinic, delta polypeptide | 11447 |
| III | 2.99 | U | CHRNG | cholinergic receptor, nicotinic, gamma polypeptide | 11449 |
| III | 2.99 | U | HAUS8 | HAUS augmin-like complex, subunit 8 | 76478 |
| III | 2.94 | U | STAC2 | SH3 and cysteine rich domain 2 | 217154 |
| III | 2.75 | U | UCHL1 | ubiquitin carboxy-terminal hydrolase L1 | 22223 |
| III | 2.65 | U | EPHX4 | epoxide hydrolase 4 | 384214 |
| III | 2.58 | U | 1700029J07RIK | unknown | 69479 |
| III | 2.57 | U | ATP13A3 | PREDICTED: ATPase type 13A3, transcript variant 1 | 224088 |
| III | 2.54 | U | PLEKHB2 | pleckstrin homology domain containing B2 | 226971 |
| III | 2.52 | U | ITGA9 | integrin A9 | 104099 |
| III | 2.49 | U | MTAP1B | microtubule-associated protein 1 B | 17755 |
| III | 2.42 | U | DUSP4 | dual specificity phosphatase 4 | 319520 |
| IV | 2.40 | D | DUSP18 | dual specificity phosphatase 18 | 75219 |
| III | 2.37 | U | HIP1R | huntingtin interacting protein 1 related | 29816 |
| III | 2.37 | U | 9330102E08RIK | unknown | 330427 |
| III | 2.34 | U | DUPD1 | dual specificity phosphatase/pro isomerase 1 | 435391 |
| III | 2.31 | U | UGCG | UDP-glucose ceramide glucosyltransferase | 22234 |
| IV | 2.31 | D | 9930105H17RIK | unknown | 100669 |
| IV | 2.27 | D | NAPEPLD | N-acyl phosphatidylethanolamine phospholipase D | 242864 |
| III | 2.27 | U | BC034090 | unknown | 207792 |
| III | 2.26 | U | HHIPL1 | hedgehog interacting protein-like 1 | 214305 |
| III | 2.26 | U | MFHAS1 | malignant fibrous histiocytoma amplified seq 1 | 52065 |
| III | 2.22 | U | MNS1 | meiosis-specific nuclear structural protein 1 | 17427 |
| III | 2.21 | U | LRP11 | low density lipoprotein receptor-related protein 11 | 237253 |
| III | 2.17 | U | C230098O21RIK | unknown | 102153 |
| III | 2.17 | U | BPIFB1 | BPI fold containing family B, member 1 | 228801 |
| III | 2.10 | U | KRT18 | keratin 18 | 16668 |
| III | 2.07 | U | VASP | vasodilator-stimulated phosphoprotein | 22323 |
| III | 2.06 | U | MYH3 | myosin, heavy polypeptide 3, skeletal muscle | 17883 |
| III | 2.05 | U | SH3RF1 | SH3 domain containing ring finger 1 | 59009 |
| III | 2.04 | U | XRRA1 | PREDICTED: X-ray radiation resistance associated 1 | 446101 |
| III | 2.04 | U | MUSK | muscle, skeletal, receptor tyrosine kinase, variant 5 | 18198 |
| III | 2.01 | U | SH3D19 | PREDICTED: SH3 domain protein D19, variant 2 | 27059 |
A: genes whose average signal is significantly higher in MuRF1 KO mice after 3d of DEN. B: genes whose average signal is significantly higher in WT mice after 3d of DEN. C: genes whose average signal is significantly higher in MuRF1 KO mice after 14d of denervation. D: genes whose average signal is significantly higher in WT mice after 14d of DEN. Genes localized near MuRF1 on chromosome 4 are excluded from analysis, and only genes up (U)- or downregulated (D) by DEN are included. Category I genes show delayed repression in MuRF1 KO mice but are equally repressed in both genotypes at 14d; category II genes are upregulated at 3d in both genotypes but remain elevated in MuRF1 KO mice at 14d; category III genes are upregulated to a lesser extent in MuRF1 KO mice than in WT mice; category IV genes are downregulated to a greater extent in MuRF1 KO mice at either 3d or 14d; category V genes are upregulated to a greater extent in MuRF1 KO mice at 3d or both 3d and 14d.
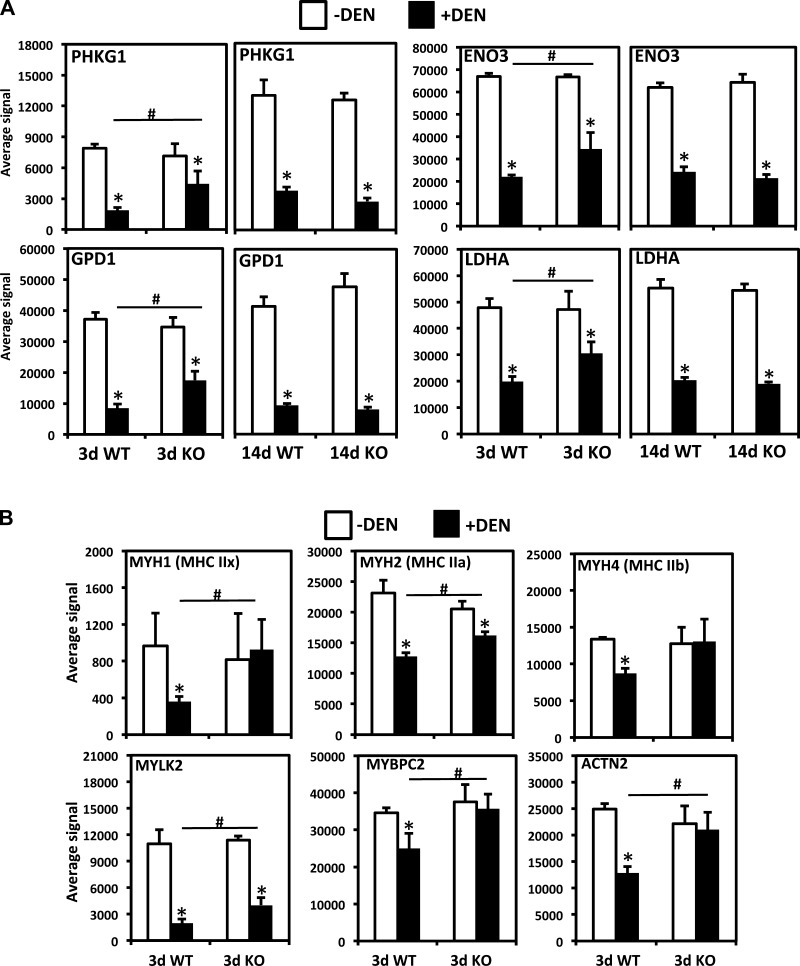
Examples of category I genes are shown in Fig. 4. We found certain metabolic enzymes and contractile apparatus genes to be regulated in this manner by using the initial stringent filter, including glycogen phosphorylase kinase γ-subunit (PHKG1), glycerol-3-phosphate dehydrogenase (GPD1), myosin heavy chain I (MYH1, or MHC IIx), and myosin light chain kinase 2 (MYLK2). Given these findings, we expanded our analysis to include additional genes involved in muscle fuel metabolism and muscle contraction with statistically significant differences in induced signals. For example, muscle-specific enolase (ENO3) and lactate dehydrogenase (LDHA) also showed significantly delayed repression in a pattern essentially identical to PHKG1 and GPD1 (Fig. 4A). Other genes encoding glycolytic and glycogen metabolism genes (including α- and β-glycogen phosphorylase kinase subunits) followed the same trend, but differences between WT and MuRF1 KO mice were not significant given the sample size, time point, and genotype (P > 0.05). We did not observe significant differences in expression of TCA cycle and electron transport chain genes, which were all repressed to a similar degree in both genotypes, at least at the time points chosen for this study. Several genes involved in contractile function, besides MYH1 and MYLK2, also showed delayed repression in MuRF1 KO mice (Fig. 4B) and included MYH2 (MHC IIa), ACTN2 (α-actinin 2), and MYH4 (MHC IIb) showed the same pattern in that it was significantly repressed in WT but not MuRF1 KO mice; however, the average signal difference in DEN WT vs. MuRF1 KO mice was not significant. All of these genes are repressed in MuRF1 KO mice at least as well as in WT mice by 14 days, similar to the metabolic enzymes shown in Fig. 4A (not shown), with the possible exception of MYBPC2, which showed a high degree of variability on the microarray at 14 days, but blunted repression was confirmed by qPCR (Table 1).
