Abstract
The progenitor zones of the embryonic mouse ventral telencephalon give rise to GABAergic and cholinergic neurons. We have shown previously that two LIM-homeodomain (LIM-HD) transcription factors, Lhx6 and Lhx8, that are downstream of Nkx2.1, are critical for the development of telencephalic GABAergic and cholinergic neurons. Here we investigate the role of Ldb1, a nuclear protein that binds directly to all LIM-HD factors, in the development of these ventral telencephalon derived neurons. We show that Ldb1 is expressed in the Nkx2.1 cell lineage during embryonic development and in mature neurons. Conditional deletion of Ldb1 causes defects in the expression of a series of genes in the ventral telencephalon and severe impairment in the tangential migration of cortical interneurons from the ventral telencephalon. Similar to the phenotypes observed in Lhx6 or Lhx8 mutant mice, the Ldb1 conditional mutants show a reduction in the number of both GABAergic and cholinergic neurons in the telencephalon. Furthermore, our analysis reveals defects in the development of the parvalbumin-positive neurons in the globus pallidus and striatum of the Ldb1 mutants. These results provide evidence that Ldb1 plays an essential role as a transcription co-regulator of Lhx6 and Lhx8 in the control of mammalian telencephalon development.
Keywords: Differentiation, Forebrain development, Interneuron, Mouse, Tangential migration
Introduction
Functioning of the mammalian telencephalon is dependent on the formation of complex circuitry by different types of neurons. Many of these neurons are derived from distinct progenitor domains in the ventral telencephalon during embryonic development. Largely through mouse mutant analysis, the molecular genetic program controlling the generation of these neurons has begun to be unveiled.
Nkx2.1 is a transcription factor expressed in the medial ganglionic eminence (MGE) and preoptic area (POA) of the developing telencephalon. Loss-of-function studies have revealed that Nkx2.1 is essential for both patterning of the MGE/POA and the specification/differentiation of a variety of MGE/POA-derived neurons, including GABAergic interneurons, GABAergic projection neurons, and cholinergic neurons (Sussel et al., 1999, Marin et al., 2000, Butt et al., 2008, Du et al., 2008, Flandin et al., 2011).
The function of Nkx2.1 in the developing telencephalon is mediated by controlling the expression of two closely related LIM-homeodomain (LIM-HD) genes, Lhx6 and Lhx8. Deletion of Nkx2.1 impairs the Lhx6 and Lhx8 expression in the ventral telencephalon (Sussel et al., 1999, Marin et al., 2000). Exogenous expression of Lhx6 restores the ability for the generation of cortical interneurons in Nkx2.1−/− MGE cells. Moreover, RNAi knockdown of Lhx6 blocks the rescue of the interneuron phenotype in the Nkx2.1−/− cells by exogenous Nkx2.1 expression (Du et al., 2008). Nkx2.1 has also been shown to bind directly to a promoter region of the Lhx6 and to activate reporter gene expression (Du et al., 2008).
Roles of Lhx6 and Lhx8 in telencephalon development have been extensively analyzed. Deletion of Lhx6 severely impairs tangential migration and specification of telencephalic GABAergic interneurons (Liodis et al., 2007, Zhao et al., 2008, Neves et al., 2012). Inactivation of Lhx8 causes defects in telencephalic cholinergic neuron development (Zhao et al., 2003, Mori et al., 2004, Fragkouli et al., 2005, Fragkouli et al., 2009, Lopes et al., 2012). Analysis of double mutants further revealed that Lhx6/Lhx8 are required to directly control Sonic hedgehog (Shh) gene expression in the MGE and to regulate the development of the globus pallidus (Flandin et al., 2011).
It has been suggested that LIM-HD proteins regulate gene transcription by forming multi-protein complexes through binding to other nuclear factors (Hobert and Westphal, 2000). Ldb1 (LIM-domain-binding protein 1) is one of such factors (Agulnick et al., 1996, Jurata et al., 1996, Bach et al., 1997). Previous mouse mutant studies have shown that Ldb1 plays important roles in controlling multiple aspects of embryogenesis ranging from early embryo patterning to limb, pancreas, and spinal cord development (Thaler et al., 2002, Mukhopadhyay et al., 2003, Tzchori et al., 2009, Hunter et al., 2013). To determine the role of Ldb1 in development of the Nkx2.1-lineage derived neurons in the telencephalon, we generated a Ldb1 conditional mutant by crossing a Ldb1 floxed mouse line (Zhao et al., 2007) with a BAC-transgenic line expressing the Cre recombinase under control of enhancer elements of the Nkx2.1 (Xu et al., 2008). Our studies show that Ldb1 is essential for the development of multiple types of neurons in the telencephalon.
Materials and Methods
Animals
Animals were maintained and handled by following the National Institutes of Health guidelines and procedures approved by the animal care and use committee of the National Institute of Child Health and Human Development.
To specifically inactivate Ldb1 function in the cells of the Nkx2.1-lineage, mice carrying one null allele of the Ldb1 gene (Ldb1+/−) (Mukhopadhyay et al., 2003) were crossed to a BAC-transgenic mouse line that expresses the Cre recombinase under control of regulatory elements of the Nkx2.1 gene (Nkx2.1-Cre+) (Xu et al., 2008). Offspring double heterozygous for the Ldb1 and the Nkx2.1-Cre alleles (Ldb1+/−; Nkx2.1-Cre+) were mated with homozygous Ldb1 floxed (Ldb1f/f) mice (Zhao et al., 2007) to generate Ldb1/Nkx2.1-Cre conditional mutant (Ldb1f/−; Nkx2.1-Cre+) and control (Ldb1f/+; Nkx2.1-Cre+, Ldb1f/+; Nkx2.1-Cre−, or Ldb1f/−; Nkx2.1-Cre−) animals for analysis. For the experiments that required genetic marking of the Nkx2.1-lineage cells, the Ldb1+/−; Nkx2.1-Cre+ mice were crossed to the Rosa26-Yellow Fluorescent Protein (YFP) reporter (R26R-YFP) line (R26+/LoxP-stop-LoxP-YFP) (Srinivas et al., 2001) for two generations to generate offspring that were heterozygous for the Ldb1-null and Nkx2.1-Cre alleles, and homozygous for the YFP reporter allele (Ldb1+/−; Nkx2.1-Cre+; R26LoxP-stop-LoxP-YFP/LoxP-stop-LoxP-YFP). These mice were mated with homozygous Ldb1 floxed (Ldb1f/f) mice to produce Ldb1 conditional mutant (Ldb1f/−; Nkx2.1-Cre+; R26+/LoxP-stop-LoxP-YFP) and control (Ldb1f/−; Nkx2.1-Cre+; R26+/LoxP-stop-LoxP-YFP) animals for analysis. The Ldb1-null, Ldb1-floxed, and the Nkx2.1-Cre+ alleles were maintained in a C57/BL6 background, whereas the R26-YFP reporter allele was in a CD-1 background. Genotyping of the various alleles was performed by PCR as previously described in the reference for each of these alleles.
Tissue preparation
To obtain embryos of specific developmental stages, female mice were checked daily for the presence of vaginal plug after mating. Noon of the day when a plug was detected was considered embryonic day (E) 0.5. Embryo heads (E12.5 to E14.5) or brains (E18.5) were fixed by immersion in 4% paraformaldehyde (PFA) in 0.1 M phosphate buffer (PB; pH 7.4) overnight at 4°C. After wash in phosphate-buffered saline (PBS; pH 7.4), the tissue was dehydrated through a series of ethanol solutions with ascending concentrations and embedded in paraffin, or cryo-protected in sucrose/PBS (pH 7.4) (15%, and then 30%) solutions, embedded in Tissue-Tek OCT compound, and frozen on dry ice.
Postnatal mice were fixed by transcardial perfusion with 4% PFA/0.1 M PB (pH 7.4). The brains were dissected and post-fixed in PFA overnight at 4°C. The brains were washed in PBS (pH 7.4), cryo-protected in sucrose (15%, and then 30%)/PBS solutions, embedded in OCT compound, and frozen on dry ice.
Immunohistochemistry
The primary antibodies and their dilutions used are as follows: rabbit anti-Ldb1 (a gift from Drs. Liqi Li and Paul Love, NICHD, Li et al., 2011, 1:2000), chick anti-Green Fluorescent Protein (GFP) (Aves Labs, GFP-1020, also reactive to YFP, 1:500), goat anti-Choline Acetyltransferase (ChAT) (Millipore, AB144, 1:250), rat anti-Somatostatin (SOM) (Millipore, MAB354, 1:150), mouse anti-Parvalbumin (PV) (Sigma, P3088, 1:2000).
