Coupling of replication fork speed and PCNA unloading to nucleosome assembly may maintain chromatin integrity during transient histone shortage.
Abstract
Correct duplication of DNA sequence and its organization into chromatin is central to genome function and stability. However, it remains unclear how cells coordinate DNA synthesis with provision of new histones for chromatin assembly to ensure chromosomal stability. In this paper, we show that replication fork speed is dependent on new histone supply and efficient nucleosome assembly. Inhibition of canonical histone biosynthesis impaired replication fork progression and reduced nucleosome occupancy on newly synthesized DNA. Replication forks initially remained stable without activation of conventional checkpoints, although prolonged histone deficiency generated DNA damage. PCNA accumulated on newly synthesized DNA in cells lacking new histones, possibly to maintain opportunity for CAF-1 recruitment and nucleosome assembly. Consistent with this, in vitro and in vivo analysis showed that PCNA unloading is delayed in the absence of nucleosome assembly. We propose that coupling of fork speed and PCNA unloading to nucleosome assembly provides a simple mechanism to adjust DNA replication and maintain chromatin integrity during transient histone shortage.
Introduction
When cells divide, the entire genome must be accurately replicated, and its chromatin landscape must be reproduced. This takes place in S phase of the cell cycle, during which thousands of replication forks traverse the chromosomes. Chromatin structure is disrupted ahead of replication forks and restored on the two new daughter DNA strands. The initial step in chromatin restoration, nucleosome assembly, relies on local recycling of parental histones along with deposition of newly synthesized histones through the Asf1–CAF-1 pathway (Alabert and Groth, 2012; Annunziato, 2012). Defects in chromatin assembly can jeopardize transmission of epigenetically defined chromatin states (Zhang et al., 2000; Alabert and Groth, 2012), and cellular aging is associated with global changes in chromatin structure that may derive from insufficient histone supply (Feser et al., 2010; O’Sullivan et al., 2010). Furthermore, two developmental disorders, Wolf–Hirschhorn syndrome and CDAI (congenital dyserythropoietic anemia type I), have recently been associated with aberrant production and delivery of new histones (Ask et al., 2012; Kerzendorfer et al., 2012). Even so, it remains largely unknown how replicating cells respond to shortage of new histones and whether specialized mechanisms have evolved to prevent loss of chromatin integrity.
The high demand for canonical histones (H3.1, H3.2, H4, H2A, H2B, and H1) throughout S phase is met by coordinated expression of multiple histone genes, giving rise to ∼75 distinct mammalian histone mRNAs (Marzluff et al., 2008). Histone biosynthesis is regulated both at the transcriptional and posttranscriptional level within the cell cycle (Marzluff et al., 2008). In addition, DNA damage signaling can promote histone mRNA degradation to prevent accumulation of toxic excess histones in mammals (Kaygun and Marzluff, 2005), and yeast checkpoint kinases participate to histone homeostasis by controlling histone degradation (Gunjan and Verreault, 2003). Histone biosynthesis is required for survival, but yeast cells can complete one round of replication in the absence of new histone production (Kim et al., 1988; Prado and Aguilera, 2005). In contrast, early work in mammalian systems using protein synthesis inhibitors led to the hypothesis that new histones are directly required for DNA synthesis (Weintraub, 1972; Seale and Simpson, 1975). Consistent with this, inhibition of histone biosynthesis has been shown to impair S-phase progression (Nelson et al., 2002; Zhao et al., 2004; Barcaroli et al., 2006). The coupling of nucleosome assembly to DNA synthesis relies on the recruitment of CAF-1 to the PCNA replication clamp (Shibahara and Stillman, 1999; Moggs et al., 2000), and both CAF-1 and the upstream chaperone Asf1 are required for DNA replication in mammalian cells (Hoek and Stillman, 2003; Ye et al., 2003; Nabatiyan and Krude, 2004; Groth et al., 2007). This may reflect a direct need for new histone supply and/or de novo nucleosome assembly, though these factors serve multiple functions at replication forks (Quivy et al., 2008). CAF-1 interacts with repair factors (Schöpf et al., 2012) and chromatin regulators, such as HP1 (Murzina et al., 1999; Quivy et al., 2004) and SETDB1 (Sarraf and Stancheva, 2004; Loyola et al., 2009), whereas Asf1 interacts with TONSL-MMS22L (Duro et al., 2010) and the replicative helicase MCM2-7 (Groth et al., 2007; Jasencakova et al., 2010). We thus decided to address directly how the supply of new histones, key chromosomal building blocks, influences DNA replication. By combining RNAi of two central factors in histone biosynthesis with advanced analysis of DNA replication and chromatin assembly, we find that new histone provision controls replication fork speed and limits PCNA unloading. Given that fork slowdown and PCNA retention offer an opportunity to recruit CAF-1 and deposit H3.1-H4 once histones are available, this provides a simple mechanism to adjust DNA replication with nucleosome assembly and prevents loss of chromatin integrity during genome duplication.
Results
Replication fork progression is dependent on new histone supply
To address how histone supply regulates DNA replication in human cells, we chose to target FLASH and SLBP, two key regulators of histone biosynthesis. FLASH orchestrates expression of canonical histone genes and participates in initial mRNA processing (Barcaroli et al., 2006; Yang et al., 2009), whereas SLBP governs stability, processing, nuclear export, and translation (Marzluff et al., 2008) of histone mRNAs by binding to their conserved stem loop structure. siRNA depletion of either FLASH or SLBP reduced histone mRNA levels, inhibited DNA replication, and prolonged S-phase progression in agreement with previous studies (Fig. S1, A–C; and not depicted; Zhao et al., 2004; Barcaroli et al., 2006). Similar results were obtained by short-term inhibition of protein synthesis by cycloheximide (CHX; Fig. S1 D), which has routinely been used as a rapid, though unspecific, means to block histone biosynthesis. To study the immediate effects of histone shortage on DNA replication, we established a synchronization strategy to follow the first S phase in which cells lack canonical histones (Fig. 1 A, top). In FLASH- and SLBP-depleted cells, the pools of soluble histone H3 and new histone H4 marked by K12 acetylation (Sobel et al., 1995; Alabert and Groth, 2012) were reduced (Fig. 1 A, bottom), and DNA replication was inhibited by ≤80% as measured by [3H]thymidine uptake (Fig. 1 B). In comparison, inhibition of replication by dNTP depletion by hydroxyurea (HU) leads to accumulation of new histone H3-H4 as shown previously (Fig. 1 A; Groth et al., 2005). The soluble pool of histone H2B was largely unaffected by FLASH and SLBP depletion (Fig. 1 A), indicating that the pool of H3.1-H4 is depleted more rapidly than the H2A-H2B pool. This argues that insufficient supply of histone H3-H4 limits S-phase progression in FLASH- and SLBP-depleted cells. Consistently, coexpression of H3.1 and H4 in an inducible cell system (Groth et al., 2007) partially rescued S-phase progression in FLASH- and SLBP-depleted cells (Fig. S1 E). Addback of histone H3-H4 was accompanied by a reduction in the soluble H2B pool (Fig. S1 F), suggesting that histone H2A-H2B becomes limiting when H3.1-H4 is provided. Thus, a full rescue of replication in FLASH- and SLBP-depleted cells most likely requires exogenous provision of all canonical histone subtypes.
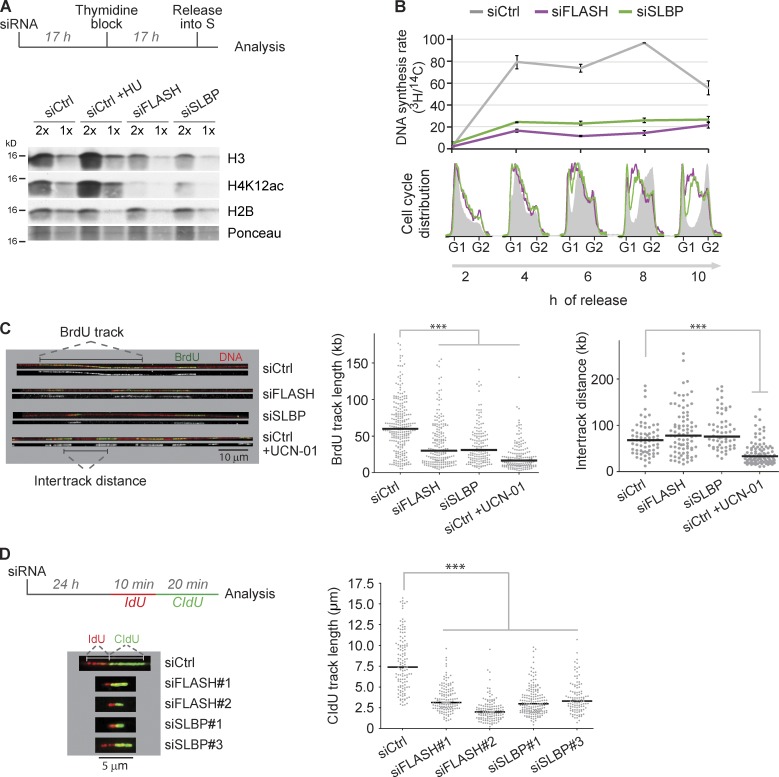
Figure 1.
Fork progression is dependent on new histone supply. (A and B) Analysis of histone supply and DNA replication rate in SLBP- and FLASH-depleted cells. (A, top) Experimental setup. U-2-OS cells were transfected with siRNAs, synchronized at the G1/S border by a thymidine block, and released into S phase. (bottom) Western blot of soluble histones in extracts harvested 6 h after release. For comparison, cells were treated 2 h with HU. (B, top) Measurement of DNA synthesis rate by [3H]thymidine pulse labeling normalized to total DNA labeled by [14C]thymidine. To label total DNA, cells were incubated with [14C]thymidine for one cell generation before siRNA treatment. After siRNA transfection, cells were synchronized as shown in A. Error bars represent the SDs of three measurements. (bottom) FACS analysis of combed DNA content. One representative experiment out of two biological replicas is shown. (C) Single-molecule analysis of DNA replication. (left) Representative images of DNA fibers from cells pulse labeled 45 min with BrdU 24 h after siRNA transfection (BrdU, green [merged] or white [alone]). (middle) Size distribution of BrdU track length. Bars represent the median. Statistics: Mann–Whitney; n > 150; ***, P < 10−3. (right) Distribution of intertrack distances. Bars represent the median. Statistics: Mann–Whitney; n > 60; ***, P < 10−3. One representative experiment out of two biological replicas is shown. (D) DNA fiber analysis of replication elongation rate. (left) Experimental design and representative images of DNA fibers. (right) Size distribution of CldU track length. Bars represent the median. Statistics: Mann–Whitney; n > 100; ***, P < 10−3. One representative experiment out of two biological replicas is shown. siCtrl, siRNA control.
