Thyroid hormone and its receptor act in concert with NRF1 to increase cellular respiration and reactive oxygen species production, leading to DNA damage and premature senescence in susceptible cells.
Abstract
There is increasing evidence that the thyroid hormone (TH) receptors (THRs) can play a role in aging, cancer and degenerative diseases. In this paper, we demonstrate that binding of TH T3 (triiodothyronine) to THRB induces senescence and deoxyribonucleic acid (DNA) damage in cultured cells and in tissues of young hyperthyroid mice. T3 induces a rapid activation of ATM (ataxia telangiectasia mutated)/PRKAA (adenosine monophosphate–activated protein kinase) signal transduction and recruitment of the NRF1 (nuclear respiratory factor 1) and THRB to the promoters of genes with a key role on mitochondrial respiration. Increased respiration leads to production of mitochondrial reactive oxygen species, which in turn causes oxidative stress and DNA double-strand breaks and triggers a DNA damage response that ultimately leads to premature senescence of susceptible cells. Our findings provide a mechanism for integrating metabolic effects of THs with the tumor suppressor activity of THRB, the effect of thyroidal status on longevity, and the occurrence of tissue damage in hyperthyroidism.
Introduction
Normal cells only proliferate for a finite number of times until they become senescent (Hayflick, 1965), a state of irreversible proliferative arrest as a consequence of the genomic damage caused by telomere erosion (d’Adda di Fagagna et al., 2003). Senescence can also be induced prematurely as a result of a persistent DNA damage response (DDR) secondary to oxidative stress that induces double-strand breaks (DSBs; Campisi and d’Adda di Fagagna, 2007) or to DNA replication stress induced by oncogenes or drugs, which lead to DSB generation and DDR activation (Serrano et al., 1997; Bartkova et al., 2006; Di Micco et al., 2006). Cellular senescence not only plays a role in aging and contributes to the appearance of age-related pathologies (Vijg and Campisi, 2008; Baker et al., 2011) but is also an important antiproliferative process that acts as a strong barrier against cellular transformation and cancer progression (Collado and Serrano, 2010).
The actions of the thyroid hormones (THs) are mediated by binding to nuclear TH receptors (THRs) that are encoded by two genes: THRA and THRB (Aranda and Pascual, 2001). One of the most prominent actions of THs is the regulation of mitochondrial function (Oppenheimer et al., 1987). Mitochondria are the major site of oxidative processes that lead to heat production and to generation of reactive oxygen species (ROS). Although it has been known for more than 100 yr that THs increase basal metabolism (Magnus-Levy, 1895) and are major regulators of mitochondrial activity and oxygen consumption (Tata et al., 1962), the molecular mechanisms underlying these effects have not yet been fully defined.
THRs can also play a role in aging. Thus, hyperthyroidism accelerates aging (Ooka and Shinkai, 1986; Buffenstein and Pinto, 2009), whereas longevity is associated with decreased thyroid function (Atzmon et al., 2009; Rozing et al., 2010; Gesing et al., 2012). In mammals, life span correlates with the metabolic rate (Mookerjee et al., 2010), which is controlled by the THs. Therefore, an increase of ROS production and oxidative stress may be involved in the acceleration of aging by the THs. There is also increasing evidence that THRs play a role in cancer. THRs can inhibit oncogenic proliferation, transformation, tumor growth, tumor invasion, and formation of metastasis (García-Silva and Aranda, 2004; Aranda et al., 2009; García-Silva et al., 2011). In addition, mutations or loss of expression of THRs are common events in some tumors (Aranda et al., 2009; Kim and Cheng, 2013), suggesting that these receptors have tumor suppressor activity. Because THs accelerate aging and THRs suppress tumor growth, in this work, we have examined the possibility that they could induce cellular senescence and found that THRB, but not THRA, can mediate T3 (triiodothyronine)-dependent premature senescence in cultured cells and in mice.
Results
T3 induces senescence and DNA damage in mouse embryonic fibroblasts (MEFs)
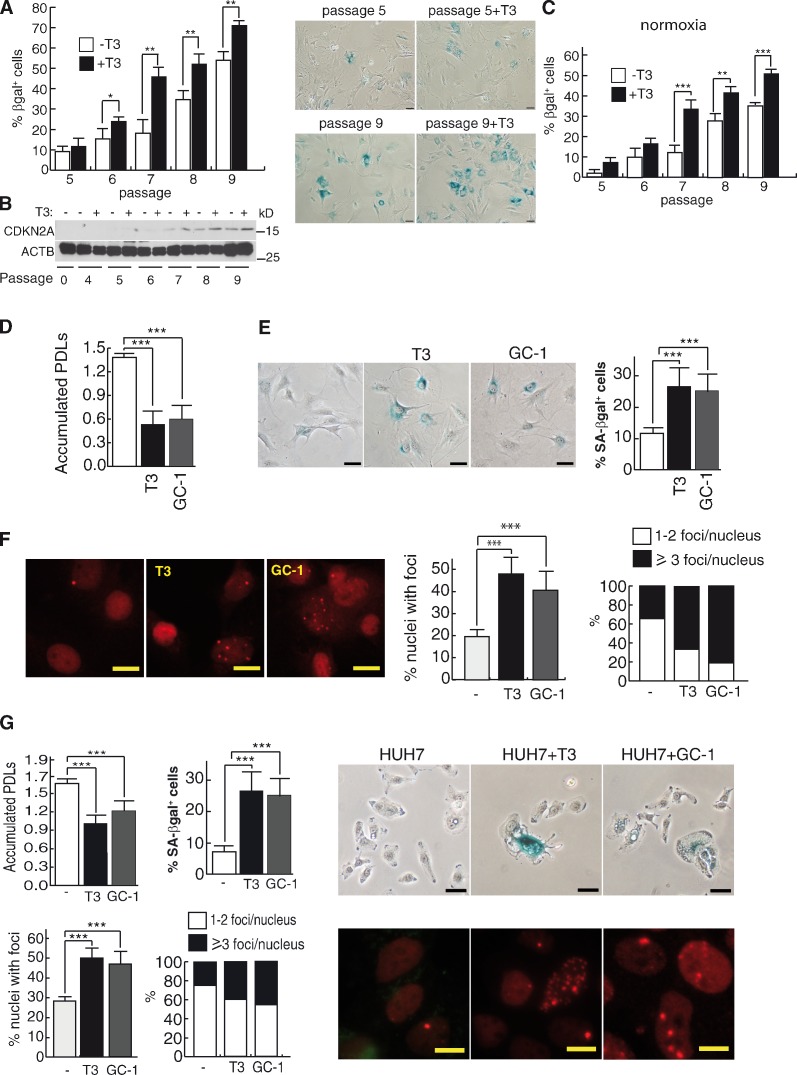
Incubation of MEFs with a physiological-relevant T3 concentration (5 nM) under 20% oxygen increases the amount of cells displaying senescence-like features, such as senescence-associated β-galactosidase (SA-βgal) activity (Fig. 1 A), and increases the levels of CDKN2A (Fig. 1 B), demonstrating that the hormone induces premature senescence. Increased senescence by T3 is also observed under normoxic conditions (Fig. 1 C). MEFs express both THRA and THRB (Fig. S1 A), but THRB appears to mediate senescence because the THRB-specific ligand GC-1, with similar affinity as T3 for the receptor (Baxter et al., 2004), induces cell growth arrest and senescence to the same extent as the natural hormone (Fig. 1, D and E). These effects were strongly enhanced after transduction with a retroviral vector encoding THRB (Fig. S1, B and C).
Figure 1.
T3 induces cellular senescence and DNA damage. (A) 5 nM T3 increases the number of SA-βgal+ cells in primary MEFs at consecutive cell passages (P < 0.0001, n = 3). Representative SA-βgal images are also shown. (B) T3 increases expression of CDKN2A. To control for the level of protein in each sample, the same samples were run on a separate gel and detected for ACTB (β-actin). (C) Percentage of SA-βgal+ cells incubated with and without T3 under normoxic conditions (3% O2). (D) Accumulated population doubling levels (PDLs) of MEFs cultured with serum replacement medium after two passages with and without T3 or GC-1 (5 nM; P < 0.0001, n = 3). (E) SA-βgal images and percentage of SA-βgal+ cells at the end of the treatment (P = 0.001, n = 3). (F) TP53BP1 foci after three passages. Percentages of cells with foci and the mean number of foci/cell are shown on the right (P = 0.0018, n = 3). (G) PDLs, percentages of SA-βgal+ cells, formation of TP53BP1 foci (P < 0,001, n = 3), and representative images of SA-βgal and TP53BP1 foci in HUH7 hepatocytes after one passage in serum replacement medium with and without T3 or GC-1. Bars: (A, E, and G, top) 20 µm; (F and G, bottom) 10 µm. Results are presented as means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Senescence is characterized by an activated DDR (Rodier et al., 2009), and incubation with T3 or GC-1 caused the appearance of discrete DNA damage foci containing TP53BP1 (a marker of DSBs), increasing both the percentage of foci-containing cells and the number of foci per cell (Fig. 1 F and Fig. S1, D and E). In contrast with T3, DNA replication stress secondary to oncogenic H-RasV12–induced senescence (OIS) produced a more potent proliferation arrest and senescence and pannuclear TP53BP1 staining. Foci induced by T3 were similar, although less abundant, than those formed in response to γ irradiation or to 600 µM H2O2 shock (Fig. S1, F–H). The effect of T3 on foci formation and senescence was quantitatively similar to that observed after treatment with 50 µM H2O2 (Fig. S1, I and J). T3 also increased TP53BP1 foci in MEFs from Thra knockout (KO) mice but not from THRB-deficient mice (Fig. S1 K). On the other hand, Thra KO MEFs showed a higher DNA damage after H2O2 treatment than those obtained from ThrbKO mice (Fig. S1 K), and both Thra and Thrb mRNAs were reduced after 6-d treatment with the DNA damaging agent camptothecin (Fig. S1 L). T3 and GC-1 also induced senescence and DNA damage in human hepatocyte HUH7 cells (Fig. 1 G), indicating the relevance of this hormonal action.
