Abstract
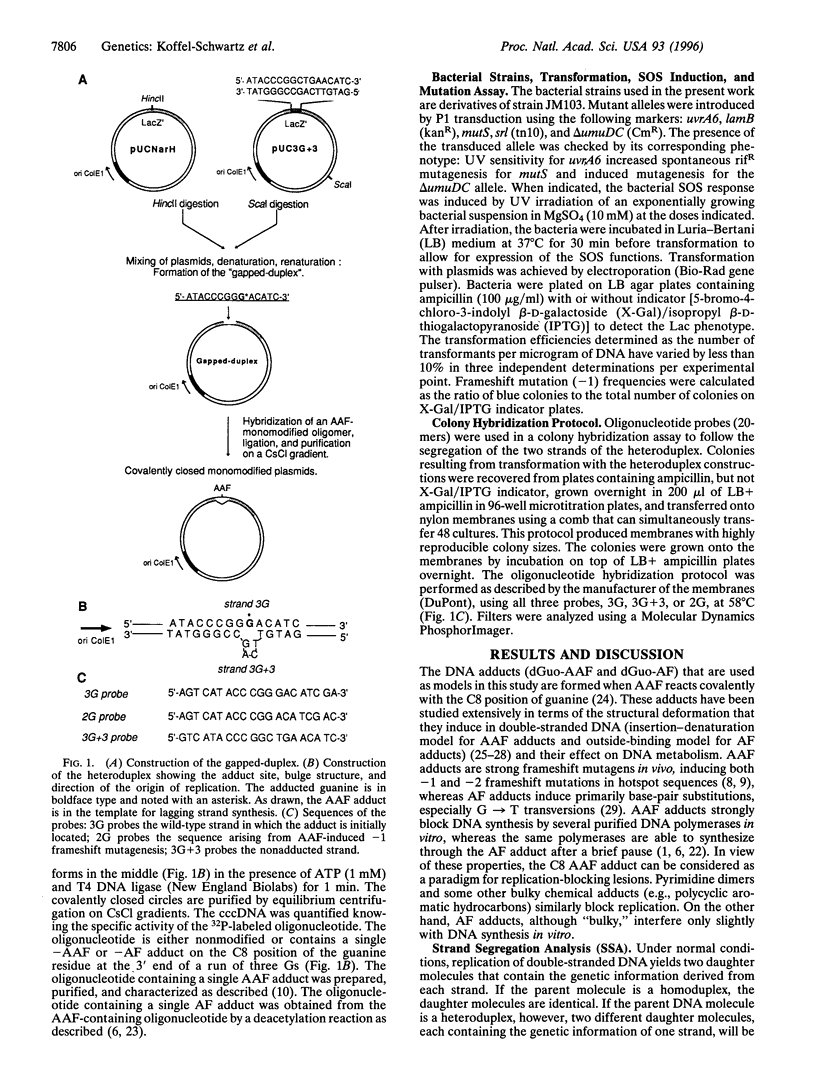
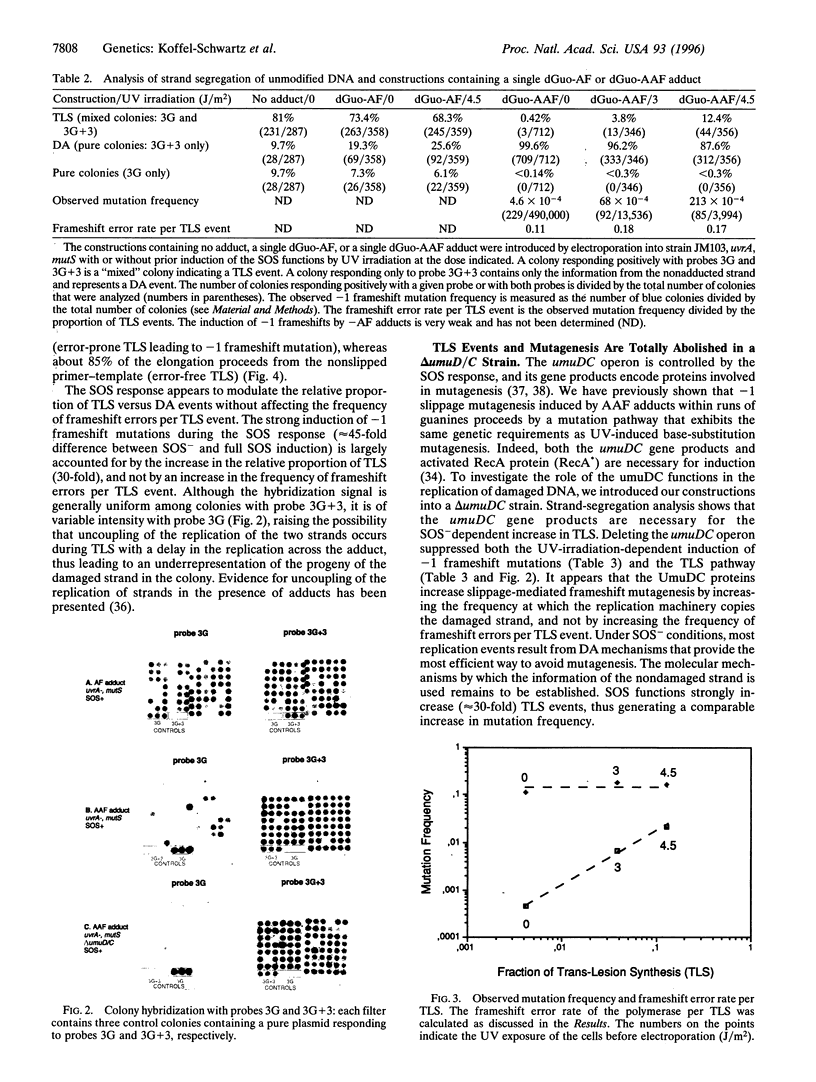
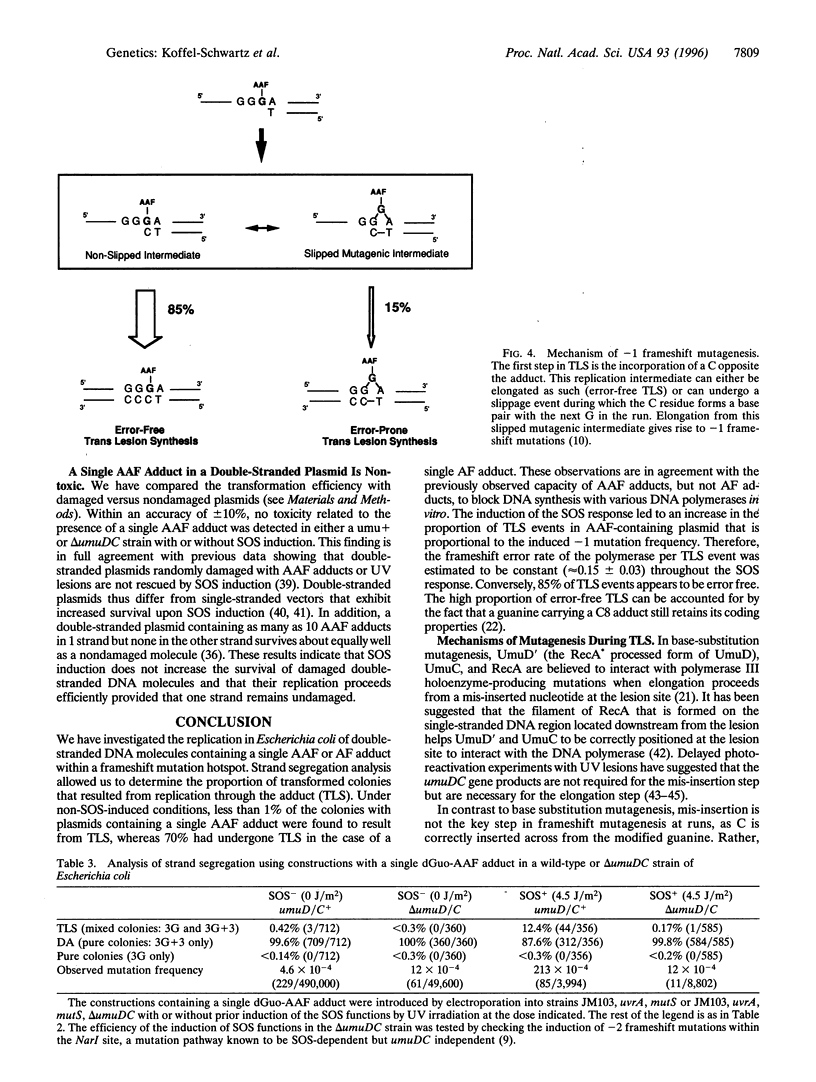
The replication of double-stranded plasmids containing a single adduct was analyzed in vivo by means of a sequence heterology that marks the two DNA strands. The single adduct was located within the sequence heterology, making it possible to distinguish trans-lesion synthesis (TLS) events from damage avoidance events in which replication did not proceed through the lesion. When the SOS system of the host bacteria is not induced, the C8-guanine adduct formed by the carcinogen N-2-acetylaminofluorene (AAF) yields less than 1% of TLS events, showing that replication does not readily proceed through the lesion. In contrast, the deacetylated adduct N-(deoxyguanosin-8-yl)-2-aminofluorene yields approximately 70% of TLS events under both SOS-induced and uninduced conditions. These results for TLS in vivo are in good agreement with the observation that AAF blocks DNA replication in vitro, whereas aminofluorene does so only weakly. Induction of the SOS response causes an increase in TLS events through the AAF adduct (approximately 13%). The increase in TLS is accompanied by a proportional increase in the frequency of AAF-induced frameshift mutations. However, the polymerase frameshift error rate per TLS event was essentially constant throughout the SOS response. In an SOS-induced delta umuD/C strain, both US events and mutagenesis are totally abolished even though there is no decrease in plasmid survival. Error-free replication evidently proceeds efficiently by means of the damage avoidance pathway. We conclude that SOS mutagenesis results from increased TLS rather than from an increased frameshift error rate of the polymerase.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banerjee S. K., Christensen R. B., Lawrence C. W., LeClerc J. E. Frequency and spectrum of mutations produced by a single cis-syn thymine-thymine cyclobutane dimer in a single-stranded vector. Proc Natl Acad Sci U S A. 1988 Nov;85(21):8141–8145. doi: 10.1073/pnas.85.21.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belguise-Valladier P., Fuchs R. P. N-2-aminofluorene and N-2 acetylaminofluorene adducts: the local sequence context of an adduct and its chemical structure determine its replication properties. J Mol Biol. 1995 Jun 23;249(5):903–913. doi: 10.1006/jmbi.1995.0347. [DOI] [PubMed] [Google Scholar]