To elucidate whether histone deficiency impairs DNA replication at the level of elongation or new origin firing, we analyzed replication of single DNA molecules by DNA combing. We labeled active replication forks by a BrdU pulse after 24 h of siRNA treatment to focus on the short-term effects of histone depletion (Fig. 1 C). For comparison, we treated cells with the Chk1 inhibitor UCN-01, which triggers firing of dormant origins and impairs fork progression (Maya-Mendoza et al., 2007; Petermann et al., 2010). Upon FLASH and SLBP depletion, the distribution of BrdU-labeled tracks shifted markedly toward shorter track lengths (Fig. S2 A). The median track length was reduced from 60 kb in controls to 30 kb in siFLASH- and siSLBP-depleted cells (P < 10−3), corresponding to a 50% reduction of fork speed (Fig. 1 C, middle). In contrast, intertrack distance did not change significantly (Fig. 1 C, right). These data indicate that histone deficiency impairs fork progression without increasing the rate of initiation. This argues that although fork speed is slowed down when cells experience a shortage of new histones, dormant origins are not fired to compensate for slow fork movement in contrast to the situation with nucleotide depletion (Anglana et al., 2003; Ge et al., 2007; Courbet et al., 2008). Consistent with slow fork progression and lack of compensatory origin firing, S phase was prolonged to >18 h upon SLBP and FLASH depletion (Fig. S1 G). Although we cannot entirely rule out that fewer forks are present in FLASH- and SLBP-depleted cells, several observations argue against this. First, intertrack distance is largely unaffected by lack of SLBP and FLASH (Fig. 1 C, right). Second, FLASH- and SLBP-depleted cells do not arrest in G1 as judged by FACS analysis (Fig. S1 B). Additionally, FLASH- and SLBP-depleted cells show normal activation of Chk1 in response to aphidicolin treatment (Fig. S3 A). Finally, to rule out that track length in the single-label DNA combing experiments was influenced by reduced initiation, we used double-pulse labeling with 5-iodo-2′-deoxyuridine (IdU) and 5-chloro-2′-deoxyuridine (CldU) to measure fork speed in a DNA fiber assay. In this assay, fork rates were also significantly reduced (about threefold) upon 24-h depletion of SLBP and FLASH (Fig. 1 D and Fig. S2 B), supporting that the supply of canonical histones controls elongation.
Slow fork progression is linked to impaired nucleosome assembly
Nucleosome assembly, the first step in chromatin restoration, takes place immediately behind the replication fork in a manner directly coupled to DNA synthesis through the recruitment of CAF-1 to PCNA (Shibahara and Stillman, 1999; Moggs et al., 2000). Newly synthesized DNA then converts from a nascent immature chromatin state, highly sensitive to nucleases, into a mature state within 15–20 min (Alabert and Groth, 2012; Annunziato, 2012). We asked whether fork slowdown in cells lacking new histones is connected to impaired chromatin assembly by measuring nuclease sensitivity of global and nascent chromatin labeled with [14C]thymidine and [3H]thymidine, respectively. Indeed, nascent chromatin from FLASH- and SLBP-depleted cells was significantly more sensitive to micrococcal nuclease (MNase) as compared with control cells (Fig. 2 A). Global chromatin was not affected, validating our approach to study the immediate effects of new histone depletion rather than secondary events caused by global changes in chromatin structure.
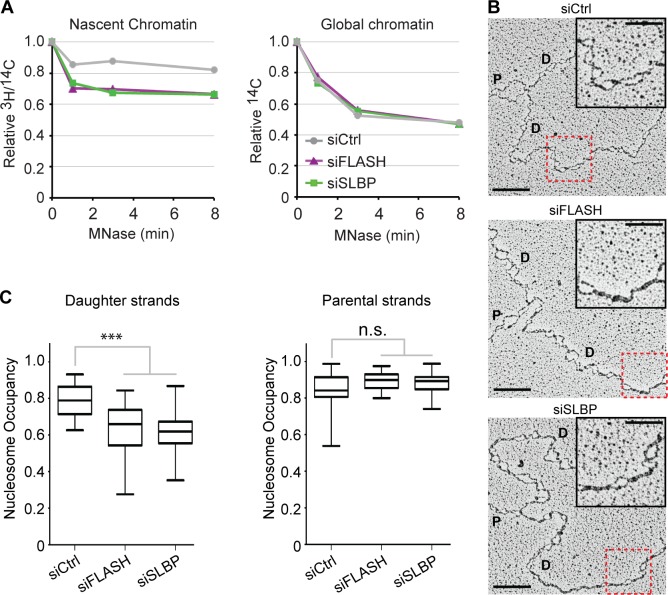
Figure 2.
Low nucleosome occupancy on daughter DNA strands. (A) MNase sensitivity of nascent and global chromatin. Cells prelabeled for one generation with [14C]thymidine were transfected, synchronized to G1/S as in Fig. 1 A, and released for 6 h. Newly synthesized DNA was labeled by a short [3H]thymidine pulse as in Fig. 1 B before nuclei were isolated and subjected to MNase digestion. Relative 3H/14C activity in undigested chromatin is displayed against MNase digestion time. One representative experiment out of two biological replicas is shown. (B and C) Analysis of psoralen–cross-linked replication intermediates by EM. (B) Representative images. P and D annotate parental and daughter strands. Bars: (main images) 200 nm (500 bp); (insets) 100 nm. The red boxes highlight the magnified insets. Full DNA molecules are shown in Fig. S2 C. (C) Nucleosome occupancy on parental and daughter DNA strands. Median is displayed. Boxes are 25–75 percentile ranges, and whiskers are 0–100 percentile ranges. Statistics: two-tailed t test; (left) n ≥ 40; ***, P < 10−3; (right) n > 20; n.s., P > 0.05. siCtrl, siRNA control.
To directly visualize nucleosome organization at forks, we took advantage of an established transmission EM-based approach (Fig. 2 B and Fig. S2, C and D; Sogo et al., 1986; Neelsen et al., 2014). Nucleosome density was markedly decreased on daughter strands in FLASH- and SLBP-depleted cells (Fig. 2 C, left) and not fully restored even at large distance (>6 kb) from the forks (Fig. S2 D), whereas it was promptly restored behind control replication forks. However, nucleosome occupancy on parental strands (Fig. 2 C, right) was not affected upon new histone depletion. Similar results were obtained by short-term inhibition of protein synthesis by CHX (Fig. S2, E and F). Thus, fork slowdown upon histone deficiency is linked to impaired nucleosome assembly and accumulation of immature chromatin behind replication forks.
Short-term histone deficiency does not challenge replisome stability
Replication stress caused by nucleotide depletion, polymerase inhibition, or UV lesions activates ATR-Chk1 checkpoint signaling as part of a program to maintain genome integrity (Cimprich and Cortez, 2008). In contrast, cells lacking new histones did not show DNA damage (γ-H2AX) or replication fork collapse (p–RPA; Fig. 3, A and B; and Fig. S3, A and B). This was true after short-term depletion of FLASH and SLBP in asynchronous cells (Fig. 3, A and B) and in the first S phase after thymidine synchronization (Fig. S3 A). Similar results were obtained in primary human fibroblasts (Fig. S3 C). As replication and chromatin assembly defects were evident under these conditions (Fig. 1 and Fig. 2), the lack of checkpoint signaling was unexpected. However, new histone deprivation did not cause a general defect in checkpoint signaling, as both ATR and ATM signaling were readily induced by aphidicolin and ionizing radiation (Fig. S3, A and B). Moreover, ATR/ATM inhibition by caffeine did not rescue DNA replication in FLASH-depleted cells (Fig. S3 D), consistent with checkpoint-independent fork slowdown. We thus conclude that impaired fork progression and lack of nucleosome assembly caused by histone deficiency do not pose an immediate challenge to replisome stability.
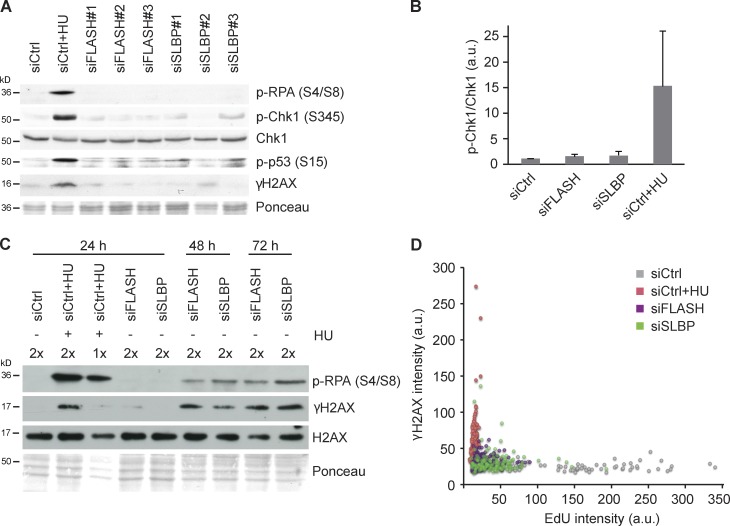
Figure 3.
Replication forks arrested by new histone deficiency remain stable and do not activate conventional checkpoints. (A) Western blot of cells treated 24 h with independent siRNAs against SLBP and FLASH. Cells treated 1 h with HU were included as a positive control. (B) Chk1 pS345 levels quantified relative to total Chk1 in cells treated as in A. n = 6. Error bars represent SDs. (C) Time course analysis of DNA damage and fork collapse markers in FLASH- and SLBP-depleted cells. Cells were treated with siRNAs for 24, 48, or 72 h and analyzed by Western blotting. (D) Dot plot of EdU and γ-H2AX intensities in S-phase cells quantified by single-cell imaging 24 h after siRNA transfection. Cells treated 1 h with HU were included as the positive control, and S-phase cells were identified by PCNA staining. Cells were pulsed with EdU for 15 min. n > 70. a.u., arbitrary unit; siCtrl, siRNA control.
Several studies have addressed the consequences of chromatin assembly defects resulting from lack of CAF-1 function (Hoek and Stillman, 2003; Ye et al., 2003; Nabatiyan and Krude, 2004; Takami et al., 2007; Quivy et al., 2008), and some of these found evidence of DNA damage and checkpoint activation (Hoek and Stillman, 2003; Ye et al., 2003; Nabatiyan and Krude, 2004). Although this could reflect additional defects in cells lacking CAF-1 as compared with those deprived of new histones, we considered that problems might develop in cells experiencing prolonged replication arrest. We thus followed the response to SLBP and FLASH depletion in a time course analysis. Indeed, at late time points, 48 and 72 h after siRNA treatment, cells with impaired histone biosynthesis showed signs of DNA damage (p-RPA and γ-H2AX; Fig. 3 C and Fig. S3 E). There was some variation in the timing and degree of this response, but these markers were generally not detectable after short-term histone depletion (Fig. 3 A) when replication fork speed was significantly reduced (Fig. 1 C). To exclude the possibility that a fraction of cells with severe replication defects experienced DNA damage at early time points, we compared replication efficiency (5-ethynyl-2′-deoxyuridine [EdU] incorporation) directly with DNA damage signaling (γ-H2AX) by quantitative single-cell imaging. This analysis showed severely impaired DNA synthesis in a large proportion of SLBP- and FLASH-depleted cells, which nevertheless remained γ-H2AX negative (Fig. 3 D). Collectively, this implies that the cellular response to histone deficiency and impaired nucleosome assembly shifts from a benign replication fork slowdown at early stages to DNA damage upon persistent defects (Hoek and Stillman, 2003; Ye et al., 2003; Nabatiyan and Krude, 2004).