Paraffin (5 µm thick) or frozen (16 µm) sections of the embryo heads (E12.5 and E14.5) or brains (E18.5) were cut and mounted onto silanized (KD Medical) or Superfrost Plus (Fisher) microscope slides. For immunofluorescent staining of frozen sections using antibodies from a non-mouse species, the sections were blocked in PBS containing 2% bovine serum albumin (BSA, Vector Laboratory) and 5% normal serum from the same species where the secondary antibody was derived. The sections were incubated overnight at 4°C in the primary antibodies diluted with PBS containing 2% of the normal serum. After washes in PBS, sections were incubated with secondary antibodies labeled with Alexa-488 or Alexa-568 (Invitrogen). The sections were washed in PBS and coverslipped with a ProLong Gold antifade reagent (Invitrogen). Staining using mouse primary antibodies was carried out with a M.O.M kit (Vector Laboratory) by following the manufacturer’s instruction. For immunofluorescent staining of paraffin sections, the sections were deparaffinized in xylene, and rehydrated through a series of ethanol solutions with descending concentrations. Sections were treated with an antigen retrieval solution (Buffer A, Electron Microscopy Sciences) using a PickCell 2100 retriever (Electron Microscopy Sciences), and then processed by following the same procedure used for the staining of frozen sections. For immunoperoxidase staining, paraffin sections were processed in procedures similar to those used for immunofluorescent staining with an additional 15-minute incubation in a hydrogen peroxide solution (0.3% in methanol) before rehydration through the ethanol. After primary antibody incubation, staining was detected by using an ABC elite kit (Vector Laboratory) and AEC solution (Millipore) as a peroxidase substrate.
Frozen sections of postnatal mouse brains were cut at a thickness of 40 µm on a cryostat and kept free-floating at 4°C in PBS. Staining of the sections was carried out in 6-well plates by procedures similar to those described above except for lengthening the primary antibody incubation to 40 hours and for adding Triton X-100 to PBS (at a concentration of 0.4%) for washing and primary antibody incubation.
Cell counting
To count immunostained neurons, comparable coronal sections (40 µm thick) of the striatum (anterior to the crossover of the anterior commissure), septum, nucleus Basalis, the anterior hippocampus, and the cortex (anterior to the crossover of the anterior commissure) from both control and Ldb1/Nkx2.1-Cre mutants were photographed under a 4x objective on a Nikon (E1000) microscope. Labeled cells within the entire regions of the striatum, septum, nucleus Basalis, and hippocampus shown on the photos were counted manually. Labeled cells in the cortex were counted only in a medial area between the midline and 1 mm lateral to the midline.
In situ hybridization
In situ RNA hybridization experiments were performed using digoxigenin labeled riboprobes on 20 µm frozen sections as previously described (Flandin et al., 2011). All riboprobes used were previously described (Flandin et al., 2011).
Electrophoretic mobility shift assay (EMSA)
EMSA was performed using the kit from Pierce. Briefly, each reaction (20µl) consisted of 2 µg nuclear extract and 1 fmole/µl of biotinylated probes, in binding buffer consisting of: 10 mM Tris pH 7.5, 50 mM KCl, 1 mM DTT, 5% glycerol, 1 mM EDTA, 50 ng/µl poly (dI-dC) (Sigma) and 50 ng/µl BSA (New England Biolabs).
LHX6, LHX8 and LDB1 proteins were generated by Fugene6 transfection of HEK 293 cells. After 48 hours, nuclear extracts were prepared using the Pierce nuclear extract kit. Biotinylated DNA probes: Probe A corresponded to the 26–64 bp of the SBE3 Shh enhancer and included LHX site A (Flandin et al., 2011); mutated probe A had the same nucleotide sequence as the wild type probe A, but the LHX site core sequence (TAATCA) was changed to TTTTTT.
Results
Expression of Ldb1 in Nkx2.1-lineage cells derived from the ventral telencephalon
To determine whether Ldb1 protein is expressed in the cells derived from the Nkx2.1-lineage in the developing telencephalon, we genetically marked these cells in the mouse embryos that carried both the Nkx2.1-Cre transgene and the R26R-YFP reporter allele; cells expressing Nkx2.1-Cre become YFP+ (Xu et al., 2008). Immunostaining revealed that at E12.5 most cells in the MGE and the POA expressed YFP (Figs. 1B, C, and data not shown). Scattered YFP+ cells were also detected within the LGE (Fig. 1D); many of these cells are likely to be MGE-derived striatal interneurons (Marin et al., 2000; Nóbrega-Pereira et al., 2008). Two streams of YFP+ cells were found in the cortical interneuron tangential migrating routes (Fig. 1D; Marin and Rubenstein 2001).
Fig.1.
Expression of Ldb1 in Nkx2.1-lineage cells in the developing mouse telencephalon. Immunofluorescent staining of coronal sections from Nkx2.1-Cre+; R26+/flox-stop-floxYFP embryos shows that Ldb1 is widely expressed in the ventral telencephalon at E12.5 (A, C), including in the Nkx2.1-lineage cells in the medial ganglionic eminence (MGE) labeled by staining of YFP (B, C). At this stage, Ldb1 is also detected in Nkx2.1-lineage (YFP+) cells that have entered the lateral ganglionic eminence (LGE) or on their way migrate to the cortex (pointed by arrowheads in D). At E18.5, Ldb1 is detected in Nkx2.1-lineage (YFP+) cells in various regions of the telencephalon including the septum (Se) (E), globus pallidus (GP) (F), cortex (Ctx) (G), and hippocampus (Hip) (H). Bar in H represents 100 µm for all panels.
Ldb1 protein was detected extensively in the developing telencephalon, including in many Nkx2.1-lineage (YFP+) cells in the MGE and LGE (Figs. 1A, C, D). By E18.5, a large number of migrating YFP+ cells had reached the cortex and hippocampus, where they continued to express Ldb1 (Figs. 1G, H). In addition, Ldb1 was detected in Nkx2.1-lineage (YFP+) cells in the ventral telencephalon, including in the septum (Fig. 1E), the striatum (not shown), and the globus pallidus (Fig. 1F).
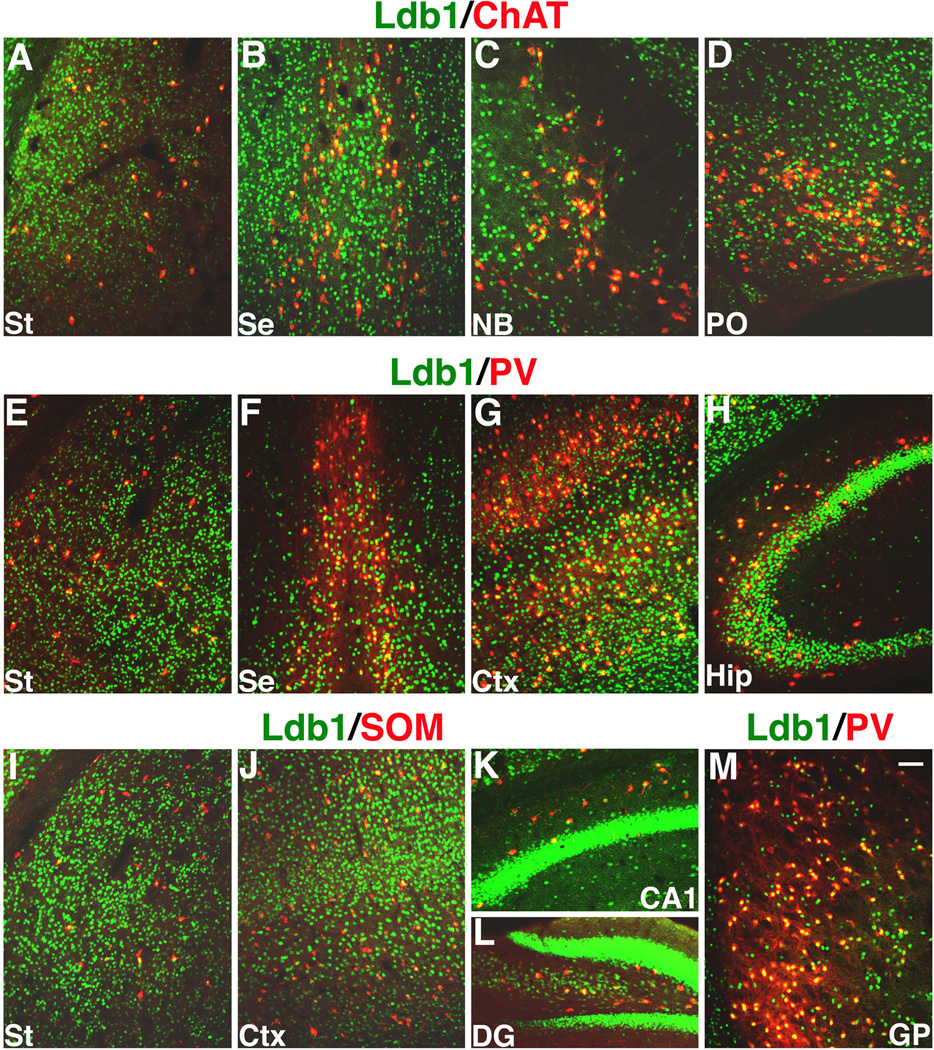
Since the Nkx2.1-lineage cells contribute to a variety of differentiated neuronal cell types in different regions of the telencephalon, we examined whether Ldb1 protein was expressed in these neurons in postnatal (P18) animals. Double immunofluorescent staining of Ldb1 and ChAT showed that Ldb1 was expressed in cholinergic interneurons in the striatum (Fig. 2A), and in cholinergic projection neurons in the septum, nucleus Basalis, and the magnocellular preoptic nucleus (Figs. 2B, C, D).