T3 did not induce senescence in MEFs obtained from Thra/ThrbKO mice that were spontaneously immortalized in culture. However, expression of THRB (but not THRA) in these cells restored the ability of T3 to induce proliferation arrest, senescence, and formation of DNA damage foci (Fig. 2, A–D). In ThrKO MEFs cultured with a serum substitute, transduction of THRB, but not THRA, increased senescence in the absence of ligand, and this action was further increased by T3 and GC-1 (Fig. 2 E). Therefore, the unliganded receptor has constitutive effects on senescence when expressed at high intracellular concentrations. The percentage of cells undergoing senescence upon THRB expression was lower than in normal MEFs, but most of the immortal cell population became senescent when cells were previously exposed to a H2O2 shock (Fig. 2 F).
Figure 2.
THRB mediates TH-induced senescence in MEFs. (A) THRB and THRA levels in immortalized MEFs from Thra/Thrb KO mice transduced with an empty vector or the receptors. (B) PDLs in cells incubated with and without T3 at three consecutive passages (P < 0.0001, n = 3). (C) Percentages of SA-βgal+ cells at passage 3 (P < 0.0001, n = 3). (D) Percentages of cells with γ-H2AFX, TP53BP1 foci, and both markers (P < 0.0001, n = 3). (E) SA-βgal+ MEFs after 3 d of treatment with T3 or GC-1 in a serum replacement medium (P < 0.0001, n = 3). Bars, 20 µm. (F) SA-βgal in MEFs subjected to a 2-h shock with 600 µM H2O2 and incubated with or without T3 during 3 or 9 d postshock (d.p.s). P1, passage 1; P3, passage 3. (P < 0.0001, n = 3). Results are presented as means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
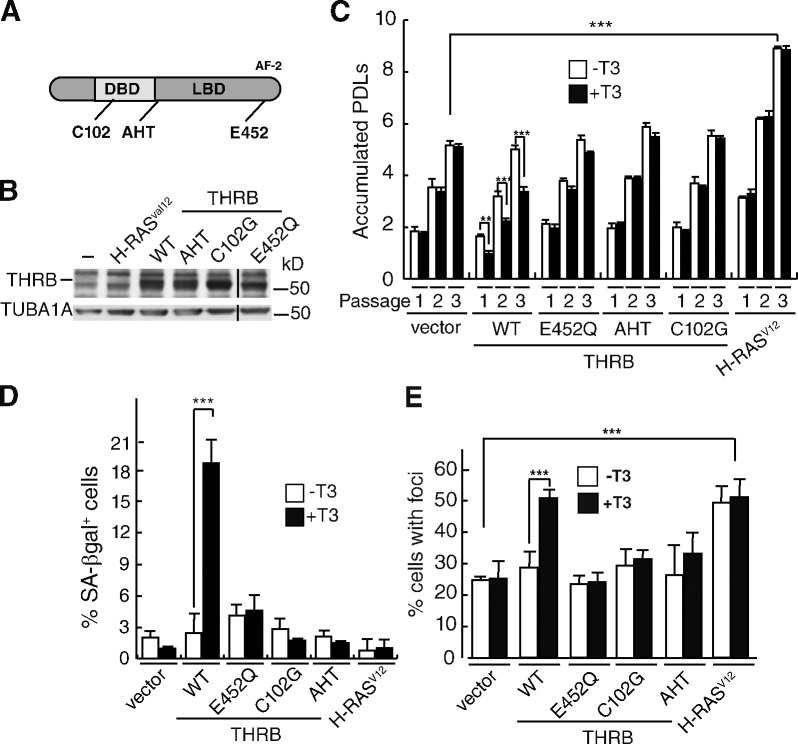
To determine the functional domains involved in T3-dependent induction of senescence, we used various THRB mutants (Fig. 3, A and B). Mutation C102G in the DNA-binding domain abolishes binding to the receptor recognition motifs. The AHT (Ala-His-Tyr) mutant is a triple substitution in the corepressor box that blocks binding of corepressors, and the mutation E452Q is in the ligand-dependent transcriptional activation domain (AF-2) and abolishes the recruitment of coactivators. Only binding of T3 to wild-type THRB inhibited proliferation and induced DNA damage and senescence (Fig. 3, C–E). It is remarkable that THRB was able to induce senescence in this immortalized cell context in which H-RasV12 enhanced proliferation rather than inducing OIS (Fig. 3, C and D).
Figure 3.
Transcriptionally active THRB mediates TH-induced senescence and DNA damage in MEFs. (A) Schematic representation of THRB, showing positions of the mutations C120G, AHT, and E452Q. DBD, DNA-binding domain; LBD, ligand-binding domain. (B) Thra/ThrbKO MEFs were transduced with vectors for the different mutants or H-RasV12, and after selection, THRB was detected by Western blotting. TUBA1A was used as a loading control. Black lines indicate the removal of an intervening lane for presentation purposes. (C) PDLs, estimated after one, two, and three passages (P < 0.0001, n = 3). (D) Percentages of SA-βgal+ cells after passage 3. (E) Percentage of cells having DNA damage foci determined from immunofluorescence of γ-H2AFX and TP53BP1 and merged images (P < 0.0001, n = 3). Results are presented as means ± SD. **, P < 0.01; ***, P < 0.001. WT, wild type.
Role of TP53 and ATM in T3-induced senescence and DNA damage
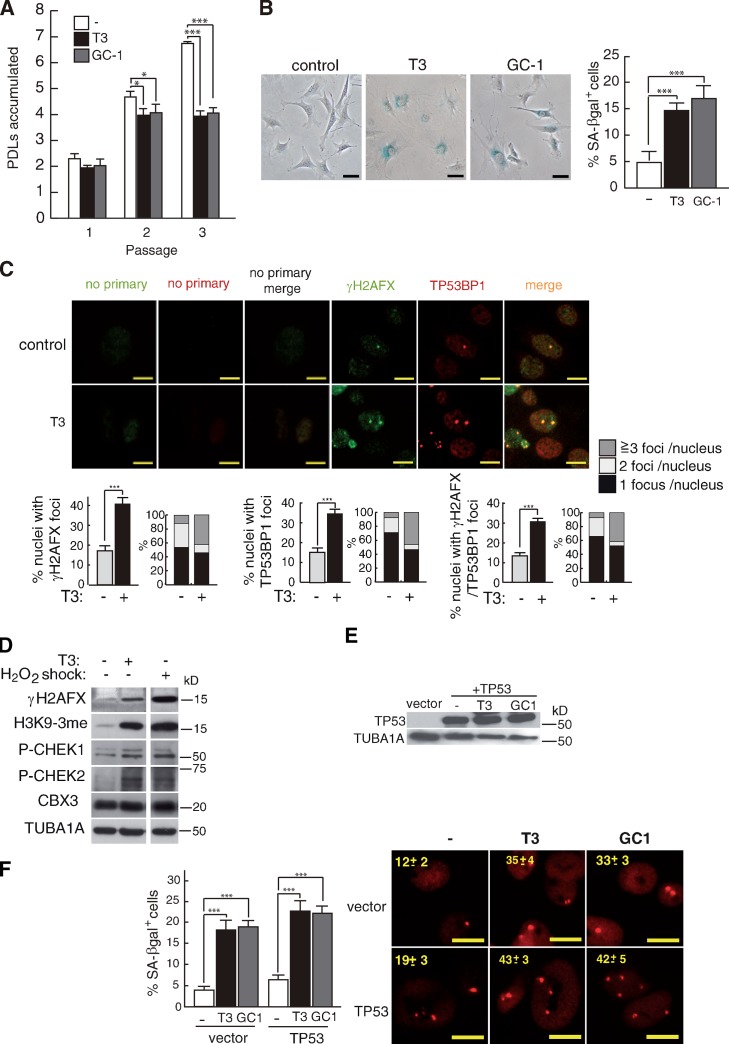
Because these proteins play a prominent role in replicative senescence (Collado and Serrano, 2006) and genomic damage (Lee and Paull, 2005), we next examined the response to T3 in TP53- and ATM-deficient MEFs. Replicative senescence-resistant MEFs obtained from TP53KO mice (Harvey et al., 1993) also underwent partial cell cycle arrest and senescence upon incubation with T3 or GC-1 (Fig. 4, A and B) and showed increased DNA damage foci with colocalization of TP53BP1 and γ-H2AFX (Fig. 4 C). Other markers of genomic DNA damage or senescence, such as phosphorylation of H2AFX, CHEK1, and CHEK2 or the levels of H3F3A K9 3me (lysine 9 histone H3 trimethylation), were also increased by T3 to levels similar to those obtained in cells subjected to H2O2 shock (Fig. 4 D). Complementation of TP53KO MEFs with an expression vector for TP53 slightly increased T3- and GC-1–induced senescence and DNA damage (Fig. 4 E), showing again that the tumor promoter is not essential for the hormonal response.
Figure 4.
T3 induces DDR and senescence in TP53KO MEFs. (A) Accumulated PDLs of TP53KO MEFs in the presence and absence of T3 or GC-1 at three consecutive passages (P < 0.0001, n = 3). (B) SA-βgal+ cells at passage 3 (P < 0.0001, n = 3). (C) γ-H2AFX and TP53BP1 immunofluorescences in TP53KO MEFs incubated with and without T3 during three passages. γ-H2AFX, TP53BP1, and merge images were quantitated, and the percentages of nuclei with foci and the distribution of cells with one, two, and more than three foci/nucleus are represented. (D) Western blot of DNA damage and heterochromatinization markers in TP53KO MEFs after three passages in the absence and presence of T3. Cells were also exposed to a 2-h shock with 600 µM H2O2, washed, and cultured during three passages. (E) TP53 levels after transfection with an empty vector or a TP53 vector. (F) Percentages of SA-βgal+ cells and formation of TP53BP1 foci in cells treated with T3 and GC-1. Bars: (B) 20 µm; (C and F) 10 µm. Results are presented as means ± SD. *, P < 0.05; ***, P < 0.001.