Knockdown of specific histone mRNAs inhibits replication
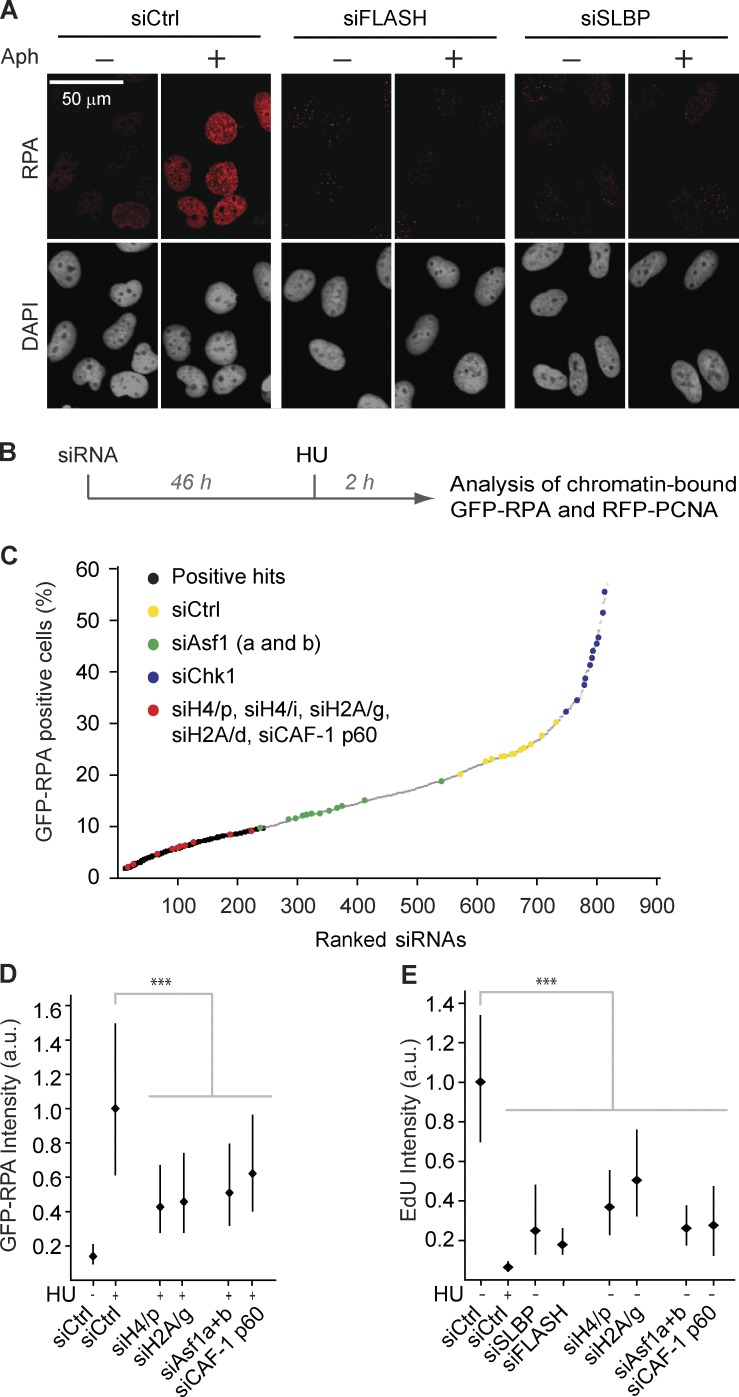
Replication forks arrested by short-term HU or aphidicolin treatment accumulate single-strand DNA (ssDNA; Cimprich and Cortez, 2008), presumably as a result of polymerase and helicase uncoupling along with additional fork-processing events (Sogo et al., 2002; Pacek and Walter, 2004; Fugger et al., 2009; Schlacher et al., 2011). We did not detect ssDNA formation at replication sites in cells depleted from SLBP and FLASH, as assessed by RPA or BrdU staining (Fig. 4 A and Fig. S3 F). This is consistent with the finding that replication forks remain stable under these conditions. However, we discovered that FLASH and SLBP depletion strongly impairs ssDNA formation in response to replication inhibitors (Fig. 4 A and Fig. S3 F). This is similar to cells lacking Asf1 (Groth et al., 2007), implying that histone shortage and lack of Asf1 arrest DNA replication in a comparable state in which polymerase and helicase uncoupling and/or fork processing is constrained. To identify other factors that likewise are required for ssDNA formation in response to deoxynucleotide (dNTP) depletion, we screened a small custom-made siRNA library targeting mainly replication and repair factors (Fig. 4 B and Table S4). To monitor ssDNA formation in S-phase cells, we used a reporter cell line expressing RFP-PCNA and GFP-RPA1 (Fig. S4 A). Two independent screens identified 55 genes, which upon knockdown impaired formation of ssDNA in response to HU treatment (Fig. 4 C; Fig. S4, B and C; and Table S5). Parallel analysis of PCNA excluded that the defect in ssDNA formation was simply caused by a low number of cells in S phase (Table S5). The hits included the ssDNA binding proteins RPA2, hSSBP1, and hSSBP2, several general replication factors (POLE, POLA, MCM3, ORC4, and CDC45) as well as potential fork-processing enzymes (Fig. S4 C and Table S5). Thus, although we had anticipated a rather narrow group of hits emerging from the screen, it appears that the ssDNA response is highly sensitive to perturbation of both replisome function and origin firing. Notably, our screen scored numerous siRNAs targeting specific histone genes as well as the histone chaperone CAF-1 p60 that interacts with Asf1 (Fig. 4 C and Table S5). We verified knockdown efficiency and the ability to block HU-induced ssDNA exposure for a set of siRNAs targeting H4, H2A, and CAF-1 p60 (Fig. 4 D and Fig. S4, D and E). Similar to FLASH and SLBP depletion, these siRNAs repressed DNA replication (Fig. 4 E). This provides direct evidence that lack of canonical histones slows down replication and blocks fork uncoupling. Our rescue experiment, in which FLASH and SLBP depletion was complemented with histone H3.1-H4, implied that the supply of both histone H3.1-H4 and H2A-H2B could be limiting for DNA replication (Fig. S1, E and F). The identification of histone H4 and H2A siRNAs in our screen is consistent with this notion, arguing that the response described here could be relevant to fluctuations in individual histone subtypes as well as global histone supply.
Figure 4.
Imbalanced histone supply impairs DNA replication. (A) Chromatin-bound RPA detected by immunofluorescence in preextracted cells. Cells were transfected, synchronized, and released into S phase for 6 h as in Fig. 1 A and treated with aphidicolin for 1 h as indicated. (B and C) siRNA screen for factors required for HU-induced RPA accumulation in chromatin. (B) Experimental setup. A reporter cell line expressing GFP-RPA1 and RFP-PCNA was transfected with a custom-made siRNA library consisting of 236 genes, targeted by three individual siRNAs. Scramble siRNA and siRNAs against Asf1 (a and b) and Chk1 were included as controls. See Fig. S4 for details. (C) Ranking of siRNAs from one representative screen according to the percentage of GFP-RPA–positive cells. (D and E) Microscopy-based high-throughput single-cell analysis of chromatin-bound RPA and EdU in siRNA-treated cells. Cells were treated 2 h with HU as indicated. Only S-phase cells positive for RFP-RCNA were analyzed. Median with interquartile range is shown. n > 7,000. Mann–Whitney: ***, P < 10−4. One representative experiment out of two biological replicas is shown. (D) Quantification of GFP-RPA intensity in PCNA-positive cells after preextraction. (E) Quantification of EdU and in PCNA-positive cells by immunofluorescence. RFP-PCNA reporter cells were pulsed 15 min with EdU. Aph, aphidicolin; a.u., arbitrary unit; siCtrl, siRNA control.
Nucleosome assembly is required for fork progression
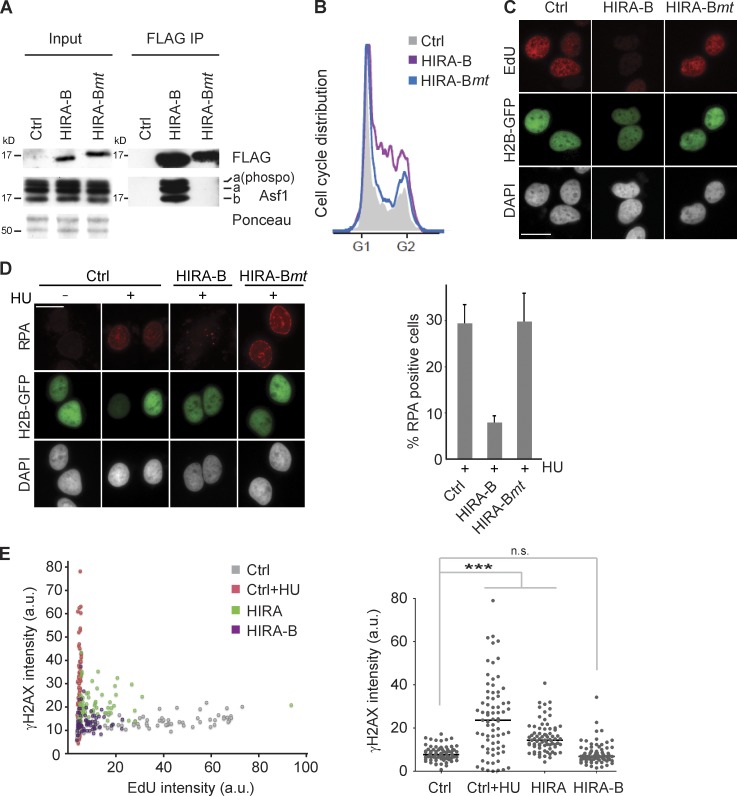
Histone pools could potentially control fork progression by regulating helicase activity, given that histone H3-H4 together with Asf1 can form a complex with MCM2-7 (Groth et al., 2007; Jasencakova et al., 2010) on chromatin. Alternatively, lack of nucleosome assembly might directly slow down fork progression. It was thus important to decipher whether histone pool size or the assembly of new DNA into nucleosomes controls fork progression. To this end, we designed a system to specifically block Asf1–CAF-1–mediated nucleosome assembly. We took advantage of the HIRA-B domain that binds with high affinity to the same pocket in Asf1 as CAF-1 p60 (Tang et al., 2006), as a means to block histone transfer from Asf1 to CAF-1. The HIRA-B domain is small (44 amino acids out of 1,017 in full-length human HIRA), highly soluble, and does not bind chromatin (unpublished data). We thus anticipate that it could work as an Asf1-blocking peptide without affecting histone expression. Of note, removal of the Asf1 protein itself by RNAi does not alter the cellular pools of histone H3-H4 (Cook et al., 2011). Overexpression of the HIRA-B domain arrested cells in S phase and inhibited DNA replication (Fig. 5, A–C). Importantly, this was entirely dependent on Asf1 binding, as mutation of three critical residues (Tang et al., 2006) in the interaction surface abrogated the effect on replication (Fig. 5, A–C). Artificial blocking of nucleosome assembly also impaired HU-induced ssDNA exposure significantly (Fig. 5 D). This is similar to the phenotype of histone deficiency and depletion of the small CAF-1 subunit p60 (Fig. 4, A, D, and E; and Fig. S3 F), required for delivery of new histones to CAF-1 (Ray-Gallet et al., 2011). To address whether replication arrest was accompanied by DNA damage, we again took advantage of single-cell imaging. In addition to the short B domain peptide, we also included full-length HIRA in the analysis as it was reported to induce checkpoint signaling (Nelson et al., 2002; Ye et al., 2003). We observed a moderate increase of γ-H2AX in HIRA-expressing cells, but γ-H2AX was not increased in cells in which DNA replication was repressed by expression of the HIRA-B domain (Fig. 5 E). The reason for this difference is unclear, but it may reflect additional functions of the full-length HIRA protein. Collectively, these data argue that direct interference with histone delivery to CAF-1 mimics the fork-pausing phenotype of histone deficiency, arguing that nucleosome assembly is required for fork progression in human cells.