Fig. 4.
Examples of delayed downregulation of metabolic and structural/contractile genes in MuRF1 KO mice after DEN. Results for 3 and 14 days post-DEN for metabolic enzymes are shown in A, only results for 3 days post-DEN for structural/contractile genes are shown in B. White bars, controls; black bars, DEN or DEX treated. #Significantly different from between groups, P < 0.05; *Different from controls in same group, P < 0.05. PHKG1, glycogen phosphorylase gamma1 subunit; GPD1, glycerol 3-phosphate dehydrogenase 1 (soluble); ENO3, muscle-specific enolase; LDHA, lactate dehydrogenase A; MYH1, myosin heavy chain IIx; MYH2, myosin heavy chain IIa; MYH4, myosin heavy chain IIb; MYLK2, myosin light chain kinase 2; MYBPC2, myosin binding protein C2 (skeletal muscle); ACTN2, α-actinin2.
The second pattern of gene expression where signals are higher in MuRF1 KO than WT mice (category II) includes two metallothionein genes (MT1 and MT2), FBXO32 (MAFbx), and GLUL (glutamine synthase), among several others. The maintained expression of MAFbx at 14 days was noted earlier and is a pattern also followed by a number of proteasome subunits (18) and consistent with elevated proteasome activity at that time point despite the significant sparing observed in MuRF1 KO mice.
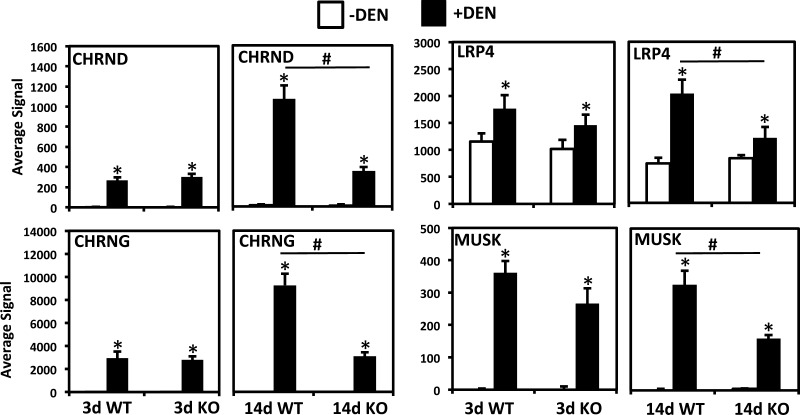
Category III genes are upregulated in WT mice as well, but in this case upregulation is blunted in MuRF1 KO mice. While some differences are clear at 3 days post-DEN such as the deubiquitinating enzyme UCHL1, the majority of genes in this category show similar upregulation between WT and KO mice early on, but while induction continues to rise in WT it is blunted in the KO at the later time point. A particularly striking group of genes in this category are associated with the neuromuscular junction (NMJ) including the embryonic AchR subunits δ and γ and MUSK, which show very low expression in innervated muscle (Fig. 5). Thus, we examined the expression of other NMJ-associated genes and found that while agrin expression was not significantly different between genotypes, its co-receptor LRP4 (and MUSK activator) (27, 65) showed blunted upregulation in MuRF1 KO mice, again significantly at 14 days after DEN.
Fig. 5.
Examples of differentially upregulated genes between WT and MuRF1 KO mice after DEN. Microarray results for selected genes upregulated by DEN; white bars, controls (−DEN); black bars, denervated (+DEN). #Significantly different from between groups, P < 0.05; *different from controls in same group, P < 0.05. CHRND and CHRNG, acetylcholine receptor subunits-δ and -γ, respectively; LRP4, leucine-rich protein 4 (agrin co-receptor); MUSK, muscle-specific tyrosine kinase.
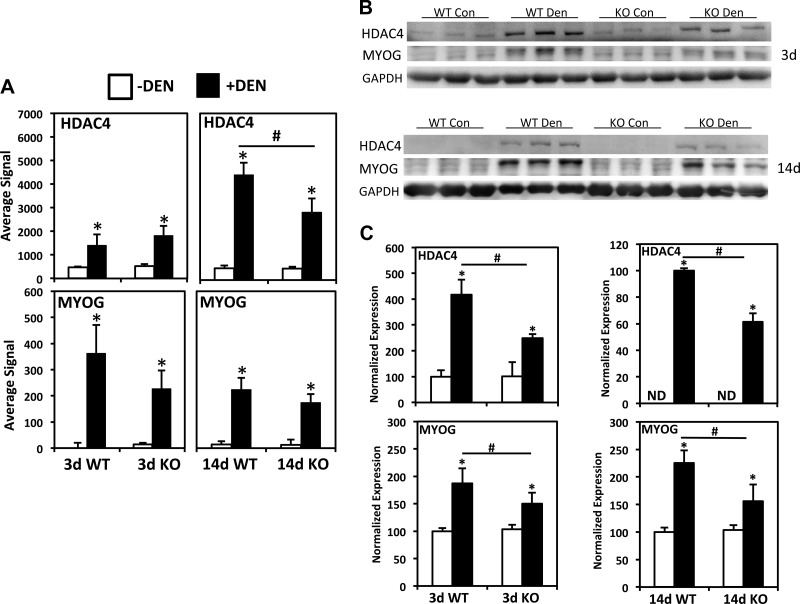
A series of recent findings has implicated an HDAC4-MYOG pathway in the upregulation of AchR subunits after DEN (12, 53, 54), as well as MuRF1 and MAFbx (41). In addition, HDAC4 has been implicated in DEN-induced repression of metabolic enzyme and contractile genes via inhibition of MEF2 activity (11, 54). Interestingly, the microarray data reveal that HDAC4 expression showed a similar pattern as category III genes (Fig. 6A). Although MYOG expression was strongly upregulated by DEN as well, mRNA levels as detected by the array and qPCR were not significantly different between genotypes (Fig. 6A, Table 1). We next performed Western blot analysis to examine HDAC4 and MYOG protein levels, and in both cases, protein expression increased as the result of DEN, but the induced levels were significantly lower in MuRF1 KO vs. WT mice at both 3 and 14 days (Fig. 6, B and C).
Fig. 6.
MuRF1 KO mice show blunted upregulation of HDAC4 and myogenin protein expression after denervation. A: microarray results for HDAC4 (top) and myogenin (MYOG, bottom) after 3 and 14 days DEN. B: Western blot of HDAC4, MYOG, and glyceraldehyde 3-phosphate dehydrogenase, showing results from controls (Con) vs. 3 day DEN WT and KO triceps surae. C: quantification of Western blot shown in B. White bars, controls (−DEN); black bars, +DEN. #Significantly different from between groups, P < 0.05, *different from controls in same group; P < 0.05.