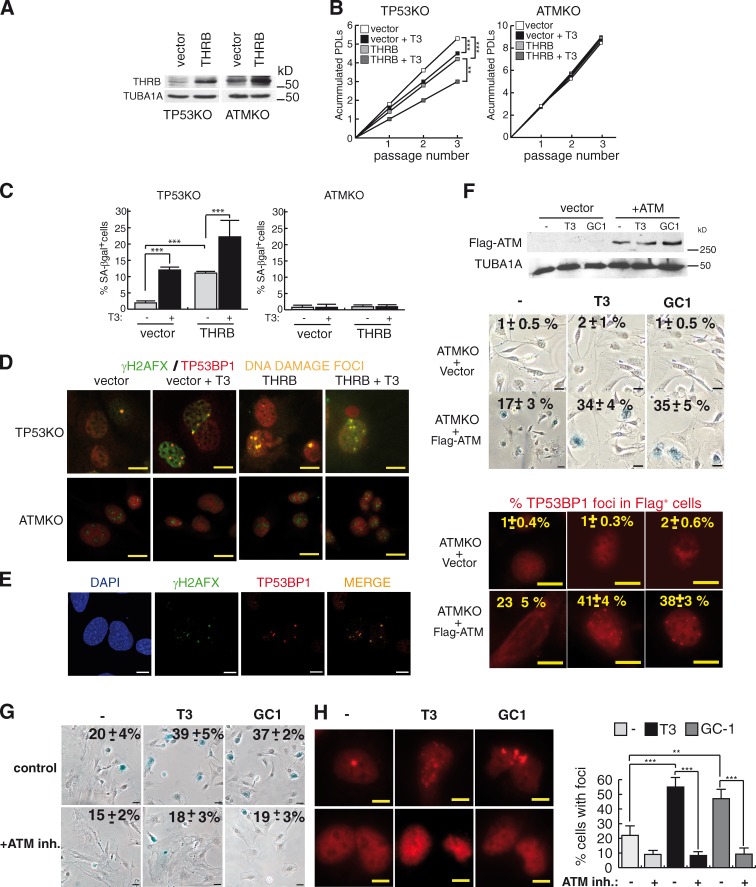
Immortalized MEFs obtained from ATMKO mice (Callén et al., 2009) express THRB (Fig. 5 A), but these cells, which, under basal conditions, essentially do not exhibit TP53BP1 foci (Ward et al., 2003), were totally resistant to T3-mediated proliferation arrest or senescence. Furthermore, transduction of TP53KO MEFs with THRB enhanced T3-dependent cell growth arrest, senescence, and DNA damage, whereas ATMKO cells were unaffected (Fig. 5, B–E). However, transfection of ATM restored T3-dependent formation of DNA damage foci and senescence in ATMKO MEFs (Fig. 5 F), whereas incubation of wild-type primary MEFs with the ATM inhibitor KU-55933 abolished the effect of T3 and GC-1 (Fig. 5, G and H), indicating the key role of this kinase for TH-dependent senescence.
Figure 5.
THRB does not induce senescence in ATMKO MEFs. (A) THRB levels after transduction of an empty vector or the receptor in TP53- and ATM-deficient cells. TUBA1A was used as a loading control. (B) Accumulated PDLs in the presence and absence of T3 at three consecutive passages after selection (P < 0.0001, n = 3). (C) SA-βgal+ cells at passage 3 (P < 0.0001, n = 3). (D) Representative merge images of double immunofluorescences of γ-H2AFX and TP53BP1 in TP53 and ATMKO MEFs incubated with and without T3 during three passages. (E) Confocal microscopy images of γ-H2AFX and TP53BP1 in TP53KO MEFs. (F) ATM levels in ATMKO MEFs transfected with an empty vector or Flag-ATM. Percentages of SA-βgal+ cells after one passage in T3 and GC-1–treated cells, and representative images of TP53BP1 cells, indicating the percentages of Flag-positive cells with foci, are shown at the bottom. (G and H) Percentages of SA-βgal+ cells (G) and TP53BP1 foci (H) in wild-type MEFs pretreated with the ATM inhibitor KU-55993 (10 µM) for 2 h and with T3 or GC-1 for one passage. Bars: (D) 10 µm; (E, F [bottom images], and H) 10 µM; (F [top images] and G) 20 µM. Results are presented as means ± SD. **, P < 0.01; ***, P < 0.001. inh., inhibitor.
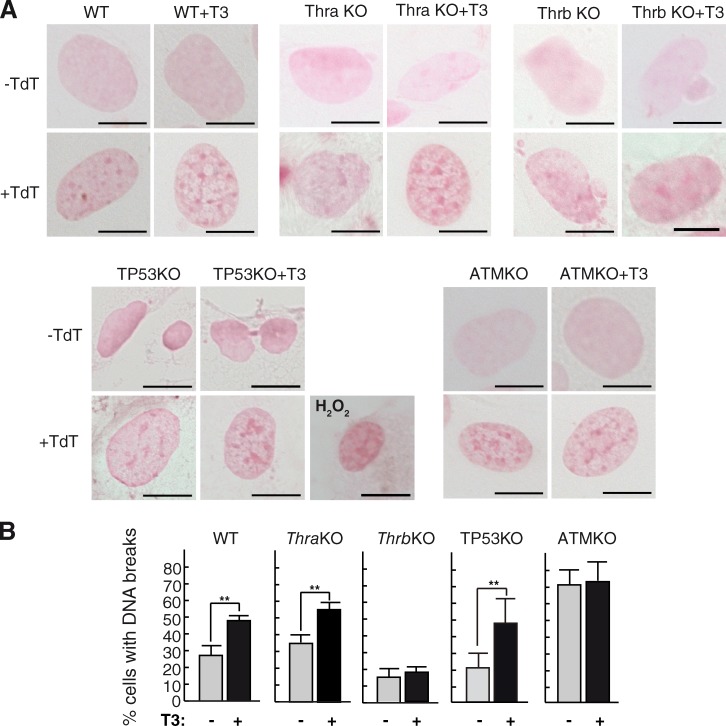
T3 induces formation of DNA strand breaks
The incorporation of deoxy-UTP (dUTP)–11-biotin by the terminal deoxynucleotidyl transferase (TdT) enzyme was used to directly assess the presence of DNA breaks (Fig. 6). The percentage of cells showing breaks was significantly enhanced by T3 both in wild-type and TP53KO MEF cells but not in ATMKO cells, which presented a higher basal level of DNA breaks. Furthermore, T3 increased DNA breaks in primary MEFs from ThraKO mice, but not from ThrbKO animals, showing again that THRB mediates the effect of T3 on genomic damage.
Figure 6.
T3 induces DNA breaks. (A) DNA strand breaks detected with the anti–biotin-HRP antibody in the presence or absence of T3 in the indicated MEF genotypes. Negative controls in the absence of TdT are also shown. Treatment with 600 µM H2O2 for 2 h was used as a positive control. Bars, 10 µm. (B) The percentages of cells with DNA strand breaks are shown. Results are presented as means ± SD. **, P < 0.01. WT, wild type.
DNA damage is not caused by defective DNA repair
We next examined the effect of T3 on repair of DNA damage in normal and TP53KO MEFs. For this purpose, cells were incubated with T3 for 24 h before γ irradiation, and TP53BP1 foci were evaluated at different times after irradiation. Irradiation induced massive foci formation, and the rate of recovery was similar in all groups, indicating that the increased genomic damage caused by T3 is not a consequence of reduced DNA repair (Fig. S2, A and B).
DNA damage is secondary to oxidative stress
We next tested whether oxidative stress could be responsible for the actions of T3 on genomic damage and senescence. As shown in Fig. 7 A, the levels of oxidized glutathione (a precise marker of the cellular oxidative status) were significantly increased by T3 in TP53KO MEFs. Because glutathione is an important sensor of ROS in cells, we measured total cellular ROS levels, finding that they were significantly increased as early as 3 h after incubation with T3, both in these cells as well as in wild-type MEFs (Fig. 7 B). As expected, ATMKO cells present elevated basal ROS levels (Okuno et al., 2012), but in these cells, T3 did not induce a further increase (Fig. 7 B). Because the mitochondria are the main source of free radicals, we also measured mitochondrial ROS levels with MitoSOX in TP53KO MEFs, observing enhanced mitochondrial ROS generation by T3 (Fig. 7 C). Furthermore, mitochondrial ROS was increased by T3 in Thr-null MEFs after expression of THRB but not THRA (Fig. 7 D). The existence of oxidative stress in T3-treated cells was confirmed by the increased expression of antioxidant defense genes, which are induced in response to ROS with the role of protecting the cells against oxidative damage (Fig. S3 A).
Figure 7.
T3 induces oxidative DNA damage. (A) Relative oxidized glutathione (Rel. GSSG) levels in TP53KO MEFs incubated with or without T3 for 3 h. (B) Cellular ROS levels measured with H2DCFDA in TP53KO, ATMKO, and wild-type MEFs treated or not treated with T3 for 3 h (P = 0.0022, n = 3). Treatment for 2 h with 1 mM H2O2 was used as a positive control. (C) Percentage of TP53KO MEFs positive for MitoSOX after incubation with T3 (P = 0.0001, n = 3). 0.1 mM paraquat (1 h) was used as a positive control. (D) Percentages of cells positive for oxidized MitoSOX in Thra/Thrb KO MEFs transduced with THRA or THRB and incubated with T3 during 3 h (P < 0.0001, n = 3). (E) Immunofluorescence of TP53BP1 and 8-OH-dG in MEFs treated for 24 h with and without T3. Merge and negative control images are also shown. (bottom) Confocal microscopy images showing colocalization of TP53BP1 and 8-OH-dG in foci. Bars, 10 µm. (F) Quantification of TP53BP1/8-OH-dG foci. (G) High resolution respirometry of intact TP53KO, wild-type, and ATMKO MEFs incubated with or without T3. Cellular oxygen flow (pmol/s × 10−6 cells) was measured under routine conditions (Cr; P < 0.0001, n = 3), inhibition by oligomycin (CrO; P = 0.0011, n = 3), uncoupling to maximum flux (CrU; P < 0.0001, n = 3), and nonmitochondrial respiration (ROX). Results are presented as means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001. WT, wild type.
An important DNA lesion secondary to oxidative stress is guanine oxidation with formation of 8′-hydroxy-2′-deoxyguanosine (8-OH-dG). We detected the presence of 8-OH-dG in nuclear foci colocalizing with TP53BP1, and the number of foci was significantly enhanced after 3 h of T3 treatment (Fig. 7, E and F). No cross-reactivity of the secondary antibodies was observed (Fig. S3 B), and T3-dependent increase of γ-H2AFX/TP53BP1 and 8-OH-dG/TP53BP1 foci was abolished by the antioxidant N-acetyl-l-cysteine (NAC; Fig. S3 C), confirming the oxidative nature of the DNA damage inflicted.