Figure 5.
Nucleosome assembly is required for fork progression. Artificial blocking of Asf1–CAF-1–mediated nucleosome assembly by transient expression of the HIRA-B domain fused to an SV40 NLS. (A) Immunoprecipitation (IP) of FLAG-HA–tagged HIRA-B and HIRA-Bmt carrying three point mutations disabling Asf1 binding. (B) Cell cycle profiles. GFP-spectrin was used to identify transfected cells by FACS. One representative experiment out of five biological replicas is shown. (C and D) EdU incorporation (C) and chromatin-bound RPA (D) detected by immunofluorescence in preextracted cells. Cells were cotransfected with H2B-GFP to identify transfected cells and treated 1 h with HU where indicated. Error bars indicate SDs of four biological replicas. Bars, 20 µm. (E) Single-cell analysis of DNA replication and DNA damage in U-2-OS cells transfected with full-length HIRA or the HIRA-B domain for 24 h. A dot plot of EdU and γ-H2AX intensities (left) and a scatter plot of γ-H2AX intensities (right) are shown. Cells treated 2 h with HU were included as a positive control. Cells were pulsed 15 min with EdU. Lines represent medians. n > 70. ***, P < 0.001. a.u., arbitrary unit; Ctrl, control.
Lack of nucleosome assembly delays PCNA unloading
To dissect how histone deficiency and lack of nucleosome assembly influence replisome composition, we monitored the chromatin-bound fraction of several replication factors by DNase I digestion. For comparison, we treated cells with HU, as the cellular responses to histone deficiency and dNTP depletion differ with respect to checkpoint activation (Fig. 3 A) and ssDNA formation (Fig. 4 A). Cells treated with HU accumulated RPA on chromatin, whereas PCNA levels decreased, consistent with previous studies (Fig. 6 A; Groth et al., 2007; Görisch et al., 2008). In contrast, PCNA accumulated on chromatin in cells lacking new histones (Fig. 6 A). Using our RFP-PCNA reporter cell line, we confirmed at the single-cell level that more PCNA is present on chromatin in cells depleted for FLASH, SLBP, CAF-1 p60, Asf1 (a and b), H2A/g, and H4/p (Fig. 6 B). A similar response was also seen in S-phase cells treated shortly with CHX (Fig. S5 B). This response likely reflects that PCNA clamps are retained on newly synthesized DNA that fails to be assembled into chromatin because (a) fork density was not increased (Fig. 1 C) and (b) replicative polymerases (polymerase δ and polymerase ε) did not accumulate in FLASH- and SLBP-depleted cells (Fig. S5 A). Of note, activation of dormant origins by short-term treatment with the Chk1 inhibitor UCN-01 is associated with accumulation of PCNA as well as DNA polymerases α and δ on chromatin (Fig. S5 B), but this was not observed in response to short-term CHX treatment (Fig. S5 B). In contrast, histone depletion moderately reduced the levels of chromatin-bound polymerase α (Fig. 6 A), suggesting that impaired nucleosome assembly might affect lagging strand synthesis.
Figure 6.
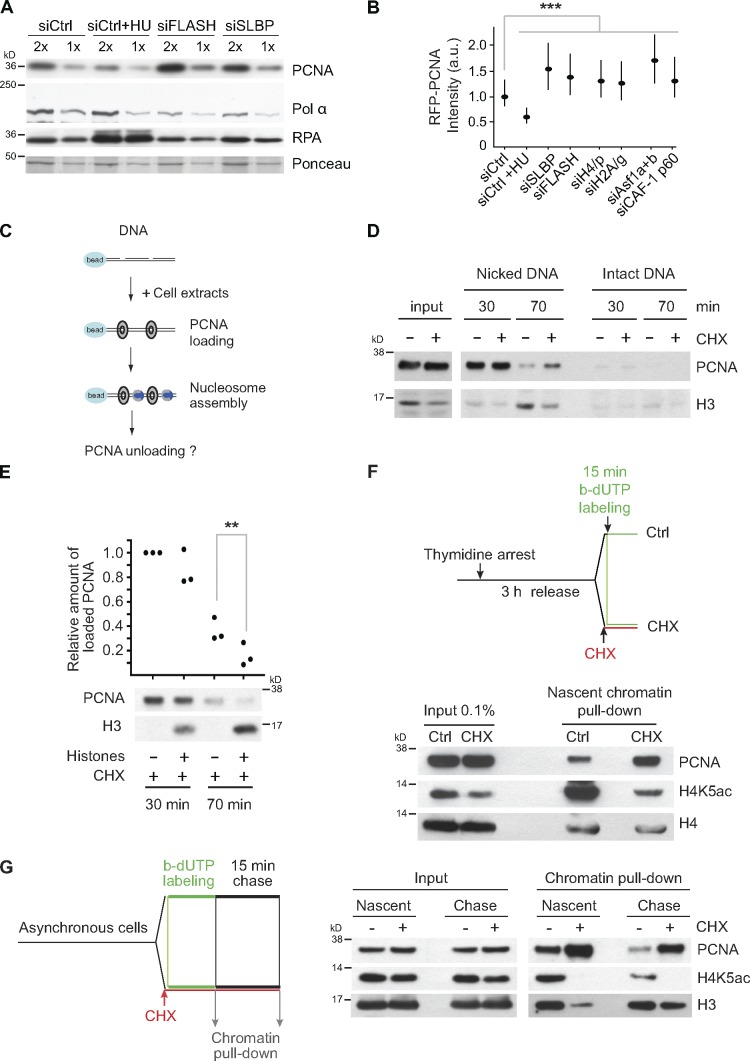
Retention of PCNA on newly synthesized DNA that fails to be assembled into nucleosomes. (A) Western blot of chromatin-bound material released by DNase I digestion. Cells were transfected, synchronized, and released into S phase for 6 h as in Fig. 1 A. Cells treated 1 h with HU were included as a control. (B) Quantification of chromatin-bound RFP-PCNA by single-cell imaging. Cells were treated 2 h with HU as a control. Median with interquartile range is shown. n > 7,000. Mann–Whitney: ***, P < 10−4. (C–E) In vitro analysis of PCNA unloading during DNA repair–coupled chromatin assembly. (C) Experimental setup. HeLa S3 cells in mid–S phase were left untreated (−) or treated (+) for 10 min with CHX to deplete the soluble histone pools (see input). Bead-coupled DNA nicked by DNase I was incubated in a chromatin assembly reaction with control extracts or histone-depleted extracts (+CHX). (D) Time course analyses of DNA-bound PCNA and histone H3 loading. Bead-coupled DNA not treated with DNase I was used as a negative control. (E) Time course analyses of DNA-bound PCNA and histone H3. Where indicated, extracts were supplemented with core histones in complex with Asf1. (top) Quantification of bound PCNA based on three independent experiments. Paired t test: **, P < 0.01. (bottom) Western blot analysis. (F) Analysis of PCNA binding on newly synthesized DNA by NCC. (top) Experimental design. HeLa S3 cells were synchronized by a thymidine block and released into mid–S phase. One part of the cells was preincubated with cycloheximide (CHX) for 2.5 min to deplete new histones before replicating DNA was labeled with b-dUTP for 15 min. (bottom) Western blot of proteins bound to biotin-tagged DNA. Acetylation of K5 marks new histone H4. (G) Pulse–chase analysis of PCNA unloading by NCC. (left) Experimental design. Newly synthesized DNA was labeled 10 min with b-dUTP in asynchronous cells treated or not treated with CHX to deplete new histones. Samples were harvested immediately or after a 15-min chase. (right) Western blot of proteins bound to biotin-tagged DNA. Acetylation of K5 marks new histone H4. a.u., arbitrary unit; siCtrl, siRNA control.
During lagging strand synthesis, each Okazaki fragment requires a PCNA clamp to recruit polymerase δ, FEN-1, and DNA ligase I. In addition, PCNA also recruits CAF-1 for deposition of H3.1-H4 (Shibahara and Stillman, 1999). We thus reasoned that the clamp might remain on chromatin to fulfill this function, explaining the accumulation of PCNA upon histone depletion. In this view, clamp release could be linked to nucleosome assembly. To test this idea, we took advantage of an in vitro repair-coupled nucleosome assembly assay (Fig. 6 C) and compared PCNA unloading from a DNA template incubated with normal extracts or extracts depleted for soluble histones by short CHX treatment (Bonner et al., 1988). PCNA was loaded efficiently onto nicked DNA irrespectively of whether the extract contained histones or not (Fig. 6 D, 30 min). At later time points, when nucleosome assembly and PCNA unloading were completed in control extracts, PCNA was still found on DNA incubated in histone-depleted extracts (70 min). Thus, lack of nucleosome assembly delays the removal of PCNA. Importantly, addition of histones in complex with Asf1 restored nucleosome assembly and accelerated PCNA unloading (Fig. 6 E). This identifies nucleosome assembly as one parameter that influences PCNA unloading and predicts that PCNA clamps linger on newly synthesized DNA when de novo nucleosome assembly is impaired. To test this directly, we took advantage of a recently developed method for purifying proteins at replication forks and in newly synthesized chromatin called nascent chromatin capture (NCC; Alabert et al., 2014). This method is based on biotin–deoxy-UTP (b-dUTP) labeling of replicating DNA, cross-linking, and isolation of labeled chromatin fragments. Using NCC, we found that only 2.5-min pretreatment with CHX is sufficient to strongly reduce deposition of new histones on newly synthesized biotin-labeled DNA without blocking DNA synthesis (Fig. 6 F). Importantly, lack of new histone deposition was accompanied by an increased PCNA occupancy on nascent chromatin (Fig. 6 F). In this setup, we are probably looking at a mean of PCNA loading and unloading. To follow this process over time, we performed a pulse–chase experiment. Here, we observed elevated levels of PCNA on DNA in CHX-treated cells both before and after the chase period (Fig. 6 G).