MuRF1 KO mice show significantly altered expression patterns after DEX treatment.
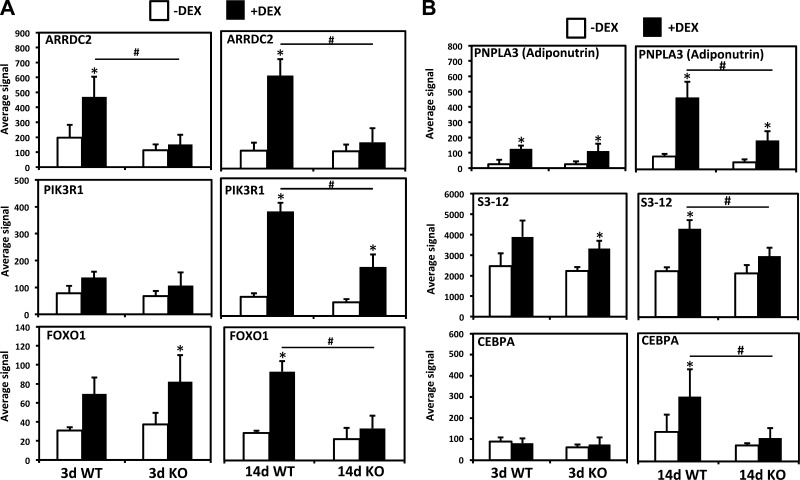
Compared with differences in DEN-induced gene expression programs, there were fewer differences in DEX-induced gene expression patterns between genotypes; for instance no genes outside of the region surrounding the MuRF1 gene on chromosome 4 were expressed at higher levels in MuRF1 KO mice than WT mice after 14 days of DEX treatment. Nevertheless, certain genes involved in signal transduction and fat metabolism were differentially regulated, with the most common pattern being a stronger induction in WT mice than MuRF1 KO mice that is most evident at 14 days (Table 7). One of the most differentially regulated genes is ARRDC2 (arrestin domain containing 2), whose induction by DEX is essentially blocked in MuRF1 KO mice at both 3 and 14 days of treatment, which was verified by qPCR (Fig. 7A, Table 1). In addition, the expression of two other important genes linked to muscle atrophy and insulin signaling, PIK3R1 (the p85 regulatory subunit of PI3 kinase) and FOXO1 (forkhead class O1 transcription factor), was strongly blunted after 14 days of DEX treatment in the MuRF1 KO when muscle sparing was evident (Fig. 7A). The FOXO1 expression pattern detected by the microarray was consistent with our previous report of reduced FOXO1 mRNA and protein induction following DEX treatment in MuRF1 KO mice (4). Another strikingly differentially regulated gene is PNPLA3, or adiponutrin, which is linked to nonalcoholic fatty liver disease (15) (Fig. 7B). PNPLA3 is considered a lipogenic gene by increasing triglyceride synthesis in liver and adipose tissue although its role in skeletal muscle is uncertain. DEPP, a gene induced by progesterone in the uterus, is also highly expressed in white adipose tissue and inversely regulated by insulin and fasting (31). Therefore, we looked for additional genes involved in lipogenesis that might be significantly differentially regulated between genotypes but did not pass our original stringent filter. Two additional examples of genes that showed blunted induction following DEX in the MuRF1 KO mice were a proadipogenic transcription factor, C/EBPα and a perilipin family member involved in lipid droplet formation (S3-12) (Fig. 7B).
Table 7.
Most differentially expressed genes between WT vs. MuRF1 KO mice after DEX treatment
| Category | Fold Difference | Effect of DEX | Gene Symbol | Definition | Gene ID |
|---|---|---|---|---|---|
| A. 3d DEX-regulated genes expressed at higher levels in MuRF1 KO mice | |||||
| V | 2.87 | U | FOXK2 | forkhead box K2 | 68837 |
| V | 2.68 | U | 1190002H23RIK | unknown | 66214 |
| V | 2.61 | U | 9230019H11RIK | unknown | 215728 |
| I | 2.53 | D | DUSP10 | dual specificity phosphatase 10 | 63953 |
| V | 2.35 | U | TMEM106B | transmembrane protein 106B | 71900 |
| I | 2.27 | D | SAP30 | sin3 associated polypeptide | 60406 |
| I | 2.20 | D | TFRC | transferrin receptor | 22042 |
| I | 2.15 | D | ACTC1 | actin, alpha, cardiac muscle 1 | 11464 |
| B. 3d DEX-regulated genes expressed at higher levels in WT mice | |||||
| III | 3.11 | U | ARRDC2 | arrestin domain containing 2 | 70807 |
| III | 2.95 | U | A730009E18RIK | unknown | 319603 |
| IV | 2.91 | D | 9430064K01RIK | unknown | 77294 |
| III | 2.54 | U | 5330406M23RIK | unknown | 76671 |
| IV | 2.51 | D | CDH4 | cadherin 4 | 12561 |
| III | 2.41 | U | CLEC2D | C-type lectin domain family 2, member d | 93694 |
| III | 2.26 | U | DPP8 | dipeptidylpeptidase 8 | 74388 |
| III | 2.17 | U | DEPP | decidual protein induced by progesterone | 213393 |
| IV | 2.09 | D | GADD45G | growth arrest and DNA-damage-inducible 45 gamma | 23882 |
| IV | 2.03 | D | 5430431A17RIK | unknown | 71368 |
| C. 14d DEX-regulated genes expressed at higher levels in WT mice | |||||
| III | 3.84 | U | RASL11B | RAS-like, family 11, member B | 68939 |
| III | 3.52 | U | ARRDC2 | arrestin domain containing 2 | 70807 |
| III | 3.00 | U | 2900002J02RIK | unknown | 70412 |
| III | 2.77 | U | STC2 | stanniocalcin 2 | 20856 |
| III | 2.77 | U | FOXO1 | forkhead box O1 | 56458 |
| III | 2.61 | U | IGFBP3 | insulin-like growth factor binding protein 3 | 16009 |
| III | 2.53 | U | GALNTL2 | UDP-N-acetyl-alpha-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 2 | 78754 |
| III | 2.51 | U | PNPLA3 | patatin-like phospholipase domain containing 3 | 116939 |
| III | 2.14 | U | PI3KR1 | PI3-kinase subunit p85-alpha | 18708 |
| III | 2.13 | U | AI607873 | expressed sequence AI607873 | 226691 |
| III | 2.01 | U | SLC43A1 | solute carrier family 43, member 1 | 72401 |
A: genes whose average signal is significantly higher in MuRF1 KO mice after 3d of DEX treatment. B: genes whose average signal is significantly higher in WT mice after 3d of DEX treatment. C: genes whose average signal is significantly higher in MuRF1 KO mice after 14d of DEX treatment. Genes localized near MuRF1 on chromosome 4 are excluded from analysis, and only genes up- or downregulated by DEX treatment are included. Category I genes show delayed repression in MuRF1 KO mice but are equally repressed in both genotypes at 14d; category II genes are upregulated at 3d in both genotypes but remain elevated in MuRF1 KO mice at 14d; category III genes are upregulated to a lesser extent in MuRF1 KO mice than in WT mice; category IV genes are downregulated to a greater extent in MuRF1 KO mice at either 3d or 14d; category V gene are upregulated to a greater extent in MuRF1 KO mice at 3d or both 3d and 14d.
Fig. 7.