Fig. 2.
Expression of Ldb1 in subtypes of neurons derived from the Nkx2.1-lineage in the postnatal (P18) mouse telencephalon. Immunofluorescent staining shows that Ldb1 is expressed in cholinergic (A–D), parvalbumin+ (E–H, M), and somatostatin+ (I–L) neurons in the various regions of the telencephalon including the striatum (St) (A, E, I), septum (Se) (B, F), globus pallidus (GP) (M), nucleus Basalis (NB) (C), preoptic area (PO) (D), cortex (Ctx) (G, J), hippocampus (Hip) (H, K), and dentate gyrus (DG) (L). Bar in M represent 100 µm for all panels.
Double staining of Ldb1/PV and Ldb1/SOM revealed that Ldb1 was also expressed in PV+ (Figs. 2E, F, G, H, M) and SOM+ (Figs. 2I, J, K, L) subtypes of GABAergic interneurons, respectively, throughout the telencephalon, including the striatum, septum, globus pallidus, neocortex and hippocampus. Thus, Ldb1 was expressed in the Nkx2.1-lineage cells in the telencephalon early in embryonic development through P18.
Deletion of Ldb1 in the ventral telencephalon of the Ldb1/Nkx2.1-Cre conditional mutant
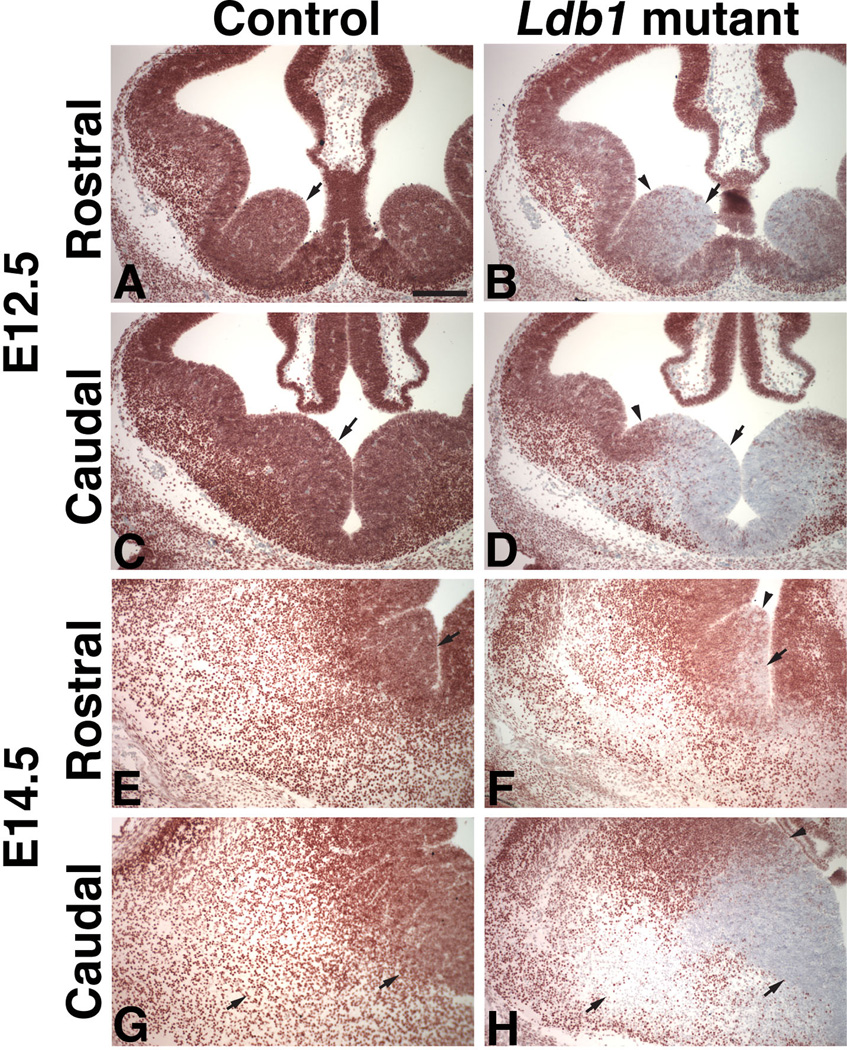
Homozygous Ldb1-null mutants die around E9.5 with severe developmental defects in the head and heart (Mukhopadhyay et al., 2003). To determine the function of Ldb1 in the development of the Nkx2.1-lineage derived cells in the telencephalon, we generated a conditional mutant (Ldb1f/−; Nkx2.1-Cre+) with a specific deletion of the Ldb1 in these cells by crossing the Ldb1 floxed mice (Ldb1f/f, Zhao et al., 2007) with mice that carry one Ldb1 null allele and an Nkx2.1-Cre transgene (Ldb1+/−; Nkx2.1-Cre+). Consistent with the result that Ldb1 was expressed in Nkx2.1-lineage cells (Fig. 1), immunostaining of Ldb1 in both E12.5 and E14.5 embryos confirmed that Ldb1 was missing in most of the cells in the MGE and the underlying POA of the Ldb1/Nkx2.1-Cre mutants (Figs. 3A–H). However, at both E12.5 and E14.5, some Ldb1+ cells were still present in the mutants, especially in the dorsal and the rostral regions of the MGE (Figs. 3B, D, F, H). The presence of these Ldb1+ cells was probably due to a previously reported lack of expression of the Cre recombinase in the dorsal and rostral regions of the MGE of the Nkx2.1-Cre transgenic mouse (Xu et al., 2008).
Fig. 3.
Conditional deletion of Ldb1 in the ventral telencephalon. Coronal sections through the telencephalon of E12.5 (A–D) and E14.5 (E–H) mouse embryos were immunostained with an antibody to Ldb1. A–B and E–F show sections from a rostral level. C–D and G–H show sections from a caudal level. Reduction of Ldb1 staining was observed in the MGE of Ldb1f/−; Nkx2.1-Cre+ conditional mutant (B, D, F, H) as compared to the control (A, C, E, G). Arrows in A–H point at regions of the MGE where Ldb1 was detected in the control (A, C, E, G), but was missing in the mutant (B, D, F, H). Arrowheads in B, D, F, H point at the dorsal region of the MGE in the mutant where Ldb1 was still detected. Bar in A represents 750 µm for all panels.
Ldb1 is required for Shh expression and in vitro binding of Lhx6 and Lhx8 to the Shh enhancer in MGE neurons
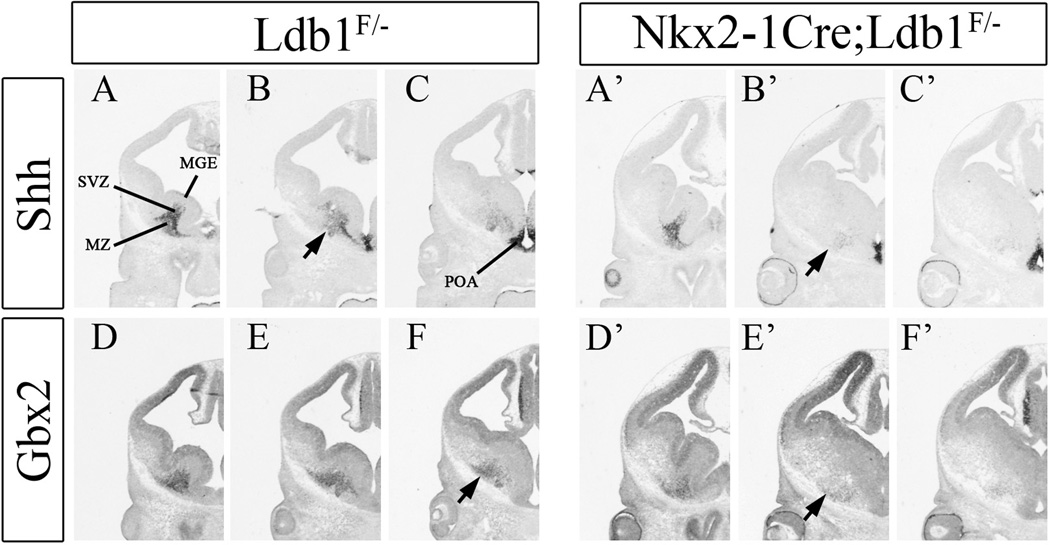
Lhx6−/−/Lhx8−/− double mutants fail to express Shh in early born neurons of the MGE (Flandin et al., 2011). Thus, we began our molecular analysis of the conditional Ldb1 mutant by studying Shh RNA expression in the ventral telencephalon. At E12.5, while Shh expression in the ventricular zone (VZ) of the POA and ventral MGE is preserved, Shh expression in MGE mantle zone (MZ) is greatly reduced, except in the rostral MGE (Figs. 4A–C, A’–C’).