Discussion
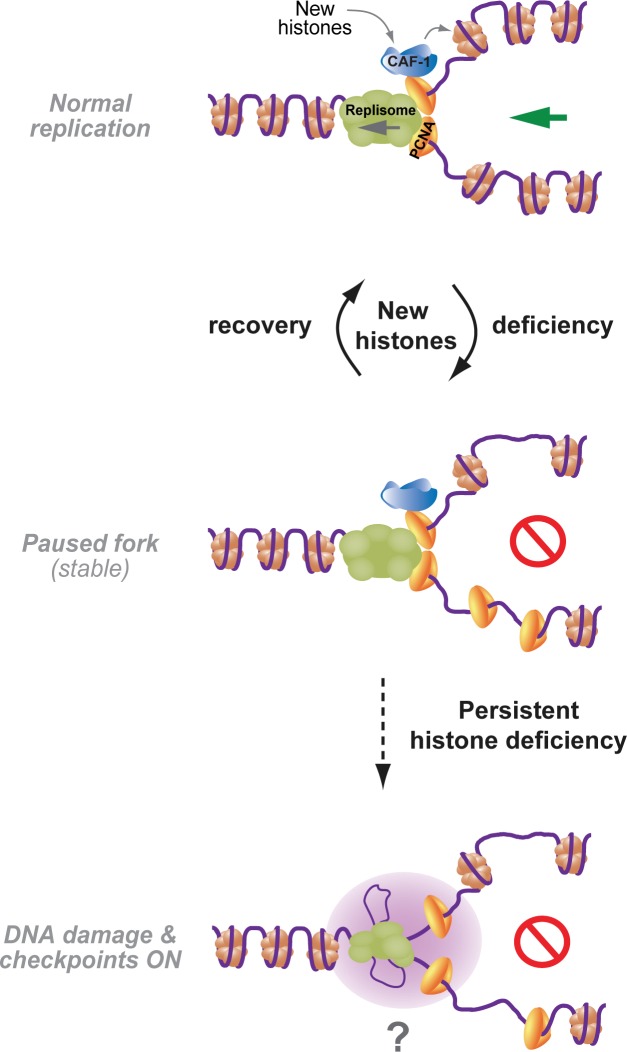
Collectively, our data show that mammalian DNA replication requires efficient provision of new histones because they control the elongation rate. We provide evidence that failure to assemble newly synthesized DNA into nucleosomes slows down replication fork speed without challenging fork stability and activating conventional checkpoints (Fig. 7). To target canonical histone biosynthesis, we independently depleted FLASH and SLBP. Although FLASH, as a transcriptional coactivator, could potentially regulate nonhistone genes, SLBP exclusively binds to the stem loop in histone mRNAs as shown by RNA immunoprecipitation–Chip (Townley-Tilson et al., 2006). Individual targeting of both FLASH and SLBP thus represents the best possible approach to study the cellular response to global deficiency of new histones in S phase. Using siRNA screening, we identified specific siRNAs that by targeting single histone H4 and H2A genes impaired DNA replication and recapitulated most phenotypes of SLBP and FLASH depletion. Even so, FLASH and SLBP depletion more robustly repress DNA replication, probably reflecting the difference between global inhibition of histone biosynthesis and targeting only one out of multiple histone genes. By targeting the ability of Asf1 to interact with and deliver histones to CAF-1, we establish that replication requires nucleosome assembly. Although this does not exclude that histones—new or old—may directly influence the replicative helicase (Groth et al., 2007), it argues that lack of nucleosome assembly can slow down fork progression. Exactly how histone deposition controls fork progression remains unclear. However, it might involve arrest or slowdown of the replicative helicase, as ssDNA formation and fork uncoupling is blocked. Recently, nucleosome assembly was shown to regulate termination of Okazaki fragment synthesis in yeast (Smith and Whitehouse, 2012), and live-cell imaging suggests that PCNA undergoes internal recycling at mammalian replication sites (Sporbert et al., 2002). Although the implications of PCNA recycling remain unknown, recent work reported that DNA synthesis is impaired in cells lacking an alternative replication factor C complex proposed to mediate PCNA unloading (Lee et al., 2013). It is thus conceivable that slow PCNA unloading could interfere with efficient lagging strand synthesis when histone deposition is delayed. However, it is equally possible that signaling pathways different from conventional checkpoints (Duch et al., 2013) and/or topological constraints could be at play. Altered replicon topology caused by lack of chromatin assembly could potentially impede fork progression as well as dormant origin firing by restraining DNA unwinding (Branzei and Foiani, 2010).
Figure 7.
Model illustrating how new histone supply controls mammalian DNA replication and genome integrity. Cells accommodate transient shortage of new histones by slowing down replication speed and maintaining PCNA on newly synthesized DNA, allowing nucleosome assembly once canonical histones become available. In contrast, persistent lack of new histones leads to DNA damage and potential loss of chromosomal stability.
We find that the cellular response to new histone deficiency differs substantially from the response to dNTP depletion. Low dNTP pools are mutagenic and thus constitute an immediate threat to genome stability (Bester et al., 2011); consequently, checkpoint activation alerts the cell to intensify dNTP production (Chabes and Thelander, 2000). At replication forks, ssDNA formation and continued primer synthesis signal activation of the checkpoint. However, checkpoint signaling inhibits histone biosynthesis (Su et al., 2004; Kaygun and Marzluff, 2005) and would therefore aggravate the problem in a situation in which cells lack new histones for nucleosome assembly. In this light, it is not surprising that short-term deprivation of new histones represses ssDNA formation and does not activate checkpoint signaling (Fig. 7). Fork slowdown, allowing time for nucleosome assembly to catch up with DNA synthesis, thus provides an attractive means for cells to tolerate fluctuations in the new histone supply. Under these conditions, replication inhibitors could induce checkpoint signaling, although there was no substantial exposure of ssDNA. This is consistent with previous work showing that primer–template junctions, not the amount of ssDNA, are limiting for checkpoint activation (Van et al., 2010). Moreover, it also argues that the forks, although slow, are functional, in line with lack of major changes in fork composition as analyzed by chromatin fractionation.
We find that new histone deficiency leads to accumulation of PCNA on newly synthesized DNA. Given that PCNA unloading from DNA in vitro is delayed in histone-depleted extracts, we envision that chromatin assembly somehow facilitates clamp release. Our data support this view but also show that PCNA unloading can proceed (although at a reduced rate) even when new histone deposition is strongly impaired. We thus speculate that parental histone recycling may also influence PCNA release. Biochemical analysis by iPOND (isolation of proteins on nascent DNA) showed a half-life of PCNA on newly synthesized DNA of <10 min (Sirbu et al., 2011), which is consistent with our results. Moreover, photobleaching and BrdU pulse–chase analysis suggested that PCNA rings might remain on chromatin for ≤20 min (Sporbert et al., 2002). It is not clear whether all PCNA rings recycle with similar kinetics or, alternatively, whether a fraction engaged in chromatin assembly might turnover more slowly. Maturation of newly assembled chromatin into a nuclease-resistant structure is estimated to take 15–20 min and thus likely commences while PCNA is present (Worcel et al., 1978; Annunziato and Seale, 1983). Consistent with this view, PCNA contributes to recruitment of many chromatin-remodeling and -modifying enzymes taking part in chromatin maturation (Alabert and Groth, 2012). Lack of new histone deposition would impair maturation, potentially explaining why histone deposition impinges on the PCNA-unloading process. In any case, retention of PCNA in immature chromatin behind the fork is probably advantageous, as it provides opportunity to recruit CAF-1 and deposit H3.1-H4 once histones are available. Once PCNA is unloaded, cells depend on HIRA to incorporate the replacement variant H3.3-H4 in a form of gap filling (Ray-Gallet et al., 2011). Given that the choice of histone variant influences chromatin structure, extensive incorporation of H3.3 may not be desirable. We hypothesize that human cells, by coupling fork speed and PCNA unloading to nucleosome assembly, gain robustness to maintain chromatin structure during transient shortage of new histones (Fig. 7). However, during persistent long-term suppression of histone biosynthesis and chromatin assembly (e.g, replicative senescence; O’Sullivan et al., 2010), this defense may succumb, leading to loss of chromatin integrity and genome instability.
Materials and methods
Cell lines, constructs, synchronization, and drug treatment
U-2-OS, HeLa S3, and TIG-3 cells were grown in DMEM containing 10% FBS and 1% penicillin/streptomycin. Cells were transfected with siRNAs at a 100-nM concentration using Oligofectamine (Invitrogen). Expression plasmids were introduced by transfection with Lipofectamine (Invitrogen) or electroporation using a GenePulser Xcell (Bio-Rad Laboratories). U-2-OS cells inducible for coexpression of H3.1-HA-FLAG and H4-GFP were previously described (Groth et al., 2007). In brief, U-2-OS Tet-Off cells were cotransfected with pBI-H3.1-Flag-HA/H4-GFP and pBABE-puromycin (puro), and a resistant single-cell clone was selected. pBI-H3.1-Flag-HA/H4-GFP was generated by PCR cloning of H3.1-Flag-HA and H4-N-EGFP into the SalI and NheI sites of pBI (Takara Bio Inc.) and verified by sequencing. The RFP-PCNA reporter cell line was generated by cotransfecting U-2-OS cells with NLS-RFP-linker-PCNA (gift from C. Green, University of Oxford, Oxford, England, UK) and pBABE-puro plasmids. After 1 µg/ml puromycin selection, a clone expressing moderate RFP-PCNA was transfected with pEGFP-hsRPA70 (gift from M.S. Wold, University of Iowa, Iowa City, Iowa; Haring et al., 2008) and selected with 250 µg/ml G418 to generate a GFP-RPA1/RFP-PCNA reporter cell line. The H2B-GFP and GFP-spectrin expression vectors were a gift from C.S. Sørensen (Biotech Research and Innovation Centre, Copenhagen, Denmark) and A. Yoneda (Biotech Research and Innovation Centre, Copenhagen, Denmark), respectively. The FLAG-HA-NLS-HIRA-B expression vector was generated in pcDNA5/flippase recognition target/TO-FLAG-HA by PCR amplification of the HIRA-B motif (aa 432–476) from human HIRA. Mutations in aa 459–461 from ADD to RRI were introduced by site-directed mutagenesis (Agilent Technologies). All constructs were sequence verified. Cells were synchronized at the G1/S transition by 2 mM thymidine (17 h) and released into fresh media containing 24 µM deoxycytidine. Cells were treated as indicated with 3 mM HU, 50 µg/ml aphidicolin, 50 µg/ml CHX, 10 mM caffeine, and 40 ng/ml nocodazole.