Because T3 induces ROS production and DNA damage, the hormone should also increase mitochondrial respiration. To demonstrate this, we measured oxygen consumption by high-resolution respirometry. Incubation with T3 significantly enhanced routine cell respiration (Cr), reflecting aerobic metabolic activity under routine culture conditions, the oligomycin-inhibited leak rate of respiration after ATP synthase inhibition (CrO), and the maximal respiratory capacity (CrU), without altering nonmitochondrial respiration (residual oxygen consumption [ROX]) in TP53KO MEFs and in wild-type MEFs. Therefore, T3 increases mitochondrial respiration, augmenting electron flux through the respiratory chain and consequently the production of superoxide anion, the most abundant mitochondrial ROS. However, in agreement with the high levels of ROS and DNA breaks found in ATMKO MEFs, these cells showed high basal levels of oxygen consumption, which were not increased by T3 (Fig. 7 G).
PRKAA and ATM are essential for mitochondrial activation and induction of DNA damage by T3
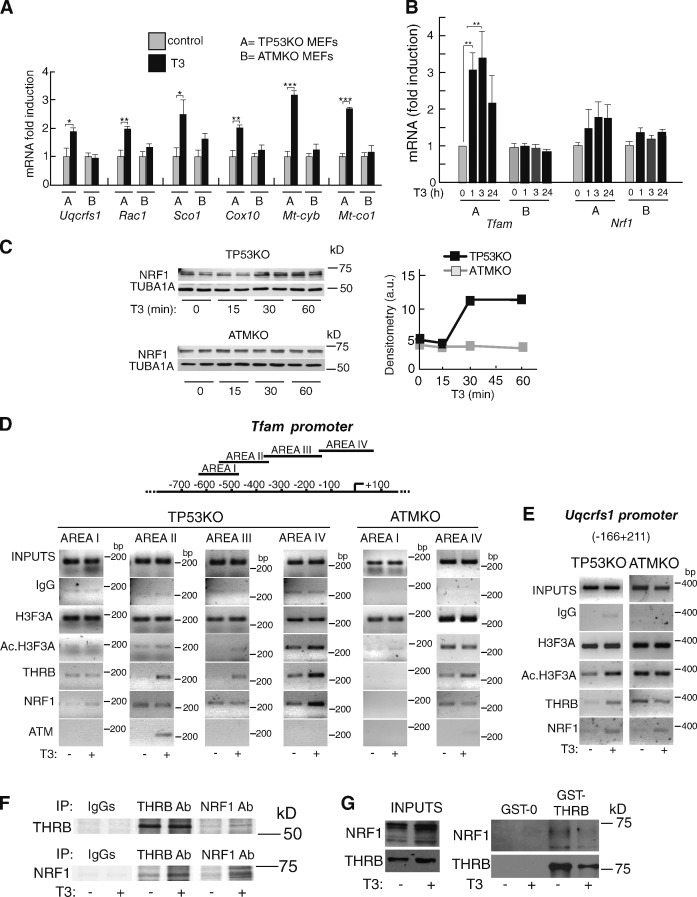
Increased electron flux should be a consequence of increased expression of respiratory chain components. Therefore, we next analyzed the effect of T3 on transcript levels of representative genes of the respiratory chain encoded by nuclear (Uqcrfs1, Rac1, Sco1, and Cox10) or mitochondrial DNA (Mt-cyb and Mt-co1). Remarkably, 1 h of incubation with T3 induced an early expression of all these genes in TP53KO but not in ATMKO MEFs (Fig. 8 A).
Figure 8.
T3 enhances mitochondrial gene expression in MEFs. (A and B) mRNA levels of representative genes of the respiratory chain in TP53KO (A) and ATMKO (B) MEFs incubated with T3 for 1 h. Results are means ± SD (P ≤ 0.0044, n = 3) and are expressed relative to the values obtained in the untreated cells. (B) mRNA levels of TFAM and NRF1 analyzed at 0, 1, 3, and 24 h of T3 treatment (P < 0.0001, n = 3). (C) Western blot of NRF1 in TP53KO and ATMKO MEFs incubated with T3 for the indicated times. TUBA1A was used as a loading control. Densitometry (NRF1/TUBA1A) in arbitrary units (a.u.) is shown on the right. (D) ChIP assays with the indicated antibodies. The depicted areas of the Tfam promoter were amplified in TP53KO cells, and the indicated areas were amplified in ATMKO MEFs. (E) ChIP assays in TP53KO and ATMKO MEFs with the proximal Uqcrfs1 promoter. Cells were treated with and without T3 for 1 h. Ac., acetylated. (F) Coimmunoprecipitation of THRB and NFR1. TP53KO MEFs were labeled in [35S]methionine/cysteine-containing medium for 16 h in the presence or absence of T3 and immunoprecipitated with control IgGs, THRB, or NRF1 antibodies as indicated. IP, immunoprecipitation. (G) GST pull-downs performed with cellular extracts of untreated and T3-treated cells and GST or GST-THRB. The inputs (10%) of the total are also shown. Results are presented as means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
NRF1 (nuclear respiratory factor 1) and TFAM (transcription factor A, mitochondrial) are the major transcription factors responsible for mitochondrial gene expression (Scarpulla, 2002). mRNA levels of Tfam, which is essential for transcription of genes of the respiratory chain, were rapidly induced by T3 (Fig. 8 B). Tfam transcription in turn depends on NRF1 binding to its promoter (Virbasius and Scarpulla, 1994). NRF1 transcripts were not induced by T3 in MEFs, but a rapid increase of NRF1 protein levels was found after T3 incubation, showing the existence of a rapid posttranscriptional regulation. NRF1 induction was also ATM dependent because no changes were found in ATMKO MEFs (Fig. 8 C).
We next performed chromatin immunoprecipitation (ChIP) assays with four overlapping fragments of the Tfam promoter to examine whether transcriptional stimulation of the Tfam gene by T3 involves increased NFR1 binding (Fig. 8 D). Incubation with T3 for 1 h caused an enrichment of histone 3 acetylation, a mark of transcriptional activation, mainly in the areas closer to the transcription start site. In addition, T3 induced NRF1 recruitment, particularly to the more proximal promoter fragment, and THRB was concomitantly recruited in a hormone-dependent manner. Neither histone acetylation nor NRF1 or THRB recruitment were found in ATM-deficient MEFs, emphasizing its importance for T3-mediated stimulation of mitochondrial gene expression. These results were extensive to other NRF1-dependent genes, such as Uqcrfs1, in which enrichment of promoter histone 3 acetylation and NRF1 and THRB recruitment in response to T3 were only observed in the presence of ATM (Fig. 8 E). T3-dependent NRF1 recruitment to the promoter could involve a direct interaction of this transcription factor with THRB, as suggested by coimmunoprecipitation experiments. Fig. 8 F shows that, as expected, the amount of coimmunoprecipitated NRF1 is higher in T3-treated cells that express higher levels of this factor. The existence of a direct interaction between THRB and NRF1 was further demonstrated in a GST pull-down assay (Fig. 8 G).
NRF1 is required for T3-dependent DNA damage because its depletion with siRNA abolished formation of γ-H2AFX/TP53BP1 foci by the hormone (Fig. 9, A and B). NRF1 undergoes posttranslational modifications that lead to increased binding to its DNA motif (Gugneja and Scarpulla, 1997). A possible mediator is PRKAA (AMP-activated protein kinase), a direct ATM target (Alexander et al., 2010), which plays a key role in mitochondrial biogenesis (Fu et al., 2008) and can be regulated by the TH (Yamauchi et al., 2008; López et al., 2010). We found a rapid phosphorylation of PRKAA by T3, and this response was markedly blunted in ATM-deficient MEFs (Fig. 9 C). Preincubation of cells with the PRKAA inhibitor compound C or with the ATM inhibitor KU-55993 abolished T3-dependent increase of NRF1 (Fig. 9, D and E), proving the central role of PRKAA and ATM in NFR1 induction by T3. ATM and PRKAA activation was also an essential step for the development of oxidative DNA damage because both inhibitors blocked formation of 8-OH-dG/TP53BP1 foci by T3 (Fig. 9, F and G). Finally, transduction of MEFs with an adenoviral vector encoding a dominant-negative form of PRKAA abolished the effects of T3 on PRKAA phosphorylation, induction of NRF1 and formation of DNA damage foci (Fig. 9, H and I), confirming that PRKAA activation plays an essential role on mitochondrial activation, ROS generation, and formation of DNA DSBs by the hormone.
Figure 9.
PKRAA and ATM activation is required for T3-dependent NRF1 induction and oxidative DNA damage. (A) Western blot of NRF1 in TP53KO MEFs treated with T3 for 3 h after transfection with control or NRF1 siRNAs 48 h before. (B) Percentage of nuclei with γ-H2AFX/TP53BP1 foci in these cells. (C) Phosphorylated and total PRKAA evaluated by Western blotting in TP53KO and ATMKO MEFs incubated with T3 for times ranging between 0 and 60 min. Densitometries (P-PRKAA/PRKAA) in arbitrary units (a.u.) are shown on the right. (D) NRF1 levels in TP53KO and wild-type MEFs preincubated for 2 h with 20 µM compound C before treatment with T3 for 3 h. Wt, wild type. (E) NRF1 in TP53KO cells preincubated with 10 µM KU-55993. TUBA1A was used as a loading control. inh., inhibitor. (F) TP53BP1/8-OH-dG immunofluorescences in cells treated as in D and E. Bars, 10 µm. (G) Quantification of TP53BP1/8-OH-dG foci in the same groups (P < 0.0001, n = 3). (H) Cells were infected with adenovirus expressing a dominant-negative (dn) PRKAA. After 24 h, cells were treated with T3 for 3 h, and the levels of NRF1, P-PRKAA, and total PRKAA were analyzed by Western blotting. Mock-infected cells were used as controls. (I) Percentages of nuclei bearing γ-H2AFX/TP53BP1 foci in cells expressing dominant-negative PRKAA. Results are presented as means ± SD. **, P < 0.01; ***, P < 0.001.