Examples of differentially upregulated genes between WT and MuRF1 KO mice after DEX treatment. A: genes linked to growth factor or adrenergic receptor signaling. B: genes linked to adipogenesis and lipid metabolism. White bars, controls (−DEX); black bars, +DEX. #Significantly different from between groups, P < 0.05; *different from controls in same group, P < 0.05. ARRDC2, arresting domain containing 2; PIK3R1, PI3 kinase regulatory subunit 1; FOXO1, forkhead transcription factor class O1.
DISCUSSION
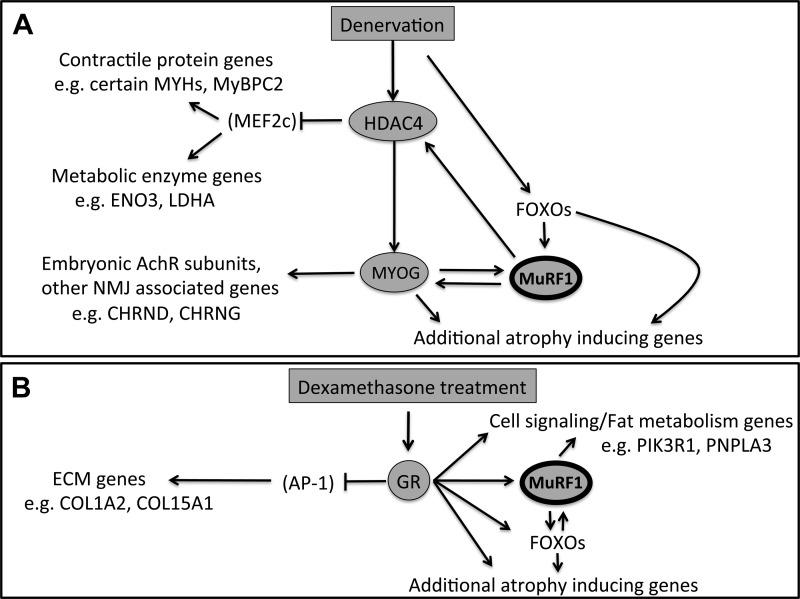
While the importance of MuRF1 in mediating skeletal muscle atrophy under divergent conditions has been established, the molecular mechanisms by which MuRF1 acts during atrophy are still not completely understood. Unequivocal determination of MuRF1 function will ultimately depend on careful examination of its potential role in an array of cellular processes, including the regulation of gene expression. In this study, we demonstrate that in the absence of MuRF1, specific sets of genes clearly show differential expression patterns upon DEN or exogenous glucocorticoid treatment. Upon DEN, MuRF1 appears to be particularly involved in full activity of the proposed HDAC4-mediated pathways of metabolic and contractile gene repression and induction of neuromuscular junction components via MYOG. The role of MuRF1 in DEX-induced gene expression is less conclusive, given the smaller number of significantly induced or repressed genes overall; nevertheless, certain genes were differentially expressed, including genes linked to muscle atrophy, cell signaling, and fat metabolism. A schematic summarizing the role of MuRF1 in gene expression pathways induced by DEN or DEX treatment is presented in Fig. 8.
Fig. 8.
Schematic of the potential role of MuRF1 in atrophy induced gene expression pathways. A: DEN-induced gene expression. DEN induces HDAC4 expression leading to increased MYOG expression, which then at least partly induces MuRF1. In turn, MuRF1 expression is necessary to support maximal HDAC4 and MYOG expression. Blunted HDAC4 expression in MuRF1 KO mice may result in decreased inhibition of MEF2c and its downstream contractile protein and metabolic enzyme gene targets. Blunted MYOG induction in MuRF1 KO mice may also lead to the subsequent blunted induction of genes linked to the neuromuscular junction, such as embryonic acetylcholine receptor subunits. Denervation also induces FOXO transcription factor expression that in parallel regulate MuRF1; unlike the HDAC4/MYOG pathway, FOXO induction appears to be independent of MuRF1 expression in this context. B: DEX-induced gene expression. DEX binding to the glucocorticoid receptor (GR) suppresses the activity of AP-1 that regulates genes encoding extracellular matrix (ECM) components like specific collagens, but in a MuRF1-independent manner. DEX-bound GRs also directly induce MuRF1 and FOXO transcription factors, which also target MuRF1; in turn MuRF1 is required for sustained DEX-induced FOXO (specifically FOXO1) expression. In addition, MuRF1 is required for full induction of specific genes encoding key cell signaling proteins such as the alpha regulatory subunit of phosphatidylinositol 3-kinase (PIK3R1) or enzymes involved in fat storage or metabolism, such as patatin-like phospholipase domain-containing protein 3 (PNPLA3, or adiponutrin). Induction of other direct GR target genes like FK506 binding protein 5 and the metallothionein 1 and 2 genes is unaffected by MuRF1 in this context, which is not shown in the schematic. COL1A2 and COL15A1, collagen 1A2 and collagen 15A1, respectively. Lines with arrowheads, activation of gene expression or transcription factor activity; lines with blunted ends, inhibition of gene expression or transcription factor activity. Factors in parentheses (MEF2c, AP-1) were not directly assayed for the influence of MuRF1 after DEN or DEX treatment.
A small number of genes show differential signals on the array between untreated WT and MuRF1 KO mice, and the majority of these genes cluster near the MuRF1 locus on chromosome 4.
Somewhat surprisingly, there were very few genes differentially expressed in control muscle, given the slight but significantly larger muscle masses in MuRF1 KO mice. The majority of the genes that showed significant differential expression were clustered near the MuRF1 locus on chromosome 4. Other studies have reported a “neighbor effect” of the KO locus on surrounding gene expression (50, 56), and in some cases this has been linked to insertion of the PGK-neomycin resistance gene cassette (52) that was used to generate the MuRF1 KO mice. We reasoned that an alternative possibility was that the apparent array signal differences might be an artifact of the way the microarray is designed, rather than reflecting real mRNA level differences. Despite several generations of backcrossing, since the MuRF1 KO locus is selected for at each round, ES cell genomic DNA flanking the knocked-in LacZ and PGK-neo construct at the MuRF1 locus will be carried along and may have single nucleotide polymorphisms that could alter probe binding kinetics that were designed based on the C57/Bl6 genome. Our prediction was that this would be most apparent when probes are located in the noncoding regions of target genes. We carefully examined one gene with a large apparent expression difference, PNRC2, and indeed, a cluster of base pair changes was found in the 3′-UTR of the PNRC2 locus from KO animals, including one in the probe binding site. While actual expression changes in genes surrounding the KO locus may occur, our results with PNRC2 provide an alternative explanation for such interpretations using arrays with a single or small number of probes. RNA-seq technology may alleviate the technical aspects of this issue (58), but true expression differences due to the neighbor effect must be accounted for in interpretation of knockout phenotypes.
The low number of differentially detected genes in WT vs. MuRF1 KO control animals may also be the result of a fairly stringent filter used to reduce false positives, since the effect in an unchallenged animal may be fairly subtle and cumulative. Consistent with our findings, however, studies using overexpressed MuRF1 in the heart and skeletal muscle revealed a relatively small number of gene expression changes and of only modest fold inductions (23, 62). Many of these changes were related to glucose and fatty acid metabolism, rather than muscle atrophy. It is interesting to note that the most highly induced gene in skeletal muscle in the MuRF1 transgenic mouse was cytosolic GPD1 (23), a key gene in both triglyceride synthesis and the glycerol 3-phosphate shuttle. In the MuRF1 KO, cytosolic GPD1 was one of those genes that showed significantly delayed repression after DEN.
Mice lacking MuRF1 show delayed repression of metabolic and structural genes and blunted upregulation of genes associated with the neuromuscular junction after DEN, which correlates with blunted HDAC4 upregulation in these mice.