DNA combing
Single-molecule analysis of DNA replication by molecular combing was performed as described in protocol 36 available from the EpiGeneSys Network of Excellence website. In brief, 24 h after siRNA transfection, U-2-OS cells were pulse labeled with 25 µM BrdU for 45 min. Cells were harvested immediately after the pulse and molded into low-melting agarose plugs. Agarose plugs were treated with proteinase K, melted at 67°C, and digested by β-agarase. DNA was combed on silanized coverslips (Genomic Vision). DNA fibers were denatured by HCl and probed by the following primary antibodies: mouse anti-ssDNA (MAB3868; EMD Millipore) and rat anti-BrdU (AbD Serotec). Coverslips were mounted in Vectashield (Vector Laboratories). Images were captured on a DeltaVision system (Applied Precision) with a U Apochromat/340 40×/1.35 NA oil objective lens and analyzed with softWoRx 5.0.0 software (Applied Precision). Measured distances were converted to kilobases by the constant stretching factor (1 µm = 2 kb).
DNA fiber assay
24 h after siRNA transfection, U-2-OS cells were labeled for 10 min with 10 µM IdU (Sigma-Aldrich) followed by 20-min labeling with 100 µM CldU (MP Biomedicals). 2 µl of cells resuspended in ice-cold PBS was deposited on a microscope slide and incubated with 7 µl of spreading buffer (200 mM Tris-HCl, pH 7.5, 0.5% SDS, and 50 mM EDTA) for 3 min. The slides were tilted 15° to stretch the DNA fibers (Bianco et al., 2012). After fixation with methanol/acetic acid (3:1), DNA was denatured with 2.5 M HCl and blocked (PBS with 1% BSA and 0.1% Triton X-100) before staining with primary (anti-CldU [AbCys SA], anti-IdU [BD], and anti-ssDNA [EMD Millipore]) and corresponding secondary antibodies conjugated with Alexa Fluor 488, 546, or 647 (all obtained from Invitrogen). Statistical analysis was performed using Prism 5 (GraphPad Software).
MNase sensitivity
Global chromatin was labeled by 0.5 pCi/ml [14C]thymidine for 24 h, and nascent chromatin was pulse labeled with 25 nCi/ml [3H]thymidine. To compensate for lower replication rate in FLASH- and SLBP-depleted cells, labeling times were adjusted to obtain similar [3H]thymidine incorporation (9 min for siRNA control, 30 min for siRNA FLASH, and 20 min for siRNA SLBP). Cells were lysed in hypotonic buffer (10 mM Tris, pH 7.4, 2.5 mM MgCl2, and 0.5% NP-40), and nuclei were isolated resuspended in digestion buffer (10 mM Tris, pH 7.4, 10 mM NaCl, 5 mM MgCl2, and 2 mM CaCl2) and subjected to 0.075 U/µl MNase digestion at 37°C. Undigested chromatin was collected by centrifugation and mixed with scintillation liquid (Ultima Gold; PerkinElmer). 14C and 3H activity was measured in a liquid scintillation counter (LS 6500; Beckman Coulter). Readings were corrected for 14C bleed through into the 3H channel.
EM analysis of genomic DNA
Transfected cells were synchronized as in Fig. 1 A. In vivo psoralen cross-linking, isolation of total genomic DNA, and enrichment of the replication intermediates and their EM visualization were performed as previously described (Lopes, 2009; Neelsen et al., 2014). In brief, cells were harvested, and genomic DNA was cross-linked by two rounds of incubation in 10 µM 4,5′,8-trimethylpsoralen and 2 min of irradiation with 366-nm UV light. Cells were lysed, and genomic DNA was isolated from the nuclei by proteinase K digestion and phenol–chloroform extraction. Purified DNA was digested with PvuII, and replication intermediates were enriched on a benzoylated naphthoylated DEAE cellulose column. EM samples were prepared by spreading the denatured DNA on carbon-coated grids and visualized by platinum rotary shadowing. Images were acquired on a microscope (Tecnai G2 Spirit; FEI) and analyzed with ImageJ (National Institutes of Health). Daughter and parental strands were identified based on the following parameters: (a) Strand symmetry: daughter strands are likely to have the same length because the genomic DNA was digested by a sequence-specific restriction enzyme; (b) Fork structure: each DNA strand from the parental duplex continues into one of the daughters. Nucleosome density is expressed by the so-called R-value, which was calculated as the combined contour length of all nucleosome bubbles in a given stretch of DNA, divided by the overall contour length of the same DNA stretch. A reduced R-value indicates a reduction in nucleosome density. More details can be found in Neelsen et al. (2014).
Robot-automated siRNA screen
The automated screen was performed using a liquid handling station (STAR; Hamilton Robotics). U-2-OS GFP-RPA1/RFP-PCNA cells were reverse transfected with a custom-made siRNA library targeting 236 genes (Applied Biosystems). After 46 h, cells were treated with 3 mM HU for 2 h, preextracted with cytoskeleton buffer (CSK: 10 mM Pipes, pH 7, 100 mM NaCl, 300 mM sucrose, and 3 mM MgCl2) containing 0.5% Triton X-100, fixed in 2% paraformaldehyde, and stained by Hoechst. Five images were acquired per well (20×/0.45 NA objective lens; IN Cell Analyzer 1000; GE Healthcare) and analyzed by IN Cell Analyzer Workstation 3.5 software. Hits were identified based on combined activities of their targeting siRNAs. Candidate genes were selected according to both the significance assessment by redundant siRNA activity (RSA) analysis (König et al., 2007) and fold change criteria based on Asf1 (a and b) depletion (Fig. S4).
High-throughput single-cell analysis
Cells were reverse transfected with siRNAs upon plating into 96-well plates. After 48 h, cells were preextracted, fixed, and stained with antibodies and DAPI. Images were acquired and analyzed as described in the previous paragraph for the robot-automated siRNA screen. Relative fluorescence intensity of GFP-RPA1, EdU, and γ-H2AX was quantified in 4,000–7,000 RFP-PCNA–positive cells.
Cell fractionation and immunoprecipitation
Soluble histones were extracted with low detergent in a hypotonic buffer (10 mM Tris, pH 7.4, 2.5 mM MgCl2, and 0.5% NP-40). For chromatin fractionation by DNase I digestion, cells were lysed in ice-cold hypotonic buffer for 5 min, and nuclei were collected by centrifugation, washed (10 mM Tris, pH 7.4, and 150 mM NaCl) and resuspended in digest buffer (see MNase sensitivity section) supplemented with 100 U DNase I/107 cells. Digest reactions were incubated 8 min at 37°C before the solubilized material was separated from core chromatin by centrifugation (13,000 g for 5 min at 4°C). For FLAG immunoprecipitation, cell extracts made in immunoprecipitation buffer (50 mM Tris, pH 7.5, 150 mM NaCl, and 0.5% NP-40) were precleared with agarose beads (SuperFlow 6; IBA) for 1 h before 2-h incubation with anti-FLAG M2-agarose beads (Sigma-Aldrich). Beads were washed six times with immunoprecipitation buffer, resuspended, and boiled for 5 min in SDS gel-loading buffer (50 mM Tris, pH 6.8, 2% SDS, and 10% glycerol). All buffers contained inhibitors 1 mM DTT, 10 µg/ml leupeptin, 10 µg/ml pepstatin, 0.1 mM PMSF, 0.2 mM sodium vanadate, 5 mM sodium fluoride, and 10 mM β-glycerolphosphate.
In vitro repair–coupled nucleosome assembly
HeLa S3 cells in mid–S phase were treated 10 min with CHX or left untreated. Cytosolic and nuclei extracts were prepared as previously described (Groth et al., 2007) except that buffers contained 0.5 mM DTT and no phosphatase inhibitors or trichostatin A. 2 vol nuclear extraction buffer was used for 1 vol nuclei.
The DNA template (pUC19) was prepared as previously described (Mello et al., 2004). Labeling reactions contained 20 µg of linearized pUC19, 7.5 µl of 0.4-mM biotin-14-deoxy-ATP (Invitrogen), 1.2 µl each of 10-mM α-thio-deoxy-TTP, α-thio-deoxy-CTP, and α-thio-deoxy-GTP (all obtained from IBA), and 10–15 U Klenow 3′ → 5′ exo− (New England Biolabs, Inc.), in a total volume of 120 µl. The reaction was incubated for 2 h at 37°C.
The biotinylated DNA template (20 µg) was coupled to 490 µl of magnetic beads (Dynabeads M-280; Invitrogen) in 2× wash/storage buffer (10 mM Tris, pH 7.5, 2 M NaCl, and 1 mM EDTA) in a final volume of 670 µl by incubation on a wheel for 16 h at room temperature. The beads were washed twice and resuspended in 2× wash/storage buffer to get 100 ng bead-linked DNA/µl. Optimal conditions for DNase I treatment producing ssDNA nicks were determined by a pilot experiment using supercoiled pUC19 monitoring the intensity of bands corresponding to circular versus linear DNA after gel electrophoresis. To introduce ssDNA nicks into bead-linked DNA, the reaction contained 10 µg of bead-linked DNA resuspended in buffer B (10 mM Hepes-KOH, pH 7.6, 50 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA, and 10% glycerol) and 0.1 U DNase I (Roche)/µl in a total volume of 650 µl. After 5-min incubation at 25°C, reactions were stopped by 200 µl of 0.5-M EDTA.
Typical in vitro nucleosome assembly reactions contained 200 µg cytosolic extracts, 20 µg nuclear extracts, 600 ng bead-linked nicked DNA, 40 mM Hepes-KOH, pH 7.8, 5 mM MgCl2, 0.5 mM DTT, 40 mM phosphocreatine, 4 µg creatine phosphokinase, and 4 mM ATP in a total volume of 50 µl. Intact bead-linked DNA (not treated with DNase I) was used as a negative control. Reactions were incubated at room temperature with horizontal rotation and stopped at the indicated times by cross-linking DNA-bound protein with glutaraldehyde (0.25% glutaraldehyde, 40 mM Hepes-KOH, pH 7.8, 40 mM KCl, and 0.05% NP-40) for 30 min on ice. The beads were then washed five times with 150 mM KCl, 40 mM Hepes-KOH, pH 7.8, and 0.2% NP-40, and bound proteins were eluted in Laemmli buffer, boiled extensively to de–cross-link, and analyzed by Western blotting.