Fig. 4.
Reduction of Shh (A–C, A’–C’) and Gbx2 (D–F, D’–F’) expression in conditional Ldb1 mutants, as determined by in situ hybridization at E12.5. Coronal telencephalic hemisections compare gene expression between control (Ldb1+/−; left) and mutant (Nkx2-1-Cre+; Ldb1f/−; right). Three planes of section are shown, with rostral at the left and caudal at the right. Arrows show reduced Shh and Gbx2 expression in the mantle zone of the MGE. Abbreviations: MGE: medial ganglionic eminence, MZ: mantle zone, POA: preoptic area, SVZ: subventricular zone.
Previously, we demonstrated that Lhx6 and Lhx8 can compensate for each other in the regulation of Shh expression in the MGE MZ, and that both proteins can bind to the Shh enhancer (SBE3 domain) in electrophoretic mobility shift assays (Flandin et al., 2011). In those assays, we included Ldb1 protein. Here we tested whether Ldb1 was required for the binding of Lhx6 and Lhx8 to the SBE3 Shh enhancer. We found that when Ldb1 was not added to nuclear extracts, Lhx6 and Lhx8 failed to bind to the SBE3 Shh enhancer (Fig. 5). Furthermore, mutating the core Lhx6/Lhx8 binding site of SBE3 Shh enhancer resulted in a strong reduction of the amount of the complex. Thus, this provides in vitro biochemical evidence that Ldb1 is required to enable Lhx6 and Lhx8 to bind the SBE3 enhancer. Lack of binding may underlie the reduced Shh expression in the MGE MZ of the Ldb1 conditional mutant (Figs. 4A–C, 4A’–C’).
Fig. 5.
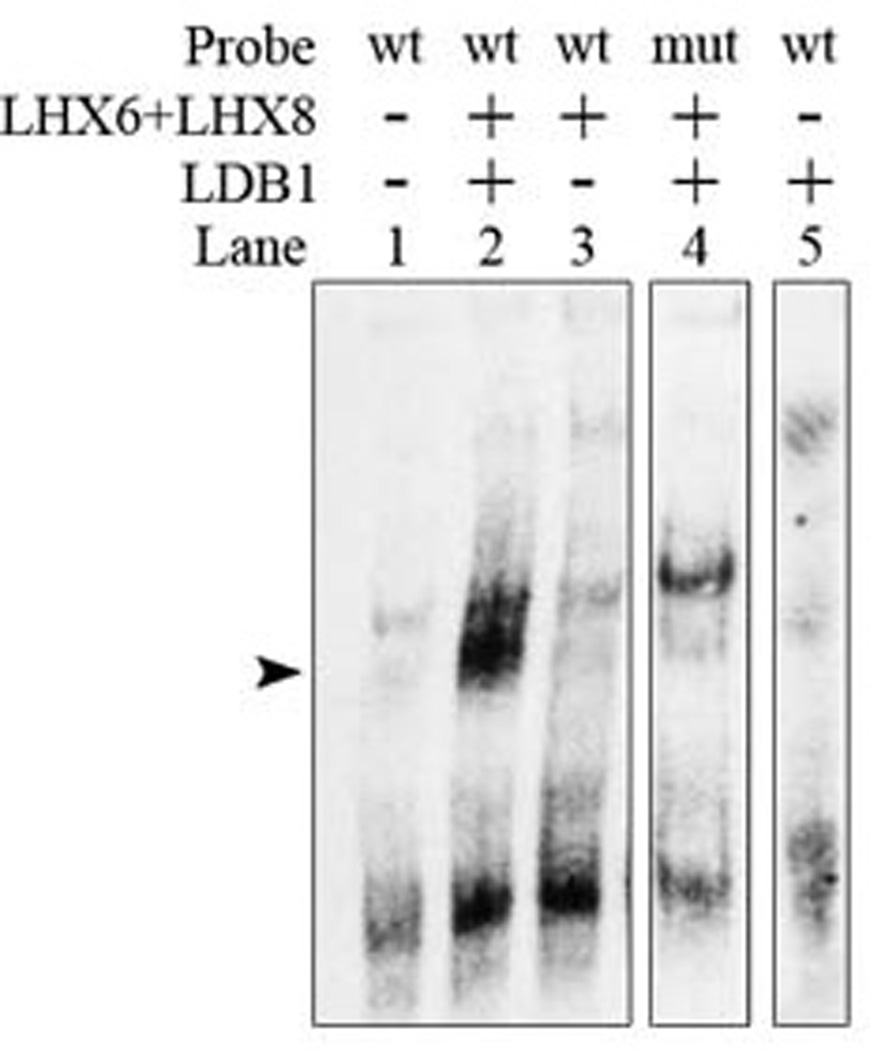
Electrophoretic mobility shift assay showing that LHX6 and LHX8 require LDB1 to bind to the SBE3 Shh enhancer. In vitro DNA binding assay, using LHX6-, LHX8- and LDB1-containing nuclear extract, results in a strong probe (wt SBE3 Shh enhancer domain) -protein complex (arrowhead; lane 2), compared to control extract (lane 1). Addition of LHX6 and LHX8 extracts without LDB1+ nuclear extra does not result in complex formation (lane 3). Mutating the LHX putative binding site of the probe (mut SBE3 Shh enhancer domain) also results in a strong reduction of the amount of complex (lane 4). LDB1 does not bind to the wt probe by itself (lane 5).
Molecular defects in the MGE and MGE-derived pallidal neurons of the Ldb1/Nkx2.1-Cre mutant
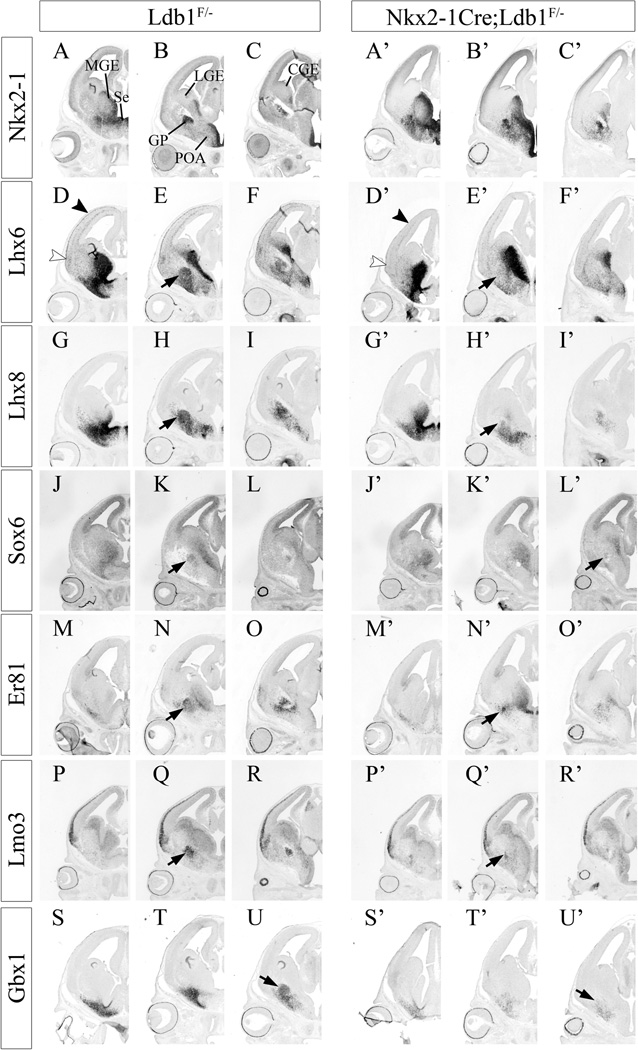
We continued our study of the conditional Ldb1 mutant by in situ hybridization analysis of expression of a series of genes in the VZ, SVZ, and MZ (including the GP) of the developing MGE. At E12.5, similar to Shh, the expression of the Gbx2 homeobox gene in the MZ of the MGE was reduced in the mutant (Figs. 4D–F, 4D’–F’). At E14.5 we found gene expression defects in the basal ganglia (Fig. 6) and in cortical interneurons (Fig. 7). There were no definitive VZ defects, although Nkx2.1 expression may be increased in both the VZ and SVZ (Figs. 6A–C, 6A’–C’). There was reduced SVZ expression of Cux2, Lmo3, Mafb and NPY (Figs. 6 and 7), whereas SVZ expression of Arx, Dlx1, Er81, ErbB4, Lhx6, Lhx8 and Sox6 appeared normal (Figs. 6 and 7). In the MZ, while there were increased Nkx2.1+ scattered cells, there were much fewer Arx, Gbx1, Lhx6, Lhx8, and Somatostatin expressing cells (Figs. 6 and 7). The GP formed, based on Nkx2.1 expression (Figs. 6A–C, 6A’–C’); however it was smaller and less compact, based on expression of Dlx1, Er81, Gbx1, Lhx6, Lhx8, Lmo3 and Sox6 (Figs. 6 and 7) (black arrow). Likewise the ventral pallidal and pallidal-septal cells expressed less Gbx1, Lhx6 and Lhx8 (Fig. 6).