The major role of MuRF1 in gene expression was only revealed during a challenge, such as after DEN or DEX treatment. Three common patterns of differential gene expression were observed between WT and MuRF1 KO mice in response to DEN. The first, which we designated category I, showed delayed repression in MuRF1 KO mice. This category includes metabolic enzymes involved in glycolysis, glycogen metabolism, and GPD1 (at the interface of glucose and fatty acid metabolism). In contrast, genes involved in oxidative metabolism, such as TCA cycle and electron transport chain components, were equally suppressed in both genotypes (not shown). MHCs IIx and IIb (and to a lesser extent IIa) also showed delayed repression in MuRF1 KO mice, along with other genes associated with the myofilaments such as MYBPC2, MYLK2, and α2 actinin. Cardiac myosin binding protein C protein (MYBPC3) was previously shown to directly interact with MuRF1 (39), and MyBPC3 levels are elevated in MuRF1 KO mice and reduced in cardiac-specific MuRF1 transgenic mice, without affecting MyBPC3 mRNA levels. Although the skeletal muscle isoform has not been reported to interact physically with MuRF1, repression of MyBPC2 mRNA levels is clearly inhibited in MuRF1 KO mice at both 3 and 14 days after DEN.
A common link with many of the genes in this category is that they are known targets of MEF2c, a transcription factor modulated by innervation state of muscle and by activity. Upon DEN, the class IIA HDAC4 translocates from the neuromuscular junction into adjacent myonuclei (12) and is thought to alter transcription of specific target genes via interaction with and inhibition of the activity of transcription factors such as MEF2c. siRNA against HDAC4 results in inhibition of DEN-induced repression of many of the same metabolic and contractile protein genes that are affected by the absence of MuRF1 shown here (11, 54), but with less effect on oxidative metabolic genes which are also not affected by the loss of MuRF1. Anaerobic and aerobic metabolism and force production are strongly suppressed in denervated muscle fibers (20); in the absence of MuRF1, delayed repression of MHC genes with maintained MYBPC2 and MYLK2 expression may yield elevated force production in denervated MuRF1 KO mice relative to their WT counterparts, a possibility that remains to be confirmed in the time frame of used these experiments. Interestingly, in C2C12 myotubes, HDAC4 knockdown results in elevated pyruvate production (54); thus MuRF1 KO mice may be able to maintain a higher rate of glycolysis relative to WT denervated muscles.
Another proposed consequence of increased HDAC4 activity upon DEN is the upregulation of MYOG and subsequent upregulation of genes associated with the neuromuscular junction on one hand (11, 12, 53, 54) and MuRF1 and MAFbx on the other (41). An important category of MuRF1 affected genes are those that are upregulated similarly in both genotypes at 3 days but are blunted at 14 days in MURF1 KO mice (category III). A set of genes with this profile encodes components of the neuromuscular junction, including embryonic AchR subunits (δ and γ), the agrin co-receptor LRP4, and MUSK. DEN induced upregulation of AchR-δ and -γ, as well as MUSK, is inhibited by HDAC4 siRNA (53). The embryonic MHC isoform, MYH3, also depends on HDAC4 and MuRF1 for full upregulation, and its upregulation was blunted in the MuRF1 KO mice. Given these possible links to HDAC4, it was gratifying to observe that the HDAC4 mRNA profile falls squarely into the same category as these category III genes. Although the difference between WT and MuRF1 KO animals is significant, it is not as dramatic as observed for AchR-δ and -γ genes for example. Interestingly, the protein levels of HDAC4 were significantly reduced in MuRF1 KO mice at both 3 and 14 days, providing a possible explanation for the observed effect at 3 days on MEF2c-dependent genes; yet these HDAC4 levels must still be sufficient to eventually support full repression of this class of genes by 14 days. Likewise, a reduced level of HDAC4 at 3 days is still enough to support the initial induction of class III genes yet only to a point where the full HDAC4 complement must be attained for full activation of these genes by 14 days. It was also interesting to note that MYOG protein levels were significantly different at 3 and 14 days, whereas the induced mRNA levels were very similar between genotypes. Again, this reduced level of protein must be sufficient for at least the early stages of the response of NMJ associated genes to DEN but not for full expression.
A recently reported alternative pathway for HDAC4-mediated gene expression changes in skeletal muscle occurs via deacetylation and activation of MEKK2 (8), a key component of MAP3 kinase signaling. Elevated MEKK2 leads to induction of AP-1 components (c-jun and c-fos), and increased AP-1 activity may also participate in MuRF1 and MAFbx induction. Besides genes associated with the NMJ, another major cluster of differentially regulated genes between WT and MuRF1 KO mice is linked to phosphorylation cascades, either as adaptor proteins or kinases/phosphatases. For example, while MAP kinases were not differentially expressed, the induction of the dual-specificity phosphatases DUSP4 and DUPD1 was much lower in MURF1 KO mice upon DEN. Both of these phosphatases have been reported to inhibit MAP kinase activity (14), and DUSP4 inhibits cardiac hypertrophy (2). An additional phosphatase, DUSP18, is repressed more robustly in MuRF1 KO mice but may target SAP/JNK and not p38 MAP kinase-mediated signaling (64). One possible function of strong DUSP4/DUPD1 upregulation during DEN may be to limit MEKK2-induced associated gene expression, since the proposed AP-1 targets MAFbx and the MuRF1 locus (monitored by LacZ expression) remain elevated at 14 days in MuRF1 KO mice. This was a pattern followed by other genes previously linked to atrophy such as MT1 and MT2, glutamine synthetase, and several proteasome subunits (18), despite the significant muscle sparing observed at that time point. In any case, defining the precise interplay among MuRF1, HDAC4, and MAP kinase signaling pathways, vs. MEF2c and MYOG-mediated pathways, in the timing and extent of muscle atrophy in different conditions will be an important area of further study.
Since HDAC4 is a candidate factor mediating the effects of MuRF1 on gene expression during DEN, as we are suggesting, how might MuRF1 affect HDAC4 expression and activity, particularly at the protein rather than transcriptional level? HDAC4 levels, like many regulatory proteins, are controlled by targeted degradation by the UPS (7) or other forms of proteolytic processing such as protein kinase A-dependent (3) or caspase activation (36). Although a specific E3 ligase has not been identified for HDAC4 polyubiquitination, MuRF1 has not been reported to date as an HDAC4 E3 ligase, and in any case, induced HDAC4 levels are lower, not higher, in MuRF1 KO mice. HDAC4 protein levels have also been shown to be controlled by various factors, including the miRNAs miR1, miR-206 (60), and miR-29, which themselves are subject to regulation by signals such as innervation state and transforming growth factor-β, among others (63). Finally, HDAC4 depends on sumoylation for nuclear import by the nuclear pore localized small ubiquitin-related modifier (SUMO) E3 ligase RanBP2 (28). Interestingly, Ubc9, a SUMO E2 ligase, and SUMO-3 (13) were among the first interacting proteins identified for MuRF1. The interplay between MuRF1 and sumoylation has not been fully explored but may play an important role in protein trafficking and activity of transcription factors and beyond. Lastly, MuRF1 may also play an indirect role in HDAC4 activity through its recently described effects on the disassembly of AchR complexes at the NMJ upon DEN (48). MuRF1 was shown to be present at the NMJ after DEN, an interesting finding given participation in the innervation state of muscle leading to altered HDAC4 levels and downstream signaling. Whether MuRF1 interacts with HDAC4 at the NMJ or otherwise indirectly promotes HDAC4 release for translocation and induction of downstream gene expression will be an interesting next step to investigate. It is also possible that MuRF1 potentiation of HDAC4 activity is part of a feed-forward process as the muscle continues to be deprived of motor neuron-mediated signaling.