For complementation experiments, Asf1–H3–H4 complexes were reconstituted in vitro before they were added to assembly reactions. 20 ng HeLa core histones (Active Motif) were incubated with 15 ng recombinant Asf1a for 20 min on ice in 10 mM Hepes, pH 7.9, 1 mM EDTA, 10 mM KCl, and 10% glycerol.
Immunocytochemistry and microscopy
Cells were either preextracted with CSK buffer with 0.5% Triton X-100 to remove soluble proteins or fixed directly with 4% formaldehyde and processed as previously described (Groth et al., 2005). For detecting ssDNA, cells were pulse labeled with 10 µM BrdU for 24 h before transfection. ssDNA was subsequently revealed by BrdU detection under nondenaturing conditions (the BrdU epitope is not detected by anti-BrdU antibodies in double-strand DNA). For detection of total BrdU incorporation in double-strand DNA, fixed cells were treated with 4 M HCl (10 min) to denature DNA before immunostaining. EdU staining was performed using EdU Alexa Fluor 488/647 high-throughput imaging (Click-iT; High Content Screening) assay kit (Invitrogen) according to the manufacturer’s instructions. The list of primary antibodies used is given in Table S1. Corresponding secondary antibodies were conjugated with Alexa Fluor 488, 568, or 594 (Invitrogen).
Images were collected using a microscope (Leitz DMRXE; Leica) with Plan Fluotar 40×/0.5–1.00 NA oil objective lens equipped with a charge-coupled device camera (DFC340 FX; Leica) or a DeltaVision system with U Apochromat/340 40×/1.35 NA oil objective lens and analyzed with softWoRx 5.0.0 software. Immersion oil (n = 1.522) was used as an imaging medium. All images in the individual panels were acquired under room temperature with the same settings and adjusted for brightness and contrast identically using Photoshop CS5 (Adobe).
Statistical analysis
The statistical tests applied in this study are stated in the figure legends. In brief, p-values in Fig. 1 (C and D), Fig. 4 (D and E), Fig. 5 E, Fig. 6 B, and Fig. S3 E were calculated by using the nonparametric Mann–Whitney test. In Fig. 2 C, Fig. S1 E, and Fig. S2 F, the two-tailed t test was applied, and in Fig. 6 E, the p-value was calculated by a paired t test.
siRNAs and quantitative PCR (qPCR) primers
Online supplemental material
Fig. S1 shows Western blot controls, cell cycle analysis, and histone gene expression in FLASH- and SLBP-depleted cells and provides evidence that conditional coexpression of H3.1 and H4 partially rescues S-phase progression in cells lacking FLASH and SLBP. Fig. S2 shows the length distribution of BrdU-labeled tracks measured by DNA combing and CldU-labeled tracks obtained from DNA fiber spreads. Fig. S3 provides further analysis of DNA damage and checkpoint signaling in U-2-OS cells and TIG-3 fibroblasts depleted for FLASH and SLBP. Fig. S4 provides a characterization of the GFP-RPA1/RFP-PCNA reporter cell line used for the siRNA screen, functional classification of the positive hits, and a validation of siRNA target specificity for selected hits. Fig. S5 provides Western blot analysis of replication fork components in FLASH- and SLBP-depleted cells as well as control cells treated with CHX, HU, and the Chk1 inhibitor UCN-01. Table S1 shows antibodies used in this study. Table S2 shows siRNA sequences used in this study. Table S3 shows quantitative RT-PCR primer sequences. Table S4 shows a list of genes used for the siRNA screen. Table S5 shows a candidate gene list from the siRNA screen. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201305017/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201305017.dv.
Supplementary Material
Acknowledgments
We thank Claus S. Sørensen, Marc S. Wold, Catherine Green, Atsuko Yoneda, and Vincenzo De Laurenzi for reagents, and Kristian Helin for comments on the manuscript. We thank the high-throughput screening facility at the Biotech Research and Innovation Centre for technical support. We also thank the Center for Microscopy and Image Analysis of the University of Zurich for technical assistance with EM and the DNA Combing Facility of Montpellier. We acknowledge Yea-Lih Lin for valuable suggestions on DNA fiber analysis.
J. Mejlvang was supported by the Danish Cancer Society. A. Groth is a European Molecular Biology Organization Young Investigator, and her laboratory is supported by a European Research Council Starting Grant (ERC2011StG), the Lundbeck Foundation, the Danish National Research Foundation (DNRF82), the Danish Cancer Society and the Danish Medical Research Council, the Novo Nordisk Foundation, Fabrikant Vilhelm Pedersen og Hustrus Mindelegat, and Seventh Framework Programme Marie Curie Actions Initial Training Networks Nucleosome4D and aDDRess. The P. Pasero laboratory is supported by the Ligue Contre le Cancer (équipe labellisée), Agence Nationale de la Recherché, and the Institut National du Cancer.
Footnotes
Abbreviations used in this paper:
- b-dUTP
- biotin–deoxy-UTP
- CHX
- cycloheximide
- CldU
- 5-chloro-2′-deoxyuridine
- dNTP
- deoxynucleotide
- EdU
- 5-ethynyl-2’-deoxyuridine
- HU
- hydroxyurea
- IdU
- 5-iodo-2′-deoxyuridine
- MNase
- micrococcal nuclease
- NCC
- nascent chromatin capture
- qPCR
- quantitative PCR
- RPA
- replication protein A
- RSA
- redundant siRNA activity
- ssDNA
- single-strand DNA
References
- Alabert C., Groth A. 2012. Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell Biol. 13:153–167 10.1038/nrm3288 [DOI] [PubMed] [Google Scholar]
- Alabert C., Bukowski-Wills J.-C., Lee S.-B., Kustatscher G., Nakamura K., Alves F.D.L., Menard P., Mejlvang J., Rappsilber J., Groth A. 2014. Nascent chromatin capture (NCC) proteomics characterize chromatin dynamics during DNA replication and identify new replication factors. Nat. Cell Biol. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anglana M., Apiou F., Bensimon A., Debatisse M. 2003. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell. 114:385–394 10.1016/S0092-8674(03)00569-5 [DOI] [PubMed] [Google Scholar]
- Annunziato A.T. 2012. Assembling chromatin: The long and winding road. Biochim. Biophys. Acta. 1819:196–210 10.1016/j.bbagrm.2011.07.005 [DOI] [PubMed] [Google Scholar]
- Annunziato A.T., Seale R.L. 1983. Histone deacetylation is required for the maturation of newly replicated chromatin. J. Biol. Chem. 258:12675–12684 [PubMed] [Google Scholar]
- Ask K., Jasencakova Z., Menard P., Feng Y., Almouzni G., Groth A. 2012. Codanin-1, mutated in the anaemic disease CDAI, regulates Asf1 function in S-phase histone supply. EMBO J. 31:2013–2023 10.1038/emboj.2012.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcaroli D., Bongiorno-Borbone L., Terrinoni A., Hofmann T.G., Rossi M., Knight R.A., Matera A.G., Melino G., De Laurenzi V. 2006. FLASH is required for histone transcription and S-phase progression. Proc. Natl. Acad. Sci. USA. 103:14808–14812 10.1073/pnas.0604227103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bester A.C., Roniger M., Oren Y.S., Im M.M., Sarni D., Chaoat M., Bensimon A., Zamir G., Shewach D.S., Kerem B. 2011. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 145:435–446 10.1016/j.cell.2011.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco J.N., Poli J., Saksouk J., Bacal J., Silva M.J., Yoshida K., Lin Y.L., Tourrière H., Lengronne A., Pasero P. 2012. Analysis of DNA replication profiles in budding yeast and mammalian cells using DNA combing. Methods. 57:149–157 10.1016/j.ymeth.2012.04.007 [DOI] [PubMed] [Google Scholar]
- Bonner W.M., Wu R.S., Panusz H.T., Muneses C. 1988. Kinetics of accumulation and depletion of soluble newly synthesized histone in the reciprocal regulation of histone and DNA synthesis. Biochemistry. 27:6542–6550 10.1021/bi00417a052 [DOI] [PubMed] [Google Scholar]
- Branzei D., Foiani M. 2010. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 11:208–219 10.1038/nrm2852 [DOI] [PubMed] [Google Scholar]
- Chabes A., Thelander L. 2000. Controlled protein degradation regulates ribonucleotide reductase activity in proliferating mammalian cells during the normal cell cycle and in response to DNA damage and replication blocks. J. Biol. Chem. 275:17747–17753 10.1074/jbc.M000799200 [DOI] [PubMed] [Google Scholar]
- Cimprich K.A., Cortez D. 2008. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9:616–627 10.1038/nrm2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook A.J., Gurard-Levin Z.A., Vassias I., Almouzni G. 2011. A specific function for the histone chaperone NASP to fine-tune a reservoir of soluble H3-H4 in the histone supply chain. Mol. Cell. 44:918–927 10.1016/j.molcel.2011.11.021 [DOI] [PubMed] [Google Scholar]
- Courbet S., Gay S., Arnoult N., Wronka G., Anglana M., Brison O., Debatisse M. 2008. Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature. 455:557–560 10.1038/nature07233 [DOI] [PubMed] [Google Scholar]
- Duch A., Felipe-Abrio I., Barroso S., Yaakov G., García-Rubio M., Aguilera A., de Nadal E., Posas F. 2013. Coordinated control of replication and transcription by a SAPK protects genomic integrity. Nature. 493:116–119 10.1038/nature11675 [DOI] [PubMed] [Google Scholar]
- Duro E., Lundin C., Ask K., Sanchez-Pulido L., MacArtney T.J., Toth R., Ponting C.P., Groth A., Helleday T., Rouse J. 2010. Identification of the MMS22L-TONSL complex that promotes homologous recombination. Mol. Cell. 40:632–644 10.1016/j.molcel.2010.10.023 [DOI] [PubMed] [Google Scholar]
- Feser J., Truong D., Das C., Carson J.J., Kieft J., Harkness T., Tyler J.K. 2010. Elevated histone expression promotes life span extension. Mol. Cell. 39:724–735 10.1016/j.molcel.2010.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugger K., Mistrik M., Danielsen J.R., Dinant C., Falck J., Bartek J., Lukas J., Mailand N. 2009. Human Fbh1 helicase contributes to genome maintenance via pro- and anti-recombinase activities. J. Cell Biol. 186:655–663 10.1083/jcb.200812138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X.Q., Jackson D.A., Blow J.J. 2007. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 21:3331–3341 10.1101/gad.457807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görisch S.M., Sporbert A., Stear J.H., Grunewald I., Nowak D., Warbrick E., Leonhardt H., Cardoso M.C. 2008. Uncoupling the replication machinery: replication fork progression in the absence of processive DNA synthesis. Cell Cycle. 7:1983–1990 10.4161/cc.7.13.6094 [DOI] [PubMed] [Google Scholar]
- Groth A., Ray-Gallet D., Quivy J.P., Lukas J., Bartek J., Almouzni G. 2005. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol. Cell. 17:301–311 10.1016/j.molcel.2004.12.018 [DOI] [PubMed] [Google Scholar]
- Groth A., Corpet A., Cook A.J., Roche D., Bartek J., Lukas J., Almouzni G. 2007. Regulation of replication fork progression through histone supply and demand. Science. 318:1928–1931 10.1126/science.1148992 [DOI] [PubMed] [Google Scholar]