THs induce senescence and oxidative DNA damage in vivo
We next examined the effect of TH treatment on senescence and DNA damage in vivo. For this, we treated wild-type, ThraKO, and ThrbKO mice with THs for 2 wk, and DNA damage was evaluated in the liver and kidney, which express preferentially THRB, and in the heart, which expresses high THRA levels (Fig. S4 A). Hyperthyroidism was confirmed by elevated expression of Deiodinase 1 mRNA, a sensitive marker of TH action (Fig. S4 B; Zavacki et al., 2005). Senescence was negligible in livers of young euthyroid mice, but hyperthyroid livers from wild-type mice showed a significant number of senescent cells and an increased number of cells expressing high CDKN2A levels (Fig. 10 A). Similar results were obtained in ThraKO mice, but not in ThrbKO mice, confirming in vivo that THRB is the receptor isoform mediating these effects of THs. T3 also induced senescence and DNA damage in normal primary hepatocyte cultures (Fig. S5 A). γ-H2AFX foci were observed in euthyroid livers of all groups, and hyperthyroidism significantly increased the percentage of cells bearing γ-H2AFX foci as well as the number of foci per cell in wild-type and ThraKO mice but not in ThrbKO mice (Fig. 10, B and C). Furthermore, immunohistochemical staining of 8-OH-dG showed that hyperthyroid livers exhibited a significantly larger number of nuclei with oxidative lesions, detected either as discrete foci or pannuclear staining (Fig. 10 D), and double immunofluorescences in the livers from euthyroid and hyperthyroid mice demonstrated colocalization of γ-H2AFX and TP53BP1 with 8-OH-dG in foci (Fig. S5 B).
Figure 10.
THRB mediates induction of senescence and DNA damage in the hyperthyroid mice liver. (A) Detection of SA-βgal and CDKN2A in livers from wild-type, ThraKO, and ThrbKO mice (six mice for each condition). Nuclear Fast Red was used as the counterstaining. (B) γ-H2AFX immunohistochemistry in the livers. Negative controls (without primary antibody) are also shown. Some foci are signaled with arrowheads. (C) Percentages of cells with foci and the mean number of foci/nucleus. (D) Detection of 8-OH-dG by immunohistochemistry in livers of euthyroid and hyperthyroid (hyperth.) wild-type mice. A negative control of a hyperthyroid liver is also shown. Representative cells (dotted squares) are shown as insets. Percentages of cells positive for 8-OH-dG is shown on the right. Bars: (A, B, and D, main images) 10 µm; (D, insets) 10 µM. wt, wild type. Results are presented as means ± SD. **, P < 0.01; ***, P < 0.001.
γ-H2AFX foci were also observed in euthyroid kidneys in all genotypes, although in a lower number than in livers, and hyperthyroidism increased the number of foci in wild-type and THRA-deficient mice but not in mice lacking THRB. However, no senescent cells were found in this organ. In contrast with the THRB target tissues, no detectable increase of DNA damage was observed in hyperthyroid hearts (Fig. S4 C), reinforcing the idea that this effect of T3 is observed in THRB but not THRA target organs.
Discussion
In this study, we define THRB as a novel inductor of cellular senescence and DNA damage. T3 induces senescence in MEFs and hepatocytes, and this action is mediated by THRB. CDKN2A and TP53 are major components of replicative and OIS (Collado and Serrano, 2006), but surprisingly, the effect of T3 appears to be largely independent of TP53 because the hormone can arrest proliferation and induce partial senescence in TP53-deficient immortal MEFs. A potential mechanism by which THRB could bypass the lack of TP53 on senescence is that T3 increased CDKN2A, suggesting that, as in the case of stress-induced senescence (Serrano et al., 1996), the hormone could work mainly through the activation of this cyclin kinase inhibitor.
T3 generates DNA damage, which is crucial in the induction of cellular senescence (d’Adda di Fagagna et al., 2003; Rodier et al., 2009). DDR involves the recruitment of γ-H2AFX and TP53BP1, a marker of DSBs (Rogakou et al., 1999; Schultz et al., 2000; Abraham, 2002), at the sites of DNA breakage. T3 mediates DSB generation that has an oxidative origin because (a) the hormone enhances the appearance of 8-OH-dG, a biomarker of oxidative DNA damage, (b) there is a strong cofocalization of 8-OH-dG with TP53BP1, showing the presence of oxidized DNA at the sites with DSBs, and (c) the antioxidant NAC prevents formation of foci containing 8-OH-dG and the DNA damage marker by T3.
T3 appears to cause persistent genomic damage because an increased number of nuclear DNA damage foci was observed at passage 3, i.e., at 9 d of treatment. However, enhanced formation of foci containing DSBs and oxidized DNA was detected after 3 h of T3 treatment, showing that this is an early hormonal action. The rapid appearance of genomic damage is compatible with the rapid increase of oxidized glutathione and mitochondrial ROS generation. ROS can induce base oxidation, abasic sites, and even both single and double DNA breaks (Wyman and Kanaar, 2006).
ROS are generated as a byproduct of mitochondrial respiration because electrons leak from the electron transport chain, converting ∼1–3% of oxygen molecules into superoxide (Chance et al., 1979). It has been known for decades that the THs regulate basal metabolic rate, heat production, and oxygen consumption (Tata et al., 1962; Oppenheimer et al., 1987), and therefore, they can increase ROS generation and cause oxidative stress (Videla, 2010). Using high-resolution respirometry, we could demonstrate for the first time that T3 induces an evident increase of oxygen consumption at different respiratory states in intact cells. This augmented mitochondrial respiration would lead to enhanced electron leak and ROS generation.
T3 stimulates expression of genes of the respiratory chain, which could lead to increased oxygen consumption. T3 mediates a rapid and important increase of Tfam, a transcription factor essential for mitochondrial gene transcription (Larsson et al., 1998), and this activation may play an important role in the mitochondrial response to the hormone. Tfam transcription depends critically on binding of nuclear respiratory factors to its promoter (Virbasius and Scarpulla, 1994), and we observed an early posttranscriptional induction of NRF1 levels by T3. Besides Tfam, many genes encoding subunits of the five respiratory complexes contain functional NRF1 binding sites within their promoters. We could demonstrate that the mechanism of stimulation of mitochondrial activity by T3 reflects a rapid chromatin remodeling, detected by increased histone acetylation and recruitment of NRF1 and THRB to the promoter region of genes such as Tfam or the respiratory gene Uqcfrs1. It is known that NRF1 phosphorylation enhances DNA binding (Gugneja and Scarpulla, 1997). PRKAA has been shown to activate NRF1 expression, increasing binding to DNA, and to stimulate mitochondrial biogenesis (Bergeron et al., 2001), and we demonstrated here the essential role of this kinase on mitochondrial T3 actions because the hormone induces a very rapid activation of PRKAA, and this activation is required for NRF1 induction and for the consequent development of oxidative DNA damage.
ATM plays a pivotal role on the detection of the genomic damage and DDR (Harper and Elledge, 2007). We observed here that T3 was unable to induce proliferation arrest and senescence in the absence of ATM, even when THRB is expressed at high levels, whereas reexpression of the kinase restored the actions of T3. ATM plays a crucial role on the activation of the mitochondrial function by T3 in MEFs. Thus, the hormone does not induce NRF1 recruitment to mitochondrial promoters in the absence of this kinase, and subsequent transcription of respiratory chain components or TFAM is not observed. This correlates with the lack of effect of T3 on ROS generation and oxidative DNA damage in ATMKO MEFs. ATM phosphorylates the α subunit of PRKAA (Suzuki et al., 2004; Sun et al., 2007; Alexander et al., 2010), and stimulation of PRKAA phosphorylation by T3 is markedly reduced in ATM-deficient MEFs. Therefore, ATM-dependent PRKAA signaling appears to be needed for the effect of T3 on mitochondrial activity, suggesting that this kinase not only directs DDR but could also be required at the first steps leading to generation of DSBs by the receptor. It is also possible that the high basal levels of ROS found in ATMKO cells have made the cells unresponsive to a further increase of ROS by T3, and as a consequence, the hormone would not induce DNA damage and senescence in these cells. In summary, these results suggest a model in which T3 binding to THRB induces a rapid nongenomic activation of PRKAA, in an ATM-dependent manner. This leads to NRF1 activation and recruitment, together with the receptor, to the promoters of key mitochondrial genes, which are essential to increase oxygen consumption. Increased respiration induces the production of mitochondrial ROS, which in turn causes oxidative stress and DSBs, triggering a persistent DDR that ultimately leads to premature senescence of susceptible cells.
The physiological importance of our findings is reinforced by the observation that administration of THs induces premature senescence in livers of young mice. This action is mediated by THRB because hyperthyroid livers from Thrb-null mice do not display senescent cells. Furthermore, these animals do not show enhanced oxidative DNA damage in contrast with the findings in wild-type and Thra-null mice. It was unexpected that oxidative DNA lesions that can be rapidly repaired by base excision repair generated DSBs in terminally differentiated nondividing hepatocytes. It is possible that other mechanisms, for example nonhomologous end joining could operate because very recent studies describe that TP53BP1 (in cooperation with RIF1) can block DSB resection and promotes nonhomologous end joining in G1 phase (Callén et al., 2013; Escribano-Díaz et al., 2013; Zimmermann et al., 2013).
In the kidneys, another THRB target tissue, TH treatment also induces formation of DNA damage foci in wild-type and ThraKO mice, although in this case, senescent cells are not observed at this age. This might be caused by the lower number of DNA damage foci observed in this organ. In contrast to the damage observed in the liver and kidney, hyperthyroidism did not induce DNA damage in the heart, a THRA target tissue (Pascual and Aranda, 2013).
Our findings provide a mechanism for integrating the well-known metabolic effects of THs with other important processes, such as cellular senescence. Cellular senescence plays an important role in aging and in the control of life span (Vijg and Campisi, 2008; Baker et al., 2011), and our results can shed light onto the mechanisms by which thyroidal status affects longevity. Hormonal regulation has been recently linked to DNA damage. Thus, mice with defects in DNA repair pathways show suppression of genes such as deiodinase 1/2 or THRs with an important role in somatotroph, lactotroph, and thyrotroph axes (Niedernhofer et al., 2006; van der Pluijm et al., 2007; Garinis et al., 2009). The current notion is that such responses are adaptive and could improve animal survival. Our results strengthen this hypothesis and show that THR expression is altered in MEFs exposed to genotoxic agents. In addition, cellular senescence also represents an important barrier against tumor development in vivo (Collado and Serrano, 2010). Because THRB represses cellular transformation and tumor progression, induction of senescence could be a critical component of the initial tumor-suppressing mechanism of this receptor. Finally, it is known that liver damage is a relatively common clinical characteristic in hyperthyroid patients (Habershon, 1874; Malik and Hodgson, 2002), and tissue damage appears after TH administration in rats (Upadhyay et al., 2004). The occurrence of oxidative DNA damage observed in liver and kidney after TH administration to mice could be involved in tissue damage observed in hyperthyroid conditions and supports the clinical use of antioxidant drugs to prevent these abnormalities (Leo et al., 2012).