Genes involved in lipid metabolism and signal transduction show blunted upregulation in MuRF1 KO mice treated with DEX.
By comparison, we also examined the effect of an MuRF1 gene deletion on the gene expression program induced by excess synthetic glucocorticoids (DEX). Our overall microarray results revealed numerous overlapping regulated genes with previous studies on glucocorticoid-induced gene expression in vivo (with prednisolone) (1) or in vitro (C2C12 myotubes) (30, 33). While fewer DEX-responsive genes than DEN-responsive genes were altered in MuRF1 KO mice, two important genes linked to muscle atrophy and insulin signaling, PI 3 kinase p85 regulatory subunit (PI3KR1) and FOXO1, showed significantly reduced levels at 14 days in MuRF1 KO mice, the latter at the protein level as well. Altering the ratio of the catalytic p110 subunit vs. the regulatory p85 subunit by glucocorticoids has been linked to inhibition of insulin and IGF-1 signaling, inhibition of Akt, and activation of FOXO transcription factors leading to induction of MuRF1 and muscle atrophy (30). Full induction of MuRF1 expression by DEX in vitro relies on the interaction of FOXO1 and the glucocorticoid receptor (57). However, other genes involved in DEX-induced atrophy that were not readily apparent from the microarray data may be affected by blunted activation of the PI3 kinase p85-FOXO1 pathway and thus may contribute the muscle sparing in MuRF1 KO mice.
In addition to PI3KR1 and FOXO1, another set of differentially regulated genes suggested that fat metabolism in muscle may be altered in DEX-treated MuRF1 KO mice. PNPLA3 (also known as adiponutrin) has been strongly implicated as a major genetic factor in development of nonalcoholic fatty liver disease (15), which is characterized by inappropriate lipid storage and whole body metabolic derangements. PNPLA family members possess triglyceride hydrolase activity in vitro but have recently been proposed to play a role in remodeling of stored triglycerides in lipid droplets as they accumulate in response to feeding (35). When we examined additional genes linked to lipid metabolism, upregulation of the proadipogenic transcription factor C/EBP-α and the lipid droplet structural protein S3–13 was blunted in MuRF1 KO mice.
Like several genes affected by the lack of MuRF1, in both DEN and DEX treatment regimens, those linked to fat metabolism are mostly upregulated after 14 days in WT animals where differences due to genotype become apparent, indicating that an effect of MuRF1 may be indirect. For example, DEX may be promoting intermyocellular fat accumulation, which is inhibited in the MuRF1 KO by the lack of myofiber atrophy. DEX treatment has been reported to have a strong potentiating effect on high fat diet-induced alterations in lipid metabolism and the accumulation of intramyocellular lipid droplets in mice, which have been linked to development of insulin resistance (19). Further studies on the metabolic alterations in DEX-treated MuRF1 KO mice, perhaps in conjunction with a high-fat diet, will be important follow-up experiments based on our array results.
Of the differentially DEX-induced genes, ARRDC2 upregulation was most striking in that it was almost completely blocked in the MuRF1 KO animals at both 3 and 14 days. While less is known about ARRDC2, another member of this novel α-arrestin domain family, ARRDC3, has been recently implicated in obesity and control of whole body energy expenditure (44). Furthermore, ARRDC3 and other ARRDC family members function as inhibitors of β-adrenergic signaling via their actions as adaptors for E3 ubiquitin ligases that internalize β2-adrenergic receptors for turnover by lysosomal degradation (42, 44). To our knowledge, this is the first description of an ARRDC family member as glucocorticoid responsive in any tissue, and may be an important new class of genes mediating glucocorticoid effects on metabolism and adrenergic receptor signaling.
In conclusion, we have demonstrated that MuRF1 KO mice show significantly altered patterns of gene expression in response to two different muscle atrophy-inducing conditions. These genes encode important metabolic, structural, and signaling pathway genes linked to muscle structure and function, as well as specific metabolic and structural changes that accompany muscle atrophy. These results suggest a new role for MuRF1 in the control of gene expression, in addition to its other proposed roles in targeted protein degradation and control of global protein synthesis. While there may be a direct effect of MuRF1 on specific transcription factors, altered gene expression may result from indirect effects of the lack of MuRF1 as well. Determining the full spectrum of the actions MuRF1 on skeletal muscle structure and function will be critical to design therapies to ameliorate the debilitating effects of muscle loss with inactivity, cachexia, and aging and to fully understand the remarkable cell biology of adult muscle plasticity at posttranslational, translational, and transcriptional levels.
GRANTS
Funds used to support this work include National Institute of Diabetes and Digestive and Kidney Diseases Grant RO1 DK-75801 to J. D. Furlow and S. C. Bodine.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: J.D.F., M.L.W., D.S.W., and S.C.B. conception and design of research; J.D.F., M.L.W., D.S.W., E.S.N., L.M.B., and A.P.R. analyzed data; J.D.F., M.L.W., D.S.W., E.S.N., L.M.B., and S.C.B. interpreted results of experiments; J.D.F. prepared figures; J.D.F. drafted manuscript; J.D.F., M.L.W., D.S.W., E.S.N., and S.C.B. edited and revised manuscript; J.D.F., M.L.W., D.S.W., E.S.N., L.M.B., A.P.R., and S.C.B. approved final version of manuscript; M.L.W., D.S.W., E.S.N., L.M.B., and A.P.R. performed experiments.
ACKNOWLEDGMENTS
The authors thank Hayung Yang, Chris Craig-Veit, Darren Hwee, and other members of the Furlow and Bodine laboratories for assistance and advice at various stages of the project. We also gratefully acknowledge the assistance and advice of the UC Davis Genome Center Gene Expression Core Facility staff.
Current addresses: E. S. Neff, Dept. of Biology, Cosumnes River College, Sacramento, CA 95823; D. S. Waddell, Dept. of Biology, Univ. of North Florida, Jacksonville, FL 32224.