Fig. 6.
Expression of genes that regulate and/or mark MGE development are reduced in conditional Ldb1 mutants, as determined by in situ hybridization at E14.5. Coronal telencephalic hemisections compare gene expression between control (Ldb1+/−; left) and mutant (Nkx2.1-Cre+; Ldb1f/−; right). Three planes of section are shown, with rostral at the left and caudal at the right. Nkx2.1 (A–C, A’–C’); Lhx6 (D–F, D’–F’), black arrowhead shows reduced Lhx6+ neocortical cortical interneurons in mutant; white arrowhead shows reduced Lhx6+ paleocortical cortical interneurons in mutant; black arrow shows reduced GP in mutant; Lhx8 (G–I, G’–I’), black arrow shows reduced GP in mutant; Sox6 (J–L, J’–L’), black arrow shows reduced GP in mutant; Er81 (Etv1; M–O, M’–O’), black arrow shows reduced GP in mutant; Lmo3 (P–R, P’–R’), black arrow shows reduced GP in mutant; Gbx1 (S–U, S’–U’); black arrow shows reduced GP in mutant. Abbreviations: CGE: caudal ganglionic eminence, GP: globus pallidus, LGE: lateral ganglionic eminence, MGE: medial ganglionic eminence, POA: preoptic area, Se: septum.
Fig. 7.
Expression of genes that regulate and/or mark cortical interneuron development are reduced in conditional Ldb1 mutants, as determined by in situ hybridization at E14.5. Coronal telencephalic hemisections compare gene expression between control (Ldb1+/−; left) and mutant (Nkx2-1-Cre+; Ldb1f/−; right). Three planes of section are shown, with rostral at the left and caudal at the right. Arx (A–C, A’–C’), black arrow and white arrowhead show reduced interneurons in the neocortex and paleocortex, respectively; Cux2 (D–F, D’–F’), black arrowhead and arrow show reduced interneurons in the neocortex and striatum, respectively; Cxcr4 (G–I, G’–I’), black arrowhead shows reduced interneurons in the neocortex; Dlx1 (J–L, J’–L’), black arrowhead and white arrowhead show reduced interneurons in the neocortex and paleocortex, respectively; ErbB4 (M–O, M’–O’), black arrowhead and arrow show reduced interneurons in the neocortex and striatum, respectively; Mafb (P–R, P’–R’), black arrowhead, white arrowhead and black arrow show reduced interneurons in the neocortex, paleocortex and striatum, respectively; Npy (S–U, S’–U’), white arrowhead and black arrow show reduced interneurons in the paleocortex and MGE, respectively; Som (V–X, V’–X’), black arrowhead and white arrowhead show reduced interneurons in the neocortex and paleocortex, respectively. Abbreviations: CGE: caudal ganglionic eminence, LGE: lateral ganglionic eminence, MGE: medial ganglionic eminence; PCx: paleocortex; Str: striatum.
Molecular defects in the MGE and MGE-derived cortical and striatal interneurons of the Ldb1/Nkx2.1-Cre mutant
Due to the severe defects in the differentiation of MGE subpallial derivatives, we analyzed development of MGE-derived cortical interneurons at E14.5, which at that stage are tangentially migrating through the cortex (Figs. 6 and 7). Expression of Arx, Cux2, Cxcr4, Erbb4, NPY and Somatostatin was greatly reduced in both the superficial and deep cortical migration streams; expression of Dlx1, Mafb and Lhx6 were preferentially reduced in the superficial stream (Figs. 6 and 7). Both neocortical and paleocortical interneurons showed reduced expression of Arx, Cux2, Cxcr4, Dlx1, Erbb4, Mafb, Lhx6, NPY and Somatostatin (black and white arrowheads, respectively).
Striatal interneuron marker expression was also reduced in the conditional Ldb1 mutant. Striatal expression of Lhx8, Er81, ErbB4, NPY and Somatostatin was not detectable (Figs. 6 and 7). Striatal expression of Nkx2.1 and Lhx6 was clearly reduced, but persistent in some cells (Fig. 6).
Defects in the distribution of the Nkx2.1-lineage cells in the developing telencephalon of the Ldb1/Nkx2.1-Cre mutant
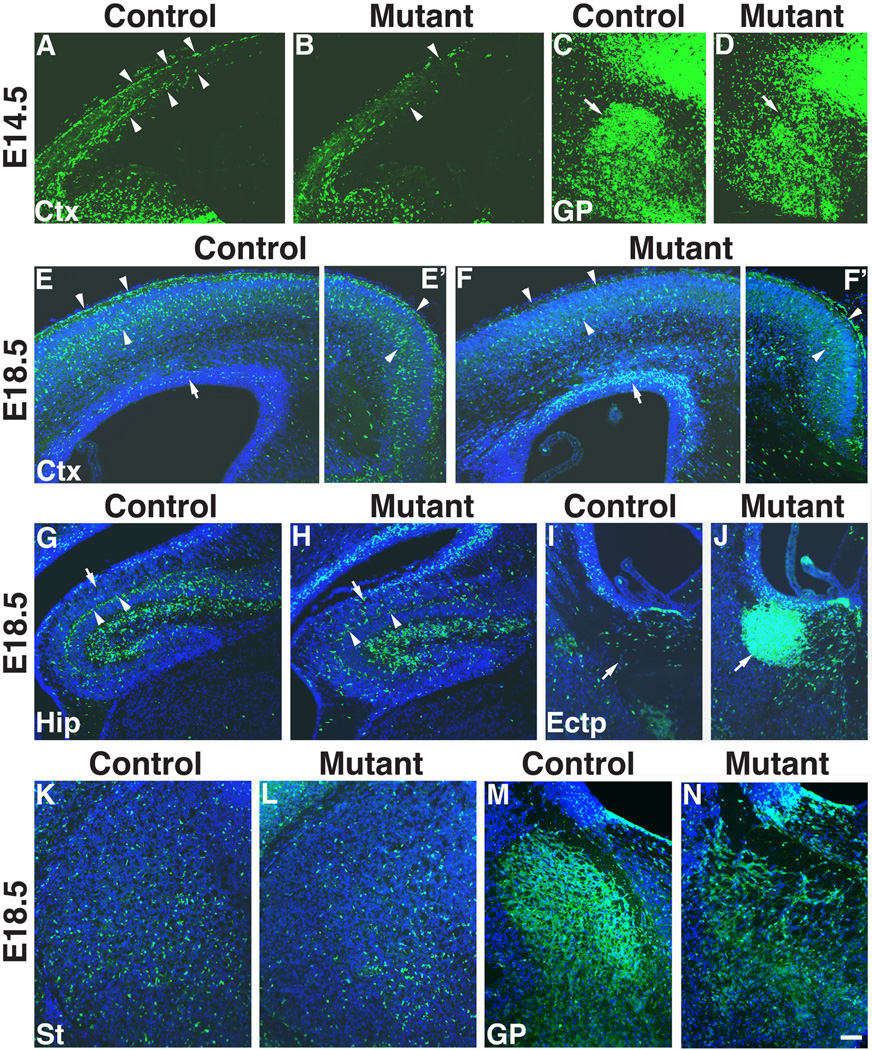
To further examine whether deletion of Ldb1 affected the migration and distribution of the cells derived from the Nkx2.1-lineage, we crossed our mice to the R26R-YFP reporter line to generate control (Ldb1f/+; Nkx2.1-Cre+; R26+/LoxP-stop-LoxP-YFP) and Ldb1 mutant mice that also carried the YFP reporter allele (Ldb1f/−; Nkx2.1-Cre+; R26+/LoxP-stop-LoxP-YFP). In these mice the YFP reporter gene was activated specifically in the Nkx2.1-lineage cells. Immunostaining for YFP at E14.5 revealed fewer migrating YFP+ cells that reached the dorsal part of the cortex in the mutant than those in the control (Figs. 8A, B), consistent with the in situ hybridization results (Fig. 6). The YFP+ cells in the GP were reduced in number and were less organized in the mutant compared to the control (Figs. 8C, D), again consistent with the gene expression data (Fig. 6). At E18.5, fewer YFP+ cells were present in the marginal zone and the cortical plate of the neocortex in the mutant compared to the control (arrowheads, Figs. 8E, E’, F, F’). In contrast, more YFP+ cells were present in the subventricular zone of the cortex in the mutant than those in the control (arrows, Figs. 8E, E’, F, F’). YFP+ cells were detected in the hippocampus of both control and Ldb1 mutant embryos. However, unlike in the control, more of the cells were scattered in the stratum oriens (arrows, Figs. 8G, H) whereas fewer of them were seen in a compact layer adjacent to the pyramidal cell layer (arrowheads, Figs. 8G, H) in the mutant.
Fig. 8.