- Belguise-Valladier P., Maki H., Sekiguchi M., Fuchs R. P. Effect of single DNA lesions on in vitro replication with DNA polymerase III holoenzyme. Comparison with other polymerases. J Mol Biol. 1994 Feb 11;236(1):151–164. doi: 10.1006/jmbi.1994.1125. [DOI] [PubMed] [Google Scholar]
- Bhanot O. S., Grevatt P. C., Donahue J. M., Gabrielides C. N., Solomon J. J. Incorporation of dA opposite N3-ethylthymidine terminates in vitro DNA synthesis. Biochemistry. 1990 Nov 13;29(45):10357–10364. doi: 10.1021/bi00497a010. [DOI] [PubMed] [Google Scholar]
- Bichara M., Fuchs R. P. DNA binding and mutation spectra of the carcinogen N-2-aminofluorene in Escherichia coli. A correlation between the conformation of the premutagenic lesion and the mutation specificity. J Mol Biol. 1985 Jun 5;183(3):341–351. doi: 10.1016/0022-2836(85)90005-1. [DOI] [PubMed] [Google Scholar]
- Bridges B. A., Dennis R. E., Munson R. J. Differential induction and repair of ultraviolet damage leading to true revesions and external suppressor mutations of an ochre codon in Escherichia coli B-r WP2. Genetics. 1967 Dec;57(4):897–908. doi: 10.1093/genetics/57.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges B. A. Mutagenesis after exposure of bacteria to ultraviolet light and delayed photoreversal. Mol Gen Genet. 1992 Jun;233(3):331–336. doi: 10.1007/BF00265428. [DOI] [PubMed] [Google Scholar]
- Bridges B. A., Woodgate R. Mutagenic repair in Escherichia coli. X. The umuC gene product may be required for replication past pyrimidine dimers but not for the coding error in UV-mutagenesis. Mol Gen Genet. 1984;196(2):364–366. doi: 10.1007/BF00328073. [DOI] [PubMed] [Google Scholar]
- Bridges B. A., Woodgate R. Mutagenic repair in Escherichia coli: products of the recA gene and of the umuD and umuC genes act at different steps in UV-induced mutagenesis. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4193–4197. doi: 10.1073/pnas.82.12.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyde S., Hingerty B. Conformation of 2-aminofluorene-modified DNA. Biopolymers. 1983 Nov;22(11):2423–2441. doi: 10.1002/bip.360221109. [DOI] [PubMed] [Google Scholar]