REFERENCES
- 1.Almon RR, DuBois DC, Yao Z, Hoffman EP, Ghimbovschi S, Jusko WJ. Microarray analysis of the temporal response of skeletal muscle to methylprednisolone: comparative analysis of two dosing regimens. Physiol Genomics 30: 282–299, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auger-Messier M, Accornero F, Goonasekera SA, Bueno OF, Lorenz JN, van Berlo JH, Willette RN, Molkentin JD. Unrestrained p38 MAPK activation in Dusp1/4 double-null mice induces cardiomyopathy. Circ Res 112: 48–56, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Backs J, Worst BC, Lehmann LH, Patrick DM, Jebessa Z, Kreusser MM, Sun Q, Chen L, Heft C, Katus HA, Olson EN. Selective repression of MEF2 activity by PKA-dependent proteolysis of HDAC4. J Cell Biol 195: 403–415, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baehr LM, Furlow JD, Bodine SC. Muscle sparing in muscle RING finger 1 null mice: response to synthetic glucocorticoids. J Physiol 589: 4759–4776, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294: 1704–1708, 2001 [DOI] [PubMed] [Google Scholar]
- 6.Centner T, Yano J, Kimura E, McElhinny AS, Pelin K, Witt CC, Bang ML, Trombitas K, Granzier H, Gregorio CC, Sorimachi H, Labeit S. Identification of muscle specific ring finger proteins as potential regulators of the titin kinase domain. J Mol Biol 306: 717–726, 2001 [DOI] [PubMed] [Google Scholar]
- 7.Cernotta N, Clocchiatti A, Florean C, Brancolini C. Ubiquitin-dependent degradation of HDAC4, a new regulator of random cell motility. Mol Biol Cell 22: 278–289, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi MC, Cohen TJ, Barrientos T, Wang B, Li M, Simmons BJ, Yang JS, Cox GA, Zhao Y, Yao TP. A direct HDAC4-MAP kinase crosstalk activates muscle atrophy program. Mol Cell 47: 122–132, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke BA, Drujan D, Willis MS, Murphy LO, Corpina RA, Burova E, Rakhilin SV, Stitt TN, Patterson C, Latres E, Glass DJ. The E3 Ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab 6: 376–385, 2007 [DOI] [PubMed] [Google Scholar]
- 10.Cohen S, Brault JJ, Gygi SP, Glass DJ, Valenzuela DM, Gartner C, Latres E, Goldberg AL. During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J Cell Biol 185: 1083–1095, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen TJ, Barrientos T, Hartman ZC, Garvey SM, Cox GA, Yao TP. The deacetylase HDAC4 controls myocyte enhancing factor-2-dependent structural gene expression in response to neural activity. FASEB J 23: 99–106, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen TJ, Waddell DS, Barrientos T, Lu Z, Feng G, Cox GA, Bodine SC, Yao TP. The histone deacetylase HDAC4 connects neural activity to muscle transcriptional reprogramming. J Biol Chem 282: 33752–33759, 2007 [DOI] [PubMed] [Google Scholar]
- 13.Dai KS, Liew CC. A novel human striated muscle RING zinc finger protein, SMRZ, interacts with SMT3b via its RING domain. J Biol Chem 276: 23992–23999, 2001 [DOI] [PubMed] [Google Scholar]
- 14.Devi YS, Seibold AM, Shehu A, Maizels E, Halperin J, Le J, Binart N, Bao L, Gibori G. Inhibition of MAPK by prolactin signaling through the short form of its receptor in the ovary and decidua: involvement of a novel phosphatase. J Biol Chem 286: 7609–7618, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubuquoy C, Burnol AF, Moldes M. PNPLA3, a genetic marker of progressive liver disease, still hiding its metabolic function? Clin Res Hepatol Gastroenterol 37: 30–35, 2013 [DOI] [PubMed] [Google Scholar]
- 16.Ebert SM, Monteys AM, Fox DK, Bongers KS, Shields BE, Malmberg SE, Davidson BL, Suneja M, Adams CM. The transcription factor ATF4 promotes skeletal myofiber atrophy during fasting. Mol Endocrinol 24: 790–799, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geng F, Wenzel S, Tansey WP. Ubiquitin and proteasomes in transcription. Annu Rev Biochem 81: 177–201, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomes AV, Waddell DS, Siu R, Stein M, Dewey S, Furlow JD, Bodine SC. Upregulation of proteasome activity in muscle RING finger 1-null mice following denervation. FASEB J 26: 2986–2999, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gounarides JS, Korach-Andre M, Killary K, Argentieri G, Turner O, Laurent D. Effect of dexamethasone on glucose tolerance and fat metabolism in a diet-induced obesity mouse model. Endocrinology 149: 758–766, 2008 [DOI] [PubMed] [Google Scholar]
- 20.Gundersen K, Leberer E, Lomo T, Pette D, Staron RS. Fibre types, calcium-sequestering proteins and metabolic enzymes in denervated and chronically stimulated muscles of the rat. J Physiol 398: 177–189, 1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hakim O, John S, Ling JQ, Biddie SC, Hoffman AR, Hager GL. Glucocorticoid receptor activation of the Ciz1-Lcn2 locus by long range interactions. J Biol Chem 284: 6048–6052, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herquel B, Ouararhni K, Khetchoumian K, Ignat M, Teletin M, Mark M, Bechade G, Van Dorsselaer A, Sanglier-Cianferani S, Hamiche A, Cammas F, Davidson I, Losson R. Transcription cofactors TRIM24, TRIM28, and TRIM33 associate to form regulatory complexes that suppress murine hepatocellular carcinoma. Proc Natl Acad Sci USA 108: 8212–8217, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirner S, Krohne C, Schuster A, Hoffmann S, Witt S, Erber R, Sticht C, Gasch A, Labeit S, Labeit D. MuRF1-dependent regulation of systemic carbohydrate metabolism as revealed from transgenic mouse studies. J Mol Biol 379: 666–677, 2008 [DOI] [PubMed] [Google Scholar]
- 24.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57, 2009 [DOI] [PubMed] [Google Scholar]
- 25.Kedar V, McDonough H, Arya R, Li HH, Rockman HA, Patterson C. Muscle specific Ring Finger I is a bona fide ubiquitin ligase that degrades cardiac troponin I. Proc Natl Acad Sci USA 52: 18135–18140, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelly EJ, Sandgren EP, Brinster RL, Palmiter RD. A pair of adjacent glucocorticoid response elements regulate expression of two mouse metallothionein genes. Proc Natl Acad Sci USA 94: 10045–10050, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim N, Stiegler AL, Cameron TO, Hallock PT, Gomez AM, Huang JH, Hubbard SR, Dustin ML, Burden SJ. Lrp4 is a receptor for Agrin and forms a complex with MuSK. Cell 135: 334–342, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirsh O, Seeler JS, Pichler A, Gast A, Muller S, Miska E, Mathieu M, Harel-Bellan A, Kouzarides T, Melchior F, Dejean A. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J 21: 2682–2691, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koyama S, Hata S, Witt CC, Ono Y, Lerche S, Ojima K, Chiba T, Doi N, Kitamura F, Tanaka K, Abe K, Witt SH, Rybin V, Gasch A, Franz T, Labeit S, Sorimachi H. Muscle RING-finger protein-1 (MuRF1) as a connector of muscle energy metabolism and protein synthesis. J Mol Biol 376: 1224–1236, 2008 [DOI] [PubMed] [Google Scholar]
- 30.Kuo T, Lew MJ, Mayba O, Harris CA, Speed TP, Wang JC. Genome-wide analysis of glucocorticoid receptor-binding sites in myotubes identifies gene networks modulating insulin signaling. Proc Natl Acad Sci USA 109: 11160–11165, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuroda Y, Kuriyama H, Kihara S, Kishida K, Maeda N, Hibuse T, Nishizawa H, Matsuda M, Funahashi T, Shimomura I. Insulin-mediated regulation of decidual protein induced by progesterone (DEPP) in adipose tissue and liver. Horm Metab Res 42: 173–177, 2010 [DOI] [PubMed] [Google Scholar]