Anti-YFP staining shows abnormal distribution of Nkx2.1-lineage (YFP+) cells in the telencephalon of Ldb1 mutant (Ldb1f/−; Nkx2.1-Cre+; R26+/flox-stop-floxYFP) compared to the control (Ldb1f/+; Nkx2.1-Cre+; R26+/flox-stop-floxYFP). At E14.5 (A–D), less YFP+ cells are present in the dorsal cortex (arrowheads) of the mutant (B) as compared to the control (A). The YFP+ cells in the globus pallidus (arrows) are less densely packed in the mutant (D) compared to the control (C). By E18.5 (E–N), in both the lateral (E, F) and the medial (E’, F’) cortex (Ctx), there are less YFP+ cells (indicated by arrowheads) in the superficial layers of the mutant (F, F’) as compared to the control (E, E’). In contrast, more YFP+ cells (indicated by arrows) are present close to the ventricular zone of the cortex in the mutant (F) compared to the control (E). G–H, YFP+ cells are present in the developing hippocampus (Hip) of the Ldb1 mutant, but the cells appear more scatted in the stratum oriens (indicated by arrows) in the mutant (H) as compared to the control (G), in which the cells form a more compact layer adjacent to the pyramidal cell layer (arrowheads). I–J, ectopic (Ectp) YFP+ cells accumulate (indicated by arrows) in the dorsal region of the MGE of the Ldb1 mutant (J) as compared to the control (I). K–L, reduction of YFP+ cells in the striatum (St) of the Ldb1 mutant (L) compared to the control (K). M–N, Disorganization of YFP+ cells in the globus pallidus (GP) of the Ldb1 mutant (N) compared to the control (M). Bar in N represents 114 µm for A–D and 100 µm for E–N.
In the ventral telencephalon the mutant had an ectopic cluster of YFP+ cells in the dorsal-lateral corner of the MGE, suggestive of a failure of migration from the progenitor zone (arrows, Figs. 8I, J). The mutant also showed a large reduction of YFP+ cells in the GP, whose internal organization appeared disrupted (Figs. 8M, N). The striatum had a moderate reduction of YFP+ cells (Figs. 8K, L). Together, these results indicate that the migration and distribution of the cells derived from the Nkx2.1-lineage were impaired in the Ldb1/Nkx2.1-Cre mutant.
Next we examined the YFP+ cells in the postnatal Ldb1/Nkx2.1-Cre mutant. At around two weeks after birth (P12 to P16), the number of YFP+ cells was reduced in the various telencephalic structures in the mutant. These included the neocortex, hippocampus, striatum, and globus pallidus (Figs. 9A–H).
Fig. 9.
Anti-YFP staining shows a reduction in number of Nkx2.1-lineage (YFP+) cells in the telencephalon of the postnatal (P16) Ldb1 mutant (Ldb1f/−; Nkx2.1-Cre+; R26+/flox-stop-floxYFP, B, D, F, H) compared to the control (Ldb1f/+; Nkx2.1-Cre+; R26+/flox-stop-floxYFP, A, C, E, G). A–B, cortex at a rostral level. C–D, striatum. E–F, cortex at a caudal level and the hippocampus. G–H, the globus pallidus. Bar in H represents 500 µm for all panels.
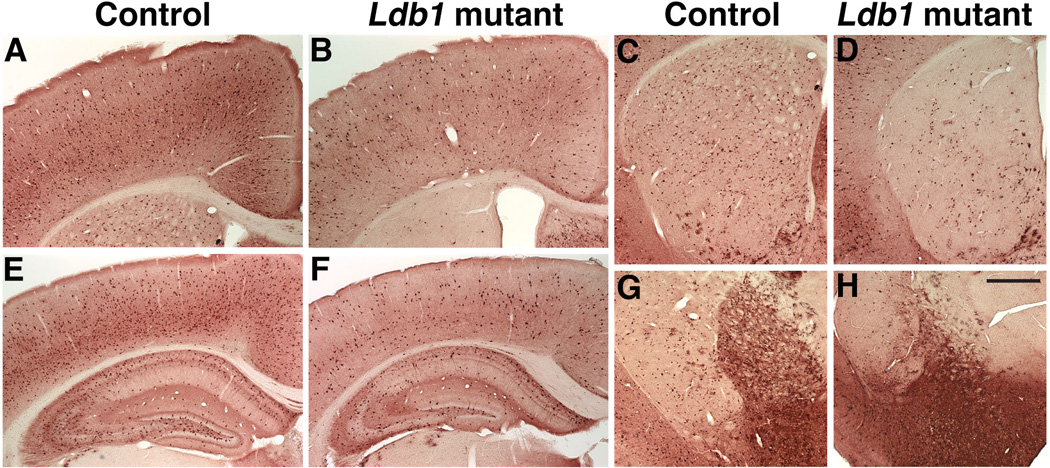
Reduction in the number of cholinergic neurons in the telencephalon of the postnatal Ldb1/Nkx2.1-Cre mutant
Previous studies have shown that Nkx2.1, and its downstream gene Lhx8, are required for the proper development of cholinergic neurons in the telencephalon (Sussel et al., 1999; Marin et al. 2000; Zhao et al. 2003; Lopes et al., 2012; Mori et al., 2004; Fragkouli et al., 2005). To determine whether Ldb1is required for the development of these neurons, we analyzed the Ldb1/Nkx2.1-Cre mutant by immunostaining of ChAT, an enzyme essential for acetylcholine synthesis. The number of cholinergic neurons was reduced in the striatum [average ± standard deviation (sd): control 190 ± 24; mutant 21 ± 5, nine sections from 3 animals for each group, P < 9 ×10−9 (Figs. 10A, B, E, F)], septum [average ± sd: control 278 ± 40; mutant 38 ± 9, three sections from 3 animals for each group, P < 0.007 (Fig. 10C, D)], and the nucleus Basalis [average ± sd: control 106 ± 15; mutant 48 ± 15, six sections from 3 animals for each group, P < 5 × 10−5 (Fig. 10G, H)] in the Ldb1 mutants at P16 and P18.
Fig. 10.
Reduction in number of cholinergic neurons in the telencephalon of the Ldb1 conditional mutant. Anti-ChAT staining shows a reduction in the number of cholinergic neurons in the striatum (A–B, E–F), the diagonal bands of the septum (C–D), and the nucleus Basalis (G–H) in a P18 Ldb1f/−; Nkx2.1-Cre+ mutant (B, D, F, H) as compared to the control (A, C, E, G). E and F show enlargement of a part of A and B, respectively. Bar in A represents 500 µm for A–D, G–H, and 200 µm for E–F.
Reduction in the number of cortical and hippocampal GABAergic interneurons in the postnatal Ldb1/Nkx2.1-Cre mutant
Lhx6 is essential for the development of the PV+ and SOM+ subclasses of the GABAergic interneurons in the cortex and hippocampus (Liodis et al., 2007; Neves et al., 2012; Zhao et al., 2008). Thus, we examined these neurons in the Ldb1/Nkx2.1-Cre mutant. Immunostaining of PV and SOM revealed that the number of PV+ and SOM+ neurons in the cortex [PV+ neurons, average ± sd: control 259 ± 23; mutant 125 ± 13, six sections from 3 animals for each group, P < 2×10−6; SOM+ neurons, average ± sd: control 138 ± 31; mutant 83 ± 21, four sections from 2 animals for each group, P < 0.04] and hippocampus [PV+ neurons, average ± sd: control 118 ± 24; mutant 26 ± 7, fourteen sections from 3 controls and twelve sections from 3 mutants, P < 8×10−10. SOM+ neurons, average ± sd: control 60 ± 18; mutant 28 ± 8, four sections from 2 animals for each group, P < 0.03] of the mutant was reduced compared to the control at P16 or P18 (Fig. 11). Thus, the reduction in number of the PV+ and SOM+ neurons in the mutant hippocampus was more pronounced than that in the cortex.
Fig. 11.

Reduction in number of GABAergic interneurons in the cortex and hippocampus of the Ldb1 conditional mutant. Immuno-staining of parvalbumin (A–D) and somatostatin (E–H) shows a reduction in number of parvalbumin+ and somatostatin+ interneurons in the cortex and hippocampus of a P18 Ldb1f/−; Nkx2.1-Cre+ mutant (B, D, F, H) as compared to the control (A, C, E, G). A, B, E, F show sections of the cortex from a rostral level. C, D, G, H show sections of the cortex and hippocampus from a caudal level. Bar in H represents 500 µm for all panels.