- Gunjan A., Verreault A. 2003. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell. 115:537–549 10.1016/S0092-8674(03)00896-1 [DOI] [PubMed] [Google Scholar]
- Haring S.J., Mason A.C., Binz S.K., Wold M.S. 2008. Cellular functions of human RPA1. Multiple roles of domains in replication, repair, and checkpoints. J. Biol. Chem. 283:19095–19111 10.1074/jbc.M800881200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek M., Stillman B. 2003. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc. Natl. Acad. Sci. USA. 100:12183–12188 10.1073/pnas.1635158100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasencakova Z., Scharf A.N., Ask K., Corpet A., Imhof A., Almouzni G., Groth A. 2010. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol. Cell. 37:736–743 10.1016/j.molcel.2010.01.033 [DOI] [PubMed] [Google Scholar]
- Kaygun H., Marzluff W.F. 2005. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 12:794–800 10.1038/nsmb972 [DOI] [PubMed] [Google Scholar]
- Kerzendorfer C., Hannes F., Colnaghi R., Abramowicz I., Carpenter G., Vermeesch J.R., O’Driscoll M. 2012. Characterizing the functional consequences of haploinsufficiency of NELF-A (WHSC2) and SLBP identifies novel cellular phenotypes in Wolf-Hirschhorn syndrome. Hum. Mol. Genet. 21:2181–2193 10.1093/hmg/dds033 [DOI] [PubMed] [Google Scholar]
- Kim U.J., Han M., Kayne P., Grunstein M. 1988. Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae. EMBO J. 7:2211–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König R., Chiang C.Y., Tu B.P., Yan S.F., DeJesus P.D., Romero A., Bergauer T., Orth A., Krueger U., Zhou Y., Chanda S.K. 2007. A probability-based approach for the analysis of large-scale RNAi screens. Nat. Methods. 4:847–849 10.1038/nmeth1089 [DOI] [PubMed] [Google Scholar]
- Lee K.Y., Fu H., Aladjem M.I., Myung K. 2013. ATAD5 regulates the lifespan of DNA replication factories by modulating PCNA level on the chromatin. J. Cell Biol. 200:31–44 10.1083/jcb.201206084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M. 2009. Electron microscopy methods for studying in vivo DNA replication intermediates. Methods Mol. Biol. 521:605–631 10.1007/978-1-60327-815-7_34 [DOI] [PubMed] [Google Scholar]
- Loyola A., Tagami H., Bonaldi T., Roche D., Quivy J.P., Imhof A., Nakatani Y., Dent S.Y., Almouzni G. 2009. The HP1alpha-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep. 10:769–775 10.1038/embor.2009.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W.F., Wagner E.J., Duronio R.J. 2008. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 9:843–854 10.1038/nrg2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maya-Mendoza A., Petermann E., Gillespie D.A., Caldecott K.W., Jackson D.A. 2007. Chk1 regulates the density of active replication origins during the vertebrate S phase. EMBO J. 26:2719–2731 10.1038/sj.emboj.7601714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello J.A., Moggs J.G., Almouzni G. 2004. Analysis of DNA repair and chromatin assembly in vitro using immobilized damaged DNA substrates. Methods Mol. Biol. 281:271–281 [DOI] [PubMed] [Google Scholar]
- Moggs J.G., Grandi P., Quivy J.P., Jónsson Z.O., Hübscher U., Becker P.B., Almouzni G. 2000. A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol. Cell. Biol. 20:1206–1218 10.1128/MCB.20.4.1206-1218.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzina N., Verreault A., Laue E., Stillman B. 1999. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol. Cell. 4:529–540 10.1016/S1097-2765(00)80204-X [DOI] [PubMed] [Google Scholar]
- Nabatiyan A., Krude T. 2004. Silencing of chromatin assembly factor 1 in human cells leads to cell death and loss of chromatin assembly during DNA synthesis. Mol. Cell. Biol. 24:2853–2862 10.1128/MCB.24.7.2853-2862.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelsen K.J., Chaudhuri A.R., Follonier C., Herrador R., Lopes M. 2014. Visualization and interpretation of eukaryotic DNA replication intermediates in vivo by electron microscopy. Methods Mol. Biol. 1094:177–208 10.1007/978-1-62703-706-8_15 [DOI] [PubMed] [Google Scholar]
- Nelson D.M., Ye X., Hall C., Santos H., Ma T., Kao G.D., Yen T.J., Harper J.W., Adams P.D. 2002. Coupling of DNA synthesis and histone synthesis in S phase independent of cyclin/cdk2 activity. Mol. Cell. Biol. 22:7459–7472 10.1128/MCB.22.21.7459-7472.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan R.J., Kubicek S., Schreiber S.L., Karlseder J. 2010. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat. Struct. Mol. Biol. 17:1218–1225 10.1038/nsmb.1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M., Walter J.C. 2004. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 23:3667–3676 10.1038/sj.emboj.7600369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E., Woodcock M., Helleday T. 2010. Chk1 promotes replication fork progression by controlling replication initiation. Proc. Natl. Acad. Sci. USA. 107:16090–16095 10.1073/pnas.1005031107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F., Aguilera A. 2005. Partial depletion of histone H4 increases homologous recombination-mediated genetic instability. Mol. Cell. Biol. 25:1526–1536 10.1128/MCB.25.4.1526-1536.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivy J.P., Roche D., Kirschner D., Tagami H., Nakatani Y., Almouzni G. 2004. A CAF-1 dependent pool of HP1 during heterochromatin duplication. EMBO J. 23:3516–3526 10.1038/sj.emboj.7600362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivy J.P., Gérard A., Cook A.J., Roche D., Almouzni G. 2008. The HP1-p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells. Nat. Struct. Mol. Biol. 15:972–979 10.1038/nsmb.1470 [DOI] [PubMed] [Google Scholar]
- Ray-Gallet D., Woolfe A., Vassias I., Pellentz C., Lacoste N., Puri A., Schultz D.C., Pchelintsev N.A., Adams P.D., Jansen L.E., Almouzni G. 2011. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol. Cell. 44:928–941 10.1016/j.molcel.2011.12.006 [DOI] [PubMed] [Google Scholar]
- Sarraf S.A., Stancheva I. 2004. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol. Cell. 15:595–605 10.1016/j.molcel.2004.06.043 [DOI] [PubMed] [Google Scholar]
- Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M. 2011. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 145:529–542 10.1016/j.cell.2011.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöpf B., Bregenhorn S., Quivy J.P., Kadyrov F.A., Almouzni G., Jiricny J. 2012. Interplay between mismatch repair and chromatin assembly. Proc. Natl. Acad. Sci. USA. 109:1895–1900 10.1073/pnas.1106696109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale R.L., Simpson R.T. 1975. Effects of cycloheximide on chromatin biosynthesis. J. Mol. Biol. 94:479–501 10.1016/0022-2836(75)90216-8 [DOI] [PubMed] [Google Scholar]
- Shibahara K., Stillman B. 1999. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 96:575–585 10.1016/S0092-8674(00)80661-3 [DOI] [PubMed] [Google Scholar]
- Sirbu B.M., Couch F.B., Feigerle J.T., Bhaskara S., Hiebert S.W., Cortez D. 2011. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 25:1320–1327 10.1101/gad.2053211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.J., Whitehouse I. 2012. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 483:434–438 10.1038/nature10895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel R.E., Cook R.G., Perry C.A., Annunziato A.T., Allis C.D. 1995. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl. Acad. Sci. USA. 92:1237–1241 10.1073/pnas.92.4.1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo J.M., Stahl H., Koller T., Knippers R. 1986. Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. J. Mol. Biol. 189:189–204 10.1016/0022-2836(86)90390-6 [DOI] [PubMed] [Google Scholar]
- Sogo J.M., Lopes M., Foiani M. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 297:599–602 10.1126/science.1074023 [DOI] [PubMed] [Google Scholar]
- Sporbert A., Gahl A., Ankerhold R., Leonhardt H., Cardoso M.C. 2002. DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters. Mol. Cell. 10:1355–1365 10.1016/S1097-2765(02)00729-3 [DOI] [PubMed] [Google Scholar]
- Su C., Gao G., Schneider S., Helt C., Weiss C., O’Reilly M.A., Bohmann D., Zhao J. 2004. DNA damage induces downregulation of histone gene expression through the G1 checkpoint pathway. EMBO J. 23:1133–1143 10.1038/sj.emboj.7600120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takami Y., Ono T., Fukagawa T., Shibahara K., Nakayama T. 2007. Essential role of chromatin assembly factor-1-mediated rapid nucleosome assembly for DNA replication and cell division in vertebrate cells. Mol. Biol. Cell. 18:129–141 10.1091/mbc.E06-05-0426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Poustovoitov M.V., Zhao K., Garfinkel M., Canutescu A., Dunbrack R., Adams P.D., Marmorstein R. 2006. Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly. Nat. Struct. Mol. Biol. 13:921–929 10.1038/nsmb1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townley-Tilson W.H., Pendergrass S.A., Marzluff W.F., Whitfield M.L. 2006. Genome-wide analysis of mRNAs bound to the histone stem-loop binding protein. RNA. 12:1853–1867 10.1261/rna.76006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van C., Yan S., Michael W.M., Waga S., Cimprich K.A. 2010. Continued primer synthesis at stalled replication forks contributes to checkpoint activation. J. Cell Biol. 189:233–246 10.1083/jcb.200909105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H. 1972. A possible role for histone in the synthesis of DNA. Nature. 240:449–453 10.1038/240449a0 [DOI] [PubMed] [Google Scholar]
- Worcel A., Han S., Wong M.L. 1978. Assembly of newly replicated chromatin. Cell. 15:969–977 10.1016/0092-8674(78)90280-5 [DOI] [PubMed] [Google Scholar]
- Yang X.C., Burch B.D., Yan Y., Marzluff W.F., Dominski Z. 2009. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3′ end processing of histone pre-mRNAs. Mol. Cell. 36:267–278 10.1016/j.molcel.2009.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Franco A.A., Santos H., Nelson D.M., Kaufman P.D., Adams P.D. 2003. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol. Cell. 11:341–351 10.1016/S1097-2765(03)00037-6 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Shibahara K., Stillman B. 2000. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature. 408:221–225 10.1038/35041601 [DOI] [PubMed] [Google Scholar]
- Zhao X., McKillop-Smith S., Müller B. 2004. The human histone gene expression regulator HBP/SLBP is required for histone and DNA synthesis, cell cycle progression and cell proliferation in mitotic cells. J. Cell Sci. 117:6043–6051 10.1242/jcs.01523 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.