Materials and methods
Mice and animal care
Wild-type mice and mice lacking the TH-binding isoforms THRA1 or THRB (ThraKO and ThrbKO, respectively), provided by B. Vennström (Karolinska Institute, Stockholm, Sweden), were made hyperthyroid by adding thyroxine (95 ng/g of mice) and T3 (25 ng/g of mice) to the drinking water for 2 wk. Experiments were performed twice in 2- and 6-mo-old mice. All experiments were performed following the regulations of the Consejo Superior de Investigaciones Científicas for animal care and handling (RD 53/2013).
Transfections, cell proliferation, and SA-βgal analysis
Cultures of MEFs were obtained at 13.5 d postcoitum. Wild-type, ThraKO, and ThrbKO MEFs were incubated ≥24 h before the experiments in medium containing 10% TH-depleted bovine fetal serum by treatment with resin AG-1-X8 (Bio-Rad Laboratories) or in serum-free medium containing Serum replacement-1 (Sigma-Aldrich) and 10 ng/ml human FGF basic. MEFs were normally grown with 20% O2, except for the experiment shown in Fig. 1 C, which was performed under normoxic conditions (3% O2). Cells were treated with T3 or with GC-1 (provided by T.S. Scanlan, Oregon Health and Science University, Portland, OR), as indicated in the corresponding figures. Immortalized MEFs from Thra/Thra double-KO mice provided by J. Samarut (Ecole Normale Supérieure de Lyon, Lyon, France), TP53KO mice (M. Serrano, Centro Nacional de Investigaciones Oncológicas, Madrid, Spain), and ATMKO mice (E. Callen, National Institutes of Health, Bethesda, MD) were also used. Primary hepatocytes were a gift from A.M. Valverde (Instituto de Investigaciones Biomédicas, Madrid, Spain). pLPCX-THRB, pLPCX-THRA, the C102G, AHT and E457Q THRB mutants (García-Silva et al., 2011), or the empty vector was used to infect MEFs by retroviral transduction. MEFs were also transduced with pWLZ-Ha-Rasval12 or pWLZ and selected with hygromycin for 5 d (García-Silva and Aranda, 2004). Transfections with control siRNA (D-001210-01-05; Thermo Fisher Scientific) and NRF1 siRNA (sc-43576; Santa Cruz Biotechnology, Inc.) were performed with Lipofectamine 2000 reagent (Invitrogen). Complementation of TP53KO and ATMKO MEFs was performed by transfection of pcDNA-TP53 or Flag-ATM vectors by the TransIT-TKO transfection method (Mirus Bio LLC). Growth curves were generated following the 3T3 protocol (Todaro and Green, 1963). Accumulated population doubling levels (PDLs) were calculated as PDL = log(Nf/N0) × 3.33, in which N0 is the number of inoculated cells, and Nf is the final number of cells obtained after 3 d. SA-βgal activity was determined as reported by Dimri et al. (1995) or with the Senescence Detection Kit (BioVision, Inc.). Micrographs were taken in a microscope (TS100F; Nikon) with a digital camera (DS-L1; Nikon), and the percentage of β-galactosidase+ MEFs was calculated after counting >200 cells.
Protein and RNA analysis
Western analysis, RNA extraction, and quantitative real-time PCR were performed as previously described (García-Silva et al., 2011) using the antibodies and primers listed under Tables S1 and S2.
Metabolic labeling and coimmunoprecipitation
Cells growing in p100 plates were incubated for 2 h in methionine/cysteine-free medium (Gibco) and labeled with 70 µCi/ml [35S]methionine/cysteine (PerkinElmer) for 16 h in medium containing glutamine and 10% dialyzed FBS in the presence or absence of 5 nM T3. The cells were washed with ice-cold PBS and lysed in triple-detergent lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.02% sodium azide, 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholate, 1 mM PMSF, 2 µg/ml pepstatin, 2 µg/ml aprotinin, 2 µg/ml leupeptin, and phosphatase inhibitors cocktail 2 and 3 [Sigma-Aldrich]). Lysates were precleared for 1 h/4°C with 50% slurry of protein A–Sepharose (CL-4B), reconstituted with single-detergent lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.02% sodium azide, 1% NP-40, 1 mM PMSF, 2 µg/ml pepstatin, 2 µg/ml aprotinin, 2 µg/ml leupeptin, and phosphatase inhibitors cocktail 2 and 3). 30 × 106 cpm per lysate and 2 µg antibody were incubated overnight at 4°C in a rotatory shaker. Immunoprecipitation complexes were incubated with 100 µl of 50% slurry of protein A–Sepharose beads for 1 h at 4°C and recovered by centrifugation at 2,000 rpm. The complexes were washed four times with single-detergent lysis buffer and resuspended in 50 µl Laemmli buffer. The samples were boiled and loaded on 8% SDS-PAGE. Gels were fixed, treated with EN3HANCE (PerkinElmer), and exposed to x-ray films under standard conditions. The antibodies used were normal IgGs, THRB (Rockland Immunochemicals, Inc.), and NRF1 (Table S1).
GST pull-down assays
GST-0 and GST-THRB fusion proteins were expressed in the bacterial strain BL21 (DE3). They were grown at 37°C in 2× YT (16 g/liter tryptone, yeast extract, and 5 g/liter NaCl, pH 7) until the absorbance reached 0.6. Then, the induction was performed at 30°C for 3 h with 0.4 mM isopropyl-1-thio-β-d-galactopyranoside. The expression of correctly sized proteins was monitored by SDS-PAGE as described previously. Cells were treated or not treated with T3 for 16 h and lysed in triple-detergent lysis buffer. 500 µg/reaction of lysate was precleared for 1 h at 4°C with 50 µl of 50% slurry of glutathione-Sepharose (CL-4B; GE Healthcare) reconstituted with single-detergent lysis. Lysates were incubated with GST fusion proteins overnight at 4°C and washed three times with single-detergent lysis buffer. Protein complexes were eluted in Laemmli buffer and loaded on 8% SDS-PAGE. Western blotting was performed with THRB and NRF1 antibodies (Table S1).
ROS analysis
Cellular ROS levels were assessed with 20 µM H2DCFDA (2′,7′-dichlorodihydrofluorescein diacetate; Sigma-Aldrich). Measurements were made in a microplate fluorometer every 2 min for 1 h at 37°C with 485/20-nm and 528/20-nm filters. Analysis of mitochondrial ROS was performed as previously described (Mukhopadhyay et al., 2007) using MitoSOX (Invitrogen) and flow cytometry.
Oxidized glutathione analysis
Oxidized glutathione measurement was performed with the Glutathione Assay Kit II (EMD Millipore).
High resolution respirometry
Respiration in intact cells was measured at 37°C by high resolution respirometry using Oxygragh-2k (Oroboros Instruments), and the DatLab4 software was used (Oroboros Instruments; Hütter et al., 2006). The protocol includes the determination in a sequential manner of the aerobic metabolic activity under routine culture conditions with the physiological substrates in culture medium (Cr), the oligomycin-inhibited leak rate of respiration after inhibition of ATP synthase with 2 µg/ml oligomycin (CrO), the maximum respiratory capacity of uncoupled mitochondria in nonpermeabilized cells (CrU), obtained by sequential addition of 0.5-µM boluses of trifluorocarbonylcyanide phenylhydrazone, and ROX, reflecting nonmitochondrial respiration, obtained after inhibition of complex I with 0.1 µM rotenone and complex III with 2.5 µM antimycin A. ROX was subtracted to calculate Cr, CrO, and CrU oxygen fluxes.
ChIP assays
Cells were plated in 150-mm dishes and the next day treated with 2.5 µM α-amanitin in serum-free medium for 2.5 h. Cells were then washed and treated with 5 nM T3 for 1 h, fixed and lysed following specifications of the ChIP Assay Kit (EMD Millipore), and sonicated in a Bioruptor (UCD-200TM; Diagenode). In each immunoprecipitation, 2–3 × 106 cells and the amount of antibodies listed in the Table S1 were used. DNA was purified, precipitated, and amplified with the primers listed in Table S3.
DNA breaks labeling
To detect DNA breaks, we developed a method based in dUTP incorporation by TdT and posterior detection by immunocytochemistry. Cells were fixed with 2% PFA for 10 min and then permeabilized with permeabilization buffer containing 0.1% Triton X-100 and 0.1% sodium citrate. After washing with PBS for 5 min, they were incubated overnight at 37°C with a 50-µl reaction mixture containing 15 U of the TdT enzyme (Promega) and 200 µM dUTP-11-biotin (Thermo Fisher Scientific). Cells were washed again and then incubated for 20 min with 1% PBS-BSA and 1 h at RT with antibiotin HRP-conjugated antibody (Table S1) diluted in 1% PBS-BSA. Revealing was performed following specifications of the DAB Substrate Kit (Vector Laboratories). Nuclei were counterstained with Nuclear Fast Red (Sigma-Aldrich), and preparations were mounted with mounting medium (DePeX; Serva). Micrographs were taken with a microscope (E90i; Nikon) with a camera (DS-Fi1) and NES Elements software (Nikon).
Indirect immunofluorescence microscopy
Cells growing in 8-well chambers (Thermo Fisher Scientific) were fixed in 2% PFA for 10 min at RT, washed with PBS, and permeabilized with 0.1% Triton X-100 and 0.1% sodium citrate. Preparations were washed with PBS and washing solution (PBS/0.25% BSA/0.1% Tween 20), blocked for 30 min with blocking solution (washing solution, 2.5% BSA, and 5% normal serum), and incubated overnight with γ-H2AFX, TP53BP1, and 8-OH-dG antibodies (Table S1). Preparations were then washed with washing solution and incubated with secondary antibodies for 1 h at RT. Nuclei were counterstained with DAPI, and samples were mounted with ProLong (Molecular Probes). Cell images were captured with a fluorescence microscopy (E90i) equipped with a camera (DS-Q1Mc; Nikon) and NES Elements software. DNA damage foci were counted from >200 cells for each experimental condition. Analysis of cofocalization was performed with a confocal microscope (LSM 710; Carl Zeiss; 63×/1.4 NA lens Plan Apochromat), using Argon (488 nm), HeNe (543 nm), and diode violet (405 nm) lasers. Images were obtained with software (ZEN 2009; Carl Zeiss).