- 32.Labeit S, Kohl CH, Witt CC, Labeit D, Jung J, Granzier H. Modulation of muscle atrophy, fatigue and MLC phosphorylation by MuRF1 as indicated by hindlimb suspension studies on MuRF1-KO mice. J Biomed Biotechnol 2010: 693741, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Latres E, Amini AR, Amini AA, Griffiths J, Martin FJ, Wei Y, Lin HC, Yancopoulos GD, Glass DJ. Insulin-like growth factor-1 (IGF-1) inversely regulates atrophy-induced genes via the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR) pathway. J Biol Chem 280: 2737–2744, 2005 [DOI] [PubMed] [Google Scholar]
- 34.Laure L, Suel L, Roudaut C, Bourg N, Ouali A, Bartoli M, Richard I, Daniele N. Cardiac ankyrin repeat protein is a marker of skeletal muscle pathological remodelling. FEBS J 276: 669–684, 2009 [DOI] [PubMed] [Google Scholar]
- 35.Li JZ, Huang Y, Karaman R, Ivanova PT, Brown HA, Roddy T, Castro-Perez J, Cohen JC, Hobbs HH. Chronic overexpression of PNPLA3I148M in mouse liver causes hepatic steatosis. J Clin Invest 122: 4130–4144, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu F, Dowling M, Yang XJ, Kao GD. Caspase-mediated specific cleavage of human histone deacetylase 4. J Biol Chem 279: 34537–34546, 2004 [DOI] [PubMed] [Google Scholar]
- 37.McElhinny AS, Kakinuma K, Sorimachi H, Labeit S, Gregorio CC. Muscle-specific RING finger-1 interacts with titin to regulate sarcomeric M-line and thick filament structure and may have nuclear functions via its interaction with glucocorticoid modulatory element binding protein-1. J Cell Biol 157: 125–136, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McLoughlin TJ, Smith SM, DeLong AD, Wang H, Unterman TG, Esser KA. FoxO1 induces apoptosis in skeletal myotubes in a DNA-binding-dependent manner. Am J Physiol Cell Physiol 297: C548–C555, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mearini G, Gedicke C, Schlossarek S, Witt CC, Kramer E, Cao P, Gomes MD, Lecker SH, Labeit S, Willis MS, Eschenhagen T, Carrier L. Atrogin-1 and MuRF1 regulate cardiac MyBP-C levels via different mechanisms. Cardiovasc Res 85: 357–366, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mittal A, Bhatnagar S, Kumar A, Lach-Trifilieff E, Wauters S, Li H, Makonchuk DY, Glass DJ. The TWEAK-Fn14 system is a critical regulator of denervation-induced skeletal muscle atrophy in mice. J Cell Biol 188: 833–849, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moresi V, Williams AH, Meadows E, Flynn JM, Potthoff MJ, McAnally J, Shelton JM, Backs J, Klein WH, Richardson JA, Bassel-Duby R, Olson EN. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell 143: 35–45, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nabhan JF, Pan H, Lu Q. Arrestin domain-containing protein 3 recruits the NEDD4 E3 ligase to mediate ubiquitination of the beta2-adrenergic receptor. EMBO Rep 11: 605–611, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ochala J, Gustafson AM, Diez ML, Renaud G, Li M, Aare S, Qaisar R, Banduseela VC, Hedstrom Y, Tang X, Dworkin B, Ford GC, Nair KS, Perera S, Gautel M, Larsson L. Preferential skeletal muscle myosin loss in response to mechanical silencing in a novel rat intensive care unit model: underlying mechanisms. J Physiol 589: 2007–2026, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patwari P, Emilsson V, Schadt EE, Chutkow WA, Lee S, Marsili A, Zhang Y, Dobrin R, Cohen DE, Larsen PR, Zavacki AM, Fong LG, Young SG, Lee RT. The arrestin domain-containing 3 protein regulates body mass and energy expenditure. Cell Metab 14: 671–683, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, Randow F, Wakatsuki S, Dikic I. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell 136: 1098–1109, 2009 [DOI] [PubMed] [Google Scholar]
- 46.Ramanathan HN, Ye Y. Cellular strategies for making monoubiquitin signals. Crit Rev Biochem Mol Biol 47: 17–28, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reed SA, Sandesara PB, Senf SM, Judge AR. Inhibition of FoxO transcriptional activity prevents muscle fiber atrophy during cachexia and induces hypertrophy. FASEB J 26: 987–1000, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rudolf R, Bogomolovas J, Strack S, Choi KR, Khan MM, Wagner A, Brohm K, Hanashima A, Gasch A, Labeit D, Labeit S. Regulation of nicotinic acetylcholine receptor turnover by MuRF1 connects muscle activity to endo/lysosomal and atrophy pathways. Age (Dordr) 35: 1663–1674, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sacheck JM, Hyatt JP, Raffaello A, Jagoe RT, Roy RR, Edgerton VR, Lecker SH, Goldberg AL. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J 21: 140–155, 2007 [DOI] [PubMed] [Google Scholar]
- 50.Schalkwyk LC, Fernandes C, Nash MW, Kurrikoff K, Vasar E, Koks S. Interpretation of knockout experiments: the congenic footprint. Genes Brain Behav 6: 299–303, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stitt TN, Drujan D, Clarke BA, Panaro FJ, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell 14: 1–14, 2004 [DOI] [PubMed] [Google Scholar]
- 52.Sugamori KS, Brenneman D, Wong S, Gaedigk A, Yu V, Abramovici H, Rozmahel R, Grant DM. Effect of arylamine acetyltransferase Nat3 gene knockout on N-acetylation in the mouse. Drug Metab Dispos 35: 1064–1070, 2007 [DOI] [PubMed] [Google Scholar]
- 53.Tang H, Goldman D. Activity-dependent gene regulation in skeletal muscle is mediated by a histone deacetylase (HDAC)-Dach2-myogenin signal transduction cascade. Proc Natl Acad Sci USA 103: 16977–16982, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang H, Macpherson P, Marvin M, Meadows E, Klein WH, Yang XJ, Goldman D. A histone deacetylase 4/myogenin positive feedback loop coordinates denervation-dependent gene induction and suppression. Mol Biol Cell 20: 1120–1131, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.U M, Shen L, Oshida T, Miyauchi J, Yamada M, Miyashita T. Identification of novel direct transcriptional targets of glucocorticoid receptor. Leukemia 18: 1850–1856, 2004 [DOI] [PubMed] [Google Scholar]
- 56.Valor LM, Grant SG. Clustered gene expression changes flank targeted gene loci in knockout mice. PLoS One 2: e1303, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waddell DS, Baehr LM, van den Brandt J, Johnsen SA, Reichardt HM, Furlow JD, Bodine SC. The glucocorticoid receptor and FOXO1 synergistically activate the skeletal muscle atrophy-associated MuRF1 gene. Am J Physiol Endocrinol Metab 295: E785–E797, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watson ML, Baehr LM, Reichardt HM, Tuckermann JP, Bodine SC, Furlow JD. A cell-autonomous role for the glucocorticoid receptor in skeletal muscle atrophy induced by systemic glucocorticoid exposure. Am J Physiol Endocrinol Metab 302: E1210–E1220, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL, Bassel-Duby R, Sanes JR, Olson EN. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 326: 1549–1554, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Willis MS, Ike C, Li L, Wang DZ, Glass DJ, Patterson C. Muscle ring finger 1, but not muscle ring finger 2, regulates cardiac hypertrophy in vivo. Circ Res 100: 456–459, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Willis MS, Schisler JC, Li L, Rodriguez JE, Hilliard EG, Charles PC, Patterson C. Cardiac muscle ring finger-1 increases susceptibility to heart failure in vivo. Circ Res 105: 80–88, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winbanks CE, Wang B, Beyer C, Koh P, White L, Kantharidis P, Gregorevic P. TGF-beta regulates miR-206 and miR-29 to control myogenic differentiation through regulation of HDAC4. J Biol Chem 286: 13805–13814, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu Q, Huang S, Sun Y, Gu S, Lu F, Dai J, Yin G, Sun L, Zheng D, Dou C, Feng C, Ji C, Xie Y, Mao Y. Dual specificity phosphotase 18, interacting with SAPK, dephosphorylates SAPK and inhibits SAPK/JNK signal pathway in vivo. Front Biosci 11: 2714–2724, 2006 [DOI] [PubMed] [Google Scholar]
- 65.Zhang B, Luo S, Wang Q, Suzuki T, Xiong WC, Mei L. LRP4 serves as a coreceptor of agrin. Neuron 60: 285–297, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]