- Chernaenko V. M., Bresler S. E. A study of the interaction of oligonucleotides with polynucleotides by the method of equilibrium gel filtration. The effect of magnesium ions and the composition of the oligonucleotides on the stability of the complex. Mol Biol. 1975 Jan;8(4):467–474. [PubMed] [Google Scholar]
- Clark J. M., Beardsley G. P. Template length, sequence context, and 3'-5' exonuclease activity modulate replicative bypass of thymine glycol lesions in vitro. Biochemistry. 1989 Jan 24;28(2):775–779. doi: 10.1021/bi00428a054. [DOI] [PubMed] [Google Scholar]
- Daune M. P., Fuchs R. P., Leng M. Structural modification and protein recognition of DNA modified by N-2-fluorenylacetamide, its 7-iodo derivative, and by N-2-fluorenamine. Natl Cancer Inst Monogr. 1981 Dec;(58):201–210. [PubMed] [Google Scholar]
- Echols H., Goodman M. F. Fidelity mechanisms in DNA replication. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- Echols H., Goodman M. F. Mutation induced by DNA damage: a many protein affair. Mutat Res. 1990 Sep-Nov;236(2-3):301–311. doi: 10.1016/0921-8777(90)90013-u. [DOI] [PubMed] [Google Scholar]
- Echols H. SOS functions, cancer and inducible evolution. Cell. 1981 Jul;25(1):1–2. doi: 10.1016/0092-8674(81)90223-3. [DOI] [PubMed] [Google Scholar]
- Frank E. G., Hauser J., Levine A. S., Woodgate R. Targeting of the UmuD, UmuD', and MucA' mutagenesis proteins to DNA by RecA protein. Proc Natl Acad Sci U S A. 1993 Sep 1;90(17):8169–8173. doi: 10.1073/pnas.90.17.8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs R. P., Schwartz N., Daune M. P. Hot spots of frameshift mutations induced by the ultimate carcinogen N-acetoxy-N-2-acetylaminofluorene. Nature. 1981 Dec 17;294(5842):657–659. doi: 10.1038/294657a0. [DOI] [PubMed] [Google Scholar]
- Fuchs R., Daune M. Physical studies on deoxyribonucleic acid after covalent binding of a carcinogen. Biochemistry. 1972 Jul 4;11(14):2659–2666. doi: 10.1021/bi00764a017. [DOI] [PubMed] [Google Scholar]