Immunohistochemistry, SA-βgal detection, and multiple antigen fluorescent labeling
Mice liver, kidney, and heart samples were either fixed in 4% buffered formalin and embedded in paraffin wax or flash frozen with liquid N2 and embedded in O.C.T. (Tissue-Tek). For γ-H2AFX, immunohistochemistry was performed on 4-µm deparaffinized, rehydrated sections. Antigen retrieval was performed in a Microwave Tender Cooker, and endogenous peroxidase activity was inhibited with 0.3% H2O2 in methanol. Permeabilization, blocking, and overnight incubation with the antibody were performed with the Vectastain ABC kits (Vector Laboratories). For 8-OH-dG staining, 10-µm-thick flash-frozen sections were fixed with 1% PFA, washed, blocked with 1% BSA, and incubated with the antibody for 3 h. For CDKN2A staining, frozen sections were fixed 2 min with ethanol, gradually rehydrated, and incubated overnight at RT with the antibody. Revealing, counterstaining, and mounting were performed as described in the previous section. To detect SA-βgal, 10-µm-thick flash-frozen liver sections were used. Sections were fixed for 2 min with ethanol and gradually rehydrated before being incubated overnight at 37°C with the SA-βgal reaction mixture. The sections were counterstained with Nuclear Fast Red and mounted. For multiple antigen labeling of liver sections, we used the double-immunofluorescent labeling protocol and reagents provided by Vector Laboratories (A-2011, A-2016, SP-2001, BA-1400, BA-5000, and Vectastain ABC kits). Sequential incubations of the antibodies were overnight and 3 h at RT for the first and second antibody, respectively. Nuclei staining, mounting, and microscopy acquisition were performed as for the indirect immunofluorescence.
Statistical analysis
Statistical significance of data were determined by applying a two-tailed Student’s t test or analysis of variance followed by the Newman–Keuls or Bonferroni test for experiments with more than two experimental groups. P < 0.05 is considered significant. Significance of analysis of variance posttest or the Student’s t test is indicated in the figures as *, P < 0.05; **, P < 0.01; and ***, P < 0.001. Statistics were calculated with the Prism 5 software (GraphPad Software). All results are presented in the figures as means ± SD.
Online supplemental material
Fig. S1 shows that THRB overexpression increases senescence and DNA damage in MEFs and compares the effect of THRB, H2O2, and H-RasV12. Fig. S2 illustrates that the increased DNA damage observed after T3 treatment is not secondary to inhibition of DNA repair. Fig. S3 demonstrates that antioxidant response genes are induced by T3, controls of secondary antibodies, and that NAC prevents T3-induced oxidative DNA damage. Fig. S4 shows expression levels of THRA and THRB in mouse tissues and proves that THs induce DNA damage in THRB, but not THRA, target tissues. Fig. S5 shows induction of senescence by T3 in primary mice hepatocytes and colocalization of DNA damage markers in the liver. Table S1 contains a list of antibodies used in this study. Table S2 contains a list of primers used for quantitative PCR. Table S3 contains a list of primers used for ChIP assays. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201305084/DC1.
Supplementary Material
Acknowledgments
We thank M. Sánchez-Prieto, C. González Páramos, and the core facilities of the Instituto de Investigaciones Biomédicas for technical help and O. Fernandez-Capetillo for advice.
This work was supported by grants from Ministerio de Economía y Competitividad (BFU2011-28958 to A. Aranda and SAF2009-11150 to A. Pascual), from the Instituto de Salud Carlos III (RD012/0036/0030 to A. Aranda; and PI 07/0167 and PI 10/0703 to R. Garesse), from the Comunidad de Madrid (S2011/BMD-2328 TIRONET to A. Aranda), and European Union grant project CRESCENDO (FP6-018652 to A. Aranda and L.M. Sachs).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used in this paper:
- 8-OH-dG
- 8′-hydroxy-2′-deoxyguanosine
- ChIP
- chromatin immunoprecipitation
- Cr
- cell respiration
- DDR
- DNA damage response
- DSB
- double-strand break
- dUTP
- deoxy-UTP
- KO
- knockout
- MEF
- mouse embryonic fibroblast
- NAC
- N-acetyl-l-cysteine
- OIS
- oncogenic H-RasV12–induced senescence
- PDL
- population doubling level
- ROS
- reactive oxygen species
- ROX
- residual oxygen consumption
- SA-βgal
- senescence-associated β-galactosidase
- TdT
- terminal deoxynucleotidyl transferase
- TH
- thyroid hormone
- THR
- TH receptor
References
- Abraham R.T. 2002. Checkpoint signalling: focusing on 53BP1. Nat. Cell Biol. 4:E277–E279 10.1038/ncb1202-e277 [DOI] [PubMed] [Google Scholar]
- Alexander A., Cai S.L., Kim J., Nanez A., Sahin M., MacLean K.H., Inoki K., Guan K.L., Shen J., Person M.D., et al. 2010. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc. Natl. Acad. Sci. USA. 107:4153–4158 10.1073/pnas.0913860107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda A., Pascual A. 2001. Nuclear hormone receptors and gene expression. Physiol. Rev. 81:1269–1304 [DOI] [PubMed] [Google Scholar]
- Aranda A., Martínez-Iglesias O., Ruiz-Llorente L., García-Carpizo V., Zambrano A. 2009. Thyroid receptor: roles in cancer. Trends Endocrinol. Metab. 20:318–324 10.1016/j.tem.2009.03.011 [DOI] [PubMed] [Google Scholar]
- Atzmon G., Barzilai N., Hollowell J.G., Surks M.I., Gabriely I. 2009. Extreme longevity is associated with increased serum thyrotropin. J. Clin. Endocrinol. Metab. 94:1251–1254 10.1210/jc.2008-2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D.J., Wijshake T., Tchkonia T., LeBrasseur N.K., Childs B.G., van de Sluis B., Kirkland J.L., van Deursen J.M. 2011. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 479:232–236 10.1038/nature10600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J., Rezaei N., Liontos M., Karakaidos P., Kletsas D., Issaeva N., Vassiliou L.V., Kolettas E., Niforou K., Zoumpourlis V.C., et al. 2006. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 444:633–637 10.1038/nature05268 [DOI] [PubMed] [Google Scholar]
- Baxter J.D., Webb P., Grover G., Scanlan T.S. 2004. Selective activation of thyroid hormone signaling pathways by GC-1: a new approach to controlling cholesterol and body weight. Trends Endocrinol. Metab. 15:154–157 10.1016/j.tem.2004.03.008 [DOI] [PubMed] [Google Scholar]
- Bergeron R., Ren J.M., Cadman K.S., Moore I.K., Perret P., Pypaert M., Young L.H., Semenkovich C.F., Shulman G.I. 2001. Chronic activation of AMP kinase results in NRF-1 activation and mitochondrial biogenesis. Am. J. Physiol. Endocrinol. Metab. 281:E1340–E1346 [DOI] [PubMed] [Google Scholar]
- Buffenstein R., Pinto M. 2009. Endocrine function in naturally long-living small mammals. Mol. Cell. Endocrinol. 299:101–111 10.1016/j.mce.2008.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callén E., Jankovic M., Wong N., Zha S., Chen H.T., Difilippantonio S., Di Virgilio M., Heidkamp G., Alt F.W., Nussenzweig A., Nussenzweig M. 2009. Essential role for DNA-PKcs in DNA double-strand break repair and apoptosis in ATM-deficient lymphocytes. Mol. Cell. 34:285–297 10.1016/j.molcel.2009.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callén E., Di Virgilio M., Kruhlak M.J., Nieto-Soler M., Wong N., Chen H.T., Faryabi R.B., Polato F., Santos M., Starnes L.M., et al. 2013. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell. 153:1266–1280 10.1016/j.cell.2013.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J., d’Adda di Fagagna F. 2007. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 8:729–740 10.1038/nrm2233 [DOI] [PubMed] [Google Scholar]
- Chance B., Sies H., Boveris A. 1979. Hydroperoxide metabolism in mammalian organs. Physiol. Rev. 59:527–605 [DOI] [PubMed] [Google Scholar]
- Collado M., Serrano M. 2006. The power and the promise of oncogene-induced senescence markers. Nat. Rev. Cancer. 6:472–476 10.1038/nrc1884 [DOI] [PubMed] [Google Scholar]
- Collado M., Serrano M. 2010. Senescence in tumours: evidence from mice and humans. Nat. Rev. Cancer. 10:51–57 10.1038/nrc2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Adda di Fagagna F., Reaper P.M., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P. 2003. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 426:194–198 10.1038/nature02118 [DOI] [PubMed] [Google Scholar]
- Di Micco R., Fumagalli M., Cicalese A., Piccinin S., Gasparini P., Luise C., Schurra C., Garre’ M., Nuciforo P.G., Bensimon A., et al. 2006. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 444:638–642 10.1038/nature05327 [DOI] [PubMed] [Google Scholar]
- Dimri G.P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E.E., Linskens M., Rubelj I., Pereira-Smith O., et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA. 92:9363–9367 10.1073/pnas.92.20.9363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano-Díaz C., Orthwein A., Fradet-Turcotte A., Xing M., Young J.T., Tkáč J., Cook M.A., Rosebrock A.P., Munro M., Canny M.D., et al. 2013. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell. 49:872–883 10.1016/j.molcel.2013.01.001 [DOI] [PubMed] [Google Scholar]
- Fu X., Wan S., Lyu Y.L., Liu L.F., Qi H. 2008. Etoposide induces ATM-dependent mitochondrial biogenesis through AMPK activation. PLoS ONE. 3:e2009 10.1371/journal.pone.0002009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Silva S., Aranda A. 2004. The thyroid hormone receptor is a suppressor of ras-mediated transcription, proliferation, and transformation. Mol. Cell. Biol. 24:7514–7523 10.1128/MCB.24.17.7514-7523.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Silva S., Martínez-Iglesias O., Ruiz-Llorente L., Aranda A. 2011. Thyroid hormone receptor β1 domains responsible for the antagonism with the ras oncogene: role of corepressors. Oncogene. 30:854–864 10.1038/onc.2010.464 [DOI] [PubMed] [Google Scholar]