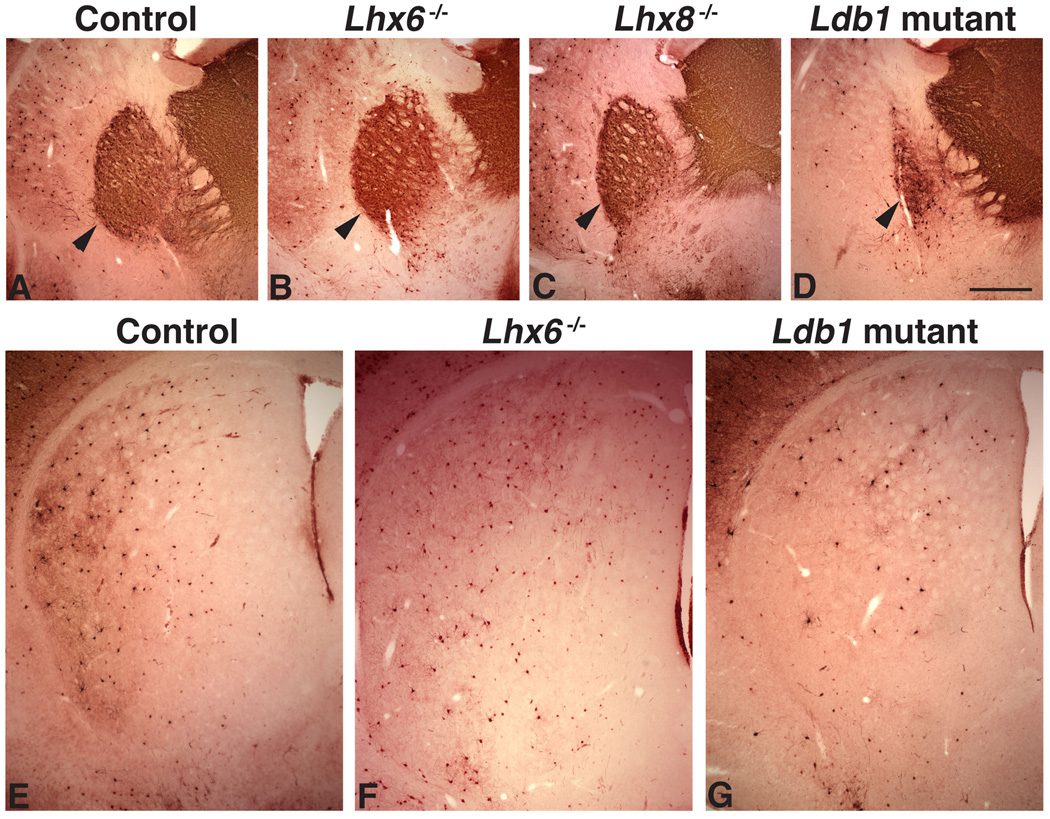
Defects in development of the globus pallidus and striatal parvalbumin+ neurons in the postnatal Ldb1/Nkx2.1-Cre mutant
Immunostaining of PV revealed additional defects in the basal telencephalon of the postnatal Ldb1/Nkx2.1-Cre mutant. Consistent with results from the molecular analysis of the defects in the E12.5 and E14.5 MGE and GP of the Ldb1 mutant embryos (Figs. 6 and 7), the GP in the postnatal Ldb1 mutant appeared irregular in shape and was smaller compared to the controls (Fig. 12 A, D). As reported previously (Zhao et al., 2003; 2008), the GP in Lhx6 or Lhx8 single mutants appeared normal (Figs. 12B, C). PV+ interneurons in the striatum were not affected in the Lhx8 mutant (Zhao et al., 2003). Although our previous study showed that the distribution of the PV+ interneurons was altered in the Lhx6 single mutant, the number of these neurons was not changed significantly (Zhao et al., 2008; Fig. 12F). However, a reduction in number of the PV+ neurons in the striatum was observed in the Ldb1/Nkx2.1-Cre mutant [average ± sd: control 117 ± 24 (16 sections from 3 animals); mutant 45 ± 10 (16 sections from 3 animals), P < 5 × 10−10] (Fig. 12G). Thus the Ldb1/Nkx2.1-Cre mutant showed a broader phenotype in the ventral telencephalon than that was observed previously in either the Lhx6 or Lhx8 single mutant.
Fig. 12.
Defect of the globus pallidus and the striatal parvalbumin interneurons in the Ldb1 conditional mutant. A–D, anti-parvalbumin staining shows that the globus pallidus (pointed by arrowheads) is severely reduced in size in a postnatal (P18) Ldb1f/−; Nkx2.1-Cre+ mutant (D) as compared to the control (A), while it appears normal in either Lhx6−/− (B) or Lhx8−/− (C) single mutant. E–G, reduction in number of parvalbumin+ interneurons in the striatum of the Ldb1 mutant (G) as compared to the control (E) and a Lhx6−/− mutant. Bar in D represents 500 µm for A–D and 370 µm for E–G.
Discussion
Previous studies have established that the LIM-HD factors Lhx6 and Lhx8 regulate the development of multiple types of neurons in the telencephalon (Zhao et al., 2003; 2008; Mori et al., 2004; Fragkouli et al., et al., 2005; 2009; Lodis et al., 2007; Lopes et al., 2012; Neves et al., 2012). However, the molecular mechanisms underlying how these transcription factors function have largely remained unclear. The nuclear factor Ldb1 has been shown to directly bind to the LIM domains of LIM-HD factors (Aglunick et al., 1996). Disruption of Ldb1 function in a number of developing systems has been shown to result in various defects similar to those observed after inactivation of certain LIM-HD factors (Aglunick et al., 1996; Mukhopadhyay et al., 2003; Zhao et al., 2007; Tzchori et al., 2009; Hunter et al., 2012). These studies indicate that Ldb1 plays an essential role in mediating the function of the LIM-HD factors.
In the present study, we showed that Ldb1 is expressed in telencephalic Nkx2.1-lineage cells, a major population that contributes to pallidal projection neurons, and striatal and pallial interneurons. To determine the function of Ldb1 in the development of these neurons, we deleted the Ldb1 in Nkx2.1-cell lineage by crossing the Ldb1-floxed mouse line with the Nkx2.1-Cre line. Inactivation of Ldb1 resulted in striking reductions in the expression of genes that control MGE neuronal development, including those that are critically involved in the transcription regulation or cell signaling. As a result, the Ldb1/Nkx2.1-Cre conditional mutant showed severe defects in the generation and migration of cortical and sub-cortical GABAergic interneurons, pallidal GABAergic projection neurons, and telencephalic cholinergic neurons.
The phenotype of the Ldb1/Nkx2.1-Cre mutant is more severe than the Lhx6 and Lhx8 single mutants, or closely phenocopies the Lhx6/Lhx8 double mutants. The developmental defects in GABAergic interneurons of the Ldb1 conditional mutant are similar to those observed previously in the Lhx6 mutant (Lodis et al., 2007; Zhao et al., 2008). Many defects in development of the cholinergic neurons are observed both in the Ldb1 conditional mutant and in the Lhx8 mutant (Zhao et al., 2003; Mori et al., 2004; Fragkouli et al., 2005). It is possible that Ldb1 mutant-dependent cholinergic defects also arise from its potential interaction with Islet-1, a LIM-HD transcription factor that like Lhx8, regulates telencephalic cholinergic neuronal development (Elshatory and Gan, 2008).
Both the Ldb1 conditional mutant and the Lhx6/Lhx8 double mutant share interneuron defects, but unlike the single mutants, also had an abnormal globus palilidus. Together these genetic results support that biochemical model that Ldb1 is an essential co-factor for the Lhx6 and Lhx8 LIM-HD proteins in regulating the development of the Nkx2.1-lineage cells in the ventral telencephalon. Moreover, EMSA analysis showed that the binding of Lhx6 or Lhx8 to the Shh gene enhancer is dependent on the presence of Ldb1. This further supports the essential role of Ldb1 as a co-factor with Lhx6 and/or Lhx8 in the transcriptional regulation of the development of the Nkx2.1-lineage derived neurons in the telencephalon.
Similar to the Lhx6/Lhx8 double mutant, the Ldb1/Nkx2.1-Cre mutant showed a reduction of the Shh expression in the MGE mantle zone. We have reported previously that a conditional Shh deletion in the MGE mantle zone can also cause defects in the generation of cortical interneurons (Flandin et al., 2011). Thus the defects in cortical interneuron development observed here in the Ldb1/Nkx2.1-Cre mutant can result from, at least partially, from the impaired Shh expression in the MGE mantle zone.
Our analysis of the Ldb1/Nkx2.1-Cre conditional mutant has also revealed interesting new phenotypes that were not previously observed in the Lhx6 and Lhx8 single or double mutants. While the MGE-derived globus pallidus appears normal in Lhx6 and Lhx8 single mutants (Zhao et al., 2003; Zhao et al., 2008; and in this study, see Fig. 12), deletion of both Lhx6 and Lhx8 leads to severe molecular and morphological defects in the globus pallidus. This indicates a redundant function of Lhx6 and Lhx8 in the development of the globus pallidus (Flandin et al., 2011). Due to severe developmental defects in the craniofacial structures, the Lhx6/Lhx8 double mutant dies neonatally, which precluded the postnatal analysis to further determine how this phenotype continues to manifest and how the later differentiated neuronal cell type such as the PV+ neurons is affected in these mutants. In this study, we observed that the Ldb1 mutant survives after birth. Thus, we were able to extend our analysis postnatally and found that PV+ projection neurons in the globus pallidus and PV+ striatal interneurons were greatly reduced.