- Garinis G.A., Uittenboogaard L.M., Stachelscheid H., Fousteri M., van Ijcken W., Breit T.M., van Steeg H., Mullenders L.H., van der Horst G.T., Brüning J.C., et al. 2009. Persistent transcription-blocking DNA lesions trigger somatic growth attenuation associated with longevity. Nat. Cell Biol. 11:604–615 10.1038/ncb1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesing A., Lewiński A., Karbownik-Lewińska M. 2012. The thyroid gland and the process of aging; what is new? Thyroid Res. 5:16 10.1186/1756-6614-5-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gugneja S., Scarpulla R.C. 1997. Serine phosphorylation within a concise amino-terminal domain in nuclear respiratory factor 1 enhances DNA binding. J. Biol. Chem. 272:18732–18739 10.1074/jbc.272.30.18732 [DOI] [PubMed] [Google Scholar]
- Habershon S.O. 1874. GUY’S HOSPITAL: Exophthalmic goiter; heart disease; jaundice; death. Lancet. 1:510–512 10.1016/S0140-6736(02)45227-0 [DOI] [Google Scholar]
- Harper J.W., Elledge S.J. 2007. The DNA damage response: ten years after. Mol. Cell. 28:739–745 10.1016/j.molcel.2007.11.015 [DOI] [PubMed] [Google Scholar]
- Harvey M., Sands A.T., Weiss R.S., Hegi M.E., Wiseman R.W., Pantazis P., Giovanella B.C., Tainsky M.A., Bradley A., Donehower L.A. 1993. In vitro growth characteristics of embryo fibroblasts isolated from p53-deficient mice. Oncogene. 8:2457–2467 [PubMed] [Google Scholar]
- Hayflick L. 1965. The limited in vitro lifetime of human diploid cell strains. Exp. Cell Res. 37:614–636 10.1016/0014-4827(65)90211-9 [DOI] [PubMed] [Google Scholar]
- Hütter E., Unterluggauer H., Garedew A., Jansen-Dürr P., Gnaiger E. 2006. High-resolution respirometry—a modern tool in aging research. Exp. Gerontol. 41:103–109 10.1016/j.exger.2005.09.011 [DOI] [PubMed] [Google Scholar]
- Kim W.G., Cheng S.Y. 2013. Thyroid hormone receptors and cancer. Biochim. Biophys. Acta. 1830:3928–3936 10.1016/j.bbagen.2012.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson N.G., Wang J., Wilhelmsson H., Oldfors A., Rustin P., Lewandoski M., Barsh G.S., Clayton D.A. 1998. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 18:231–236 10.1038/ng0398-231 [DOI] [PubMed] [Google Scholar]
- Lee J.H., Paull T.T. 2005. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 308:551–554 10.1126/science.1108297 [DOI] [PubMed] [Google Scholar]
- Leo M., Marcocci C., Pinchera A., Nardi M., Megna L., Rocchi R., Latrofa F., Altea M.A., Mazzi B., Sisti E., et al. 2012. Outcome of Graves’ orbitopathy after total thyroid ablation and glucocorticoid treatment: follow-up of a randomized clinical trial. J. Clin. Endocrinol. Metab. 97:E44–E48 10.1210/jc.2011-2077 [DOI] [PubMed] [Google Scholar]
- López M., Varela L., Vázquez M.J., Rodríguez-Cuenca S., González C.R., Velagapudi V.R., Morgan D.A., Schoenmakers E., Agassandian K., Lage R., et al. 2010. Hypothalamic AMPK and fatty acid metabolism mediate thyroid regulation of energy balance. Nat. Med. 16:1001–1008 10.1038/nm.2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnus-Levy A. 1895. Über den respiratorischen gaswechsel unter dem einfluss der thyroidea sowie unter verschiedenen pathologischen zuständen. Berl. Klin. Wochenschr. 32:650–652 [Google Scholar]
- Malik R., Hodgson H. 2002. The relationship between the thyroid gland and the liver. QJM. 95:559–569 10.1093/qjmed/95.9.559 [DOI] [PubMed] [Google Scholar]
- Mookerjee S.A., Divakaruni A.S., Jastroch M., Brand M.D. 2010. Mitochondrial uncoupling and lifespan. Mech. Ageing Dev. 131:463–472 10.1016/j.mad.2010.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay P., Rajesh M., Yoshihiro K., Haskó G., Pacher P. 2007. Simple quantitative detection of mitochondrial superoxide production in live cells. Biochem. Biophys. Res. Commun. 358:203–208 10.1016/j.bbrc.2007.04.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer L.J., Garinis G.A., Raams A., Lalai A.S., Robinson A.R., Appeldoorn E., Odijk H., Oostendorp R., Ahmad A., van Leeuwen W., et al. 2006. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 444:1038–1043 10.1038/nature05456 [DOI] [PubMed] [Google Scholar]
- Okuno Y., Nakamura-Ishizu A., Otsu K., Suda T., Kubota Y. 2012. Pathological neoangiogenesis depends on oxidative stress regulation by ATM. Nat. Med. 18:1208–1216 10.1038/nm.2846 [DOI] [PubMed] [Google Scholar]
- Ooka H., Shinkai T. 1986. Effects of chronic hyperthyroidism on the lifespan of the rat. Mech. Ageing Dev. 33:275–282 10.1016/0047-6374(86)90052-7 [DOI] [PubMed] [Google Scholar]
- Oppenheimer J.H., Schwartz H.L., Mariash C.N., Kinlaw W.B., Wong N.C., Freake H.C. 1987. Advances in our understanding of thyroid hormone action at the cellular level. Endocr. Rev. 8:288–308 10.1210/edrv-8-3-288 [DOI] [PubMed] [Google Scholar]
- Pascual A., Aranda A. 2013. Thyroid hormone receptors, cell growth and differentiation. Biochim. Biophys. Acta. 1830:3908–3916 10.1016/j.bbagen.2012.03.012 [DOI] [PubMed] [Google Scholar]
- Rodier F., Coppé J.P., Patil C.K., Hoeijmakers W.A., Muñoz D.P., Raza S.R., Freund A., Campeau E., Davalos A.R., Campisi J. 2009. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 11:973–979 10.1038/ncb1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou E.P., Boon C., Redon C., Bonner W.M. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905–916 10.1083/jcb.146.5.905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozing M.P., Houwing-Duistermaat J.J., Slagboom P.E., Beekman M., Frölich M., de Craen A.J., Westendorp R.G., van Heemst D. 2010. Familial longevity is associated with decreased thyroid function. J. Clin. Endocrinol. Metab. 95:4979–4984 10.1210/jc.2010-0875 [DOI] [PubMed] [Google Scholar]
- Scarpulla R.C. 2002. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim. Biophys. Acta. 1576:1–14 10.1016/S0167-4781(02)00343-3 [DOI] [PubMed] [Google Scholar]
- Schultz L.B., Chehab N.H., Malikzay A., Halazonetis T.D. 2000. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 151:1381–1390 10.1083/jcb.151.7.1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M., Lee H., Chin L., Cordon-Cardo C., Beach D., DePinho R.A. 1996. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 85:27–37 10.1016/S0092-8674(00)81079-X [DOI] [PubMed] [Google Scholar]
- Serrano M., Lin A.W., McCurrach M.E., Beach D., Lowe S.W. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 88:593–602 10.1016/S0092-8674(00)81902-9 [DOI] [PubMed] [Google Scholar]
- Sun Y., Connors K.E., Yang D.Q. 2007. AICAR induces phosphorylation of AMPK in an ATM-dependent, LKB1-independent manner. Mol. Cell. Biochem. 306:239–245 10.1007/s11010-007-9575-6 [DOI] [PubMed] [Google Scholar]
- Suzuki A., Kusakai G., Kishimoto A., Shimojo Y., Ogura T., Lavin M.F., Esumi H. 2004. IGF-1 phosphorylates AMPK-alpha subunit in ATM-dependent and LKB1-independent manner. Biochem. Biophys. Res. Commun. 324:986–992 10.1016/j.bbrc.2004.09.145 [DOI] [PubMed] [Google Scholar]
- Tata J.R., Ernster L., Lindberg O. 1962. Control of basal metabolic rate by thyroid hormones and cellular function. Nature. 193:1058–1060 10.1038/1931058a0 [DOI] [PubMed] [Google Scholar]
- Todaro G.J., Green H. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17:299–313 10.1083/jcb.17.2.299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay G., Singh R., Kumar A., Kumar S., Kapoor A., Godbole M.M. 2004. Severe hyperthyroidism induces mitochondria-mediated apoptosis in rat liver. Hepatology. 39:1120–1130 10.1002/hep.20085 [DOI] [PubMed] [Google Scholar]
- van der Pluijm I., Garinis G.A., Brandt R.M., Gorgels T.G., Wijnhoven S.W., Diderich K.E., de Wit J., Mitchell J.R., van Oostrom C., Beems R., et al. 2007. Impaired genome maintenance suppresses the growth hormone—insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLoS Biol. 5:e2 10.1371/journal.pbio.0050002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Videla L.A. 2010. Hormetic responses of thyroid hormone calorigenesis in the liver: Association with oxidative stress. IUBMB Life. 62:460–466 [DOI] [PubMed] [Google Scholar]
- Vijg J., Campisi J. 2008. Puzzles, promises and a cure for ageing. Nature. 454:1065–1071 10.1038/nature07216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virbasius J.V., Scarpulla R.C. 1994. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc. Natl. Acad. Sci. USA. 91:1309–1313 10.1073/pnas.91.4.1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward I.M., Minn K., Jorda K.G., Chen J. 2003. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J. Biol. Chem. 278:19579–19582 10.1074/jbc.C300117200 [DOI] [PubMed] [Google Scholar]
- Wyman C., Kanaar R. 2006. DNA double-strand break repair: all’s well that ends well. Annu. Rev. Genet. 40:363–383 10.1146/annurev.genet.40.110405.090451 [DOI] [PubMed] [Google Scholar]
- Yamauchi M., Kambe F., Cao X., Lu X., Kozaki Y., Oiso Y., Seo H. 2008. Thyroid hormone activates adenosine 5′-monophosphate-activated protein kinase via intracellular calcium mobilization and activation of calcium/calmodulin-dependent protein kinase kinase-beta. Mol. Endocrinol. 22:893–903 10.1210/me.2007-0249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavacki A.M., Ying H., Christoffolete M.A., Aerts G., So E., Harney J.W., Cheng S.Y., Larsen P.R., Bianco A.C. 2005. Type 1 iodothyronine deiodinase is a sensitive marker of peripheral thyroid status in the mouse. Endocrinology. 146:1568–1575 10.1210/en.2004-1392 [DOI] [PubMed] [Google Scholar]
- Zimmermann M., Lottersberger F., Buonomo S.B., Sfeir A., de Lange T. 2013. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science. 339:700–704 10.1126/science.1231573 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.