The defects in the development of the GABAergic neurons in the cortex of the Ldb1/Nkx2.1-Cre mutant appeared less severe compared to the Lhx6 mutant. This is probably in part due to an incomplete deletion of the Ldb1 in the MGE, especially in the rostral-dorsal region of the MGE (Fig. 3), where not all of the Nkx2.1+ cells express the Cre recombinase in the Nkx2.1-Cre BAC transgenic mice (Xu et al., 2008). In support of this idea, double immunofluorscent staining reveled that many of the remaining PV+ cells in the cortex and striatum of the postnatal Ldb1 mutants were Ldb1+ (Supplemental Fig. S1). Double staining of ChAT and Ldb1 also showed that the remaining ChAT+ cells in the striatum of the Ldb1 mutants were Ldb1+ (Supplemental Fig. S2). However, double staining of PV/Ldb1 or ChAT/Ldb1 also revealed that the remaining PV+ or ChAT+ cells in the globus pallidus (Supplemental Fig. S1) or in the septum and nucleus Basalis (Supplemental Fig. S2) of the Ldb1 mutants were Ldb1−. It is possible that the conditional Ldb1 deletion might take place relatively later during the embryonic development in contrast to a germline gene deletion. Some of the neuron precursor cells might retain the Ldb1 function and proceed with the development for a certain period before the gene was inactivated. Also, Ldb2, a gene closely related to Ldb1 (Aglunick et al., 1996), may compensate for the loss of Ldb1, as shown recently in early development of the limb (Narkis et al., 2012).
Supplementary Material
Highlights.
Ldb1 is expressed in the Nkx2.1lineage in the developing ventral telencephalon.
Ldb1 controls expression of a series of genes critical for telencephalon development.
Ldb1 regulates the tangential migration of ventral telencephalon derived neurons.
Ldb1 is essential for the generation of a variety of telencephalic neurons.
Acknowledgements
We would like to thank Dr. Stewart Anderson for sharing the Nkx2.1-Cre mice, and Drs. Liqi Li and Paul Love for sharing the Ldb1 antibody. This work was supported by the research grants to JLRR from Nina Ireland, Weston Havens Foundation, NIMH R01 MH081880, and NIMH R37 MH049428, and funds from the NIH intramural research program to HW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interests.
References
- Agulnick AD, Taira M, Breen JJ, Tanaka T, Dawid IB, Westphal H. Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature. 1996;384:270–272. doi: 10.1038/384270a0. [DOI] [PubMed] [Google Scholar]
- Bach I, Carriere C, Ostendorff HP, Andersen B, Rosenfeld MG. A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and Otx homeodomain proteins. Genes & development. 1997;11:1370–1380. doi: 10.1101/gad.11.11.1370. [DOI] [PubMed] [Google Scholar]
- Butt SJ, Sousa VH, Fuccillo MV, Hjerling-Leffler J, Miyoshi G, Kimura S, Fishell G. The requirement of Nkx2-1 in the temporal specification of cortical interneuron subtypes. Neuron. 2008;59:722–732. doi: 10.1016/j.neuron.2008.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T, Xu Q, Ocbina PJ, Anderson SA. NKX2.1 specifies cortical interneuron fate by activating Lhx6. Development. 2008;135:1559–1567. doi: 10.1242/dev.015123. [DOI] [PubMed] [Google Scholar]
- Elshatory Y, Gan L. The LIM-homeobox gene Islet-1 is required for the development of restricted forebrain cholinergic neurons. J Neurosci. 2008;28:3291–3297. doi: 10.1523/JNEUROSCI.5730-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flandin P, Zhao Y, Vogt D, Jeong J, Long J, Potter G, Westphal H, Rubenstein JL. Lhx6 and Lhx8 coordinately induce neuronal expression of Shh that controls the generation of interneuron progenitors. Neuron. 2011;70:939–950. doi: 10.1016/j.neuron.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkouli A, Hearn C, Errington M, Cooke S, Grigoriou M, Bliss T, Stylianopoulou F, Pachnis V. Loss of forebrain cholinergic neurons and impairment in spatial learning and memory in LHX7-deficient mice. The European journal of neuroscience. 2005;21:2923–2938. doi: 10.1111/j.1460-9568.2005.04141.x. [DOI] [PubMed] [Google Scholar]
- Fragkouli A, van Wijk NV, Lopes R, Kessaris N, Pachnis V. LIM homeodomain transcription factor-dependent specification of bipotential MGE progenitors into cholinergic and GABAergic striatal interneurons. Development. 2009;136:3841–3851. doi: 10.1242/dev.038083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O, Westphal H. Functions of LIM-homeobox genes. Trends in genetics : TIG. 2000;16:75–83. doi: 10.1016/s0168-9525(99)01883-1. [DOI] [PubMed] [Google Scholar]
- Hunter CS, Dixit S, Cohen T, Ediger B, Wilcox C, Ferreira M, Westphal H, Stein R, May CL. Islet alpha-, beta-, and delta-Cell Development Is Controlled by the Ldb1 Coregulator, Acting Primarily With the Islet-1 Transcription Factor. Diabetes. 2013;62:875–886. doi: 10.2337/db12-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurata LW, Kenny DA, Gill GN. Nuclear LIM interactor, a rhombotin and LIM homeodomain interacting protein, is expressed early in neuronal development. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:11693–11698. doi: 10.1073/pnas.93.21.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liodis P, Denaxa M, Grigoriou M, Akufo-Addo C, Yanagawa Y, Pachnis V. Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:3078–3089. doi: 10.1523/JNEUROSCI.3055-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes R, Verhey van Wijk N, Neves G, Pachnis V. Transcription factor LIM homeobox 7 (Lhx7) maintains subtype identity of cholinergic interneurons in the mammalian striatum. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3119–3124. doi: 10.1073/pnas.1109251109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Anderson SA, Rubenstein JL. Origin and molecular specification of striatal interneurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2000;20:6063–6076. doi: 10.1523/JNEUROSCI.20-16-06063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Yuxing Z, Takaki H, Takeuchi M, Iseki K, Hagino S, Kitanaka J, Takemura M, Misawa H, Ikawa M, Okabe M, Wanaka A. The LIM homeobox gene, L3/Lhx8, is necessary for proper development of basal forebrain cholinergic neurons. The European journal of neuroscience. 2004;19:3129–3141. doi: 10.1111/j.0953-816X.2004.03415.x. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay M, Teufel A, Yamashita T, Agulnick AD, Chen L, Downs KM, Schindler A, Grinberg A, Huang SP, Dorward D, Westphal H. Functional ablation of the mouse Ldb1 gene results in severe patterning defects during gastrulation. Development. 2003;130:495–505. doi: 10.1242/dev.00225. [DOI] [PubMed] [Google Scholar]
- Narkis G, Tzchori I, Cohen T, Holtz A, Wier E, Westphal H. Isl1 and Ldb co-regulators of transcription are essential early determinants of mouse limb development. Developmental dynamics : an official publication of the American Association of Anatomists. 2012;241:787–791. doi: 10.1002/dvdy.23761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves G, Shah MM, Liodis P, Achimastou A, Denaxa M, Roalfe G, Sesay A, Walker MC, Pachnis V. The LIM Homeodomain Protein Lhx6 Regulates Maturation of Interneurons and Network Excitability in the Mammalian Cortex. Cereb Cortex. 2012 doi: 10.1093/cercor/bhs159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L, Marin O, Kimura S, Rubenstein JL. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126:3359–3370. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- Thaler JP, Lee SK, Jurata LW, Gill GN, Pfaff SL. LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell. 2002;110:237–249. doi: 10.1016/s0092-8674(02)00823-1. [DOI] [PubMed] [Google Scholar]
- Tzchori I, Day TF, Carolan PJ, Zhao Y, Wassif CA, Li L, Lewandoski M, Gorivodsky M, Love PE, Porter FD, Westphal H, Yang Y. LIM homeobox transcription factors integrate signaling events that control three-dimensional limb patterning and growth. Development. 2009;136:1375–1385. doi: 10.1242/dev.026476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Tam M, Anderson SA. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. The Journal of comparative neurology. 2008;506:16–29. doi: 10.1002/cne.21529. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Flandin P, Long JE, Cuesta MD, Westphal H, Rubenstein JL. Distinct molecular pathways for development of telencephalic interneuron subtypes revealed through analysis of Lhx6 mutants. The Journal of comparative neurology. 2008;510:79–99. doi: 10.1002/cne.21772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Kwan KM, Mailloux CM, Lee WK, Grinberg A, Wurst W, Behringer RR, Westphal H. LIM-homeodomain proteins Lhx1 and Lhx5, and their cofactor Ldb1, control Purkinje cell differentiation in the developing cerebellum. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13182–13186. doi: 10.1073/pnas.0705464104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Marin O, Hermesz E, Powell A, Flames N, Palkovits M, Rubenstein JL, Westphal H. The LIM-homeobox gene Lhx8 is required for the development of many cholinergic neurons in the mouse forebrain. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9005–9010. doi: 10.1073/pnas.1537759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.