- Garcia A., Lambert I. B., Fuchs R. P. DNA adduct-induced stabilization of slipped frameshift intermediates within repetitive sequences: implications for mutagenesis. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):5989–5993. doi: 10.1073/pnas.90.13.5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogol E. P., Young M. C., Kubasek W. L., Jarvis T. C., von Hippel P. H. Cryoelectron microscopic visualization of functional subassemblies of the bacteriophage T4 DNA replication complex. J Mol Biol. 1992 Mar 20;224(2):395–412. doi: 10.1016/0022-2836(92)91003-8. [DOI] [PubMed] [Google Scholar]
- Higgins N. P., Kato K., Strauss B. A model for replication repair in mammalian cells. J Mol Biol. 1976 Mar 5;101(3):417–425. doi: 10.1016/0022-2836(76)90156-x. [DOI] [PubMed] [Google Scholar]
- Kato T., Shinoura Y. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol Gen Genet. 1977 Nov 14;156(2):121–131. doi: 10.1007/BF00283484. [DOI] [PubMed] [Google Scholar]
- Koffel-Schwartz N., Maenhaut-Michel G., Fuchs R. P. Specific strand loss in N-2-acetylaminofluorene-modified DNA. J Mol Biol. 1987 Feb 20;193(4):651–659. doi: 10.1016/0022-2836(87)90348-2. [DOI] [PubMed] [Google Scholar]
- Koffel-Schwartz N., Verdier J. M., Bichara M., Freund A. M., Daune M. P., Fuchs R. P. Carcinogen-induced mutation spectrum in wild-type, uvrA and umuC strains of Escherichia coli. Strain specificity and mutation-prone sequences. J Mol Biol. 1984 Jul 25;177(1):33–51. doi: 10.1016/0022-2836(84)90056-1. [DOI] [PubMed] [Google Scholar]
- Kriek E., Miller J. A., Juhl U., Miller E. C. 8-(N-2-fluorenylacetamido)guanosine, an arylamidation reaction product of guanosine and the carcinogen N-acetoxy-N-2-fluorenylacetamide in neutral solution. Biochemistry. 1967 Jan;6(1):177–182. doi: 10.1021/bi00853a029. [DOI] [PubMed] [Google Scholar]
- Lambert I. B., Napolitano R. L., Fuchs R. P. Carcinogen-induced frameshift mutagenesis in repetitive sequences. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1310–1314. doi: 10.1073/pnas.89.4.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClerc J. E., Borden A., Lawrence C. W. The thymine-thymine pyrimidine-pyrimidone(6-4) ultraviolet light photoproduct is highly mutagenic and specifically induces 3' thymine-to-cytosine transitions in Escherichia coli. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9685–9689. doi: 10.1073/pnas.88.21.9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann A. R. Postreplication repair of DNA in ultraviolet-irradiated mammalian cells. J Mol Biol. 1972 May 28;66(3):319–337. doi: 10.1016/0022-2836(72)90418-4. [DOI] [PubMed] [Google Scholar]
- Lindsley J. E., Fuchs R. P. Use of single-turnover kinetics to study bulky adduct bypass by T7 DNA polymerase. Biochemistry. 1994 Jan 25;33(3):764–772. doi: 10.1021/bi00169a018. [DOI] [PubMed] [Google Scholar]
- Livneh Z., Cohen-Fix O., Skaliter R., Elizur T. Replication of damaged DNA and the molecular mechanism of ultraviolet light mutagenesis. Crit Rev Biochem Mol Biol. 1993;28(6):465–513. doi: 10.3109/10409239309085136. [DOI] [PubMed] [Google Scholar]
- Maenhaut-Michel G., Janel-Bintz R., Fuchs R. P. A umuDC-independent SOS pathway for frameshift mutagenesis. Mol Gen Genet. 1992 Nov;235(2-3):373–380. doi: 10.1007/BF00279383. [DOI] [PubMed] [Google Scholar]
- Michaels M. L., Johnson D. L., Reid T. M., King C. M., Romano L. J. Evidence for in vitro translesion DNA synthesis past a site-specific aminofluorene adduct. J Biol Chem. 1987 Oct 25;262(30):14648–14654. [PubMed] [Google Scholar]
- Milhé C., Dhalluin C., Fuchs R. P., Lefèvre J. F. NMR evidence of the stabilisation by the carcinogen N-2-acetylaminofluorene of a frameshift mutagenesis intermediate. Nucleic Acids Res. 1994 Nov 11;22(22):4646–4652. doi: 10.1093/nar/22.22.4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. A. Carcinogenesis by chemicals: an overview--G. H. A. Clowes memorial lecture. Cancer Res. 1970 Mar;30(3):559–576. [PubMed] [Google Scholar]
- Moore P. D., Bose K. K., Rabkin S. D., Strauss B. S. Sites of termination of in vitro DNA synthesis on ultraviolet- and N-acetylaminofluorene-treated phi X174 templates by prokaryotic and eukaryotic DNA polymerases. Proc Natl Acad Sci U S A. 1981 Jan;78(1):110–114. doi: 10.1073/pnas.78.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano R. L., Lambert I. B., Fuchs R. P. DNA sequence determinants of carcinogen-induced frameshift mutagenesis. Biochemistry. 1994 Feb 15;33(6):1311–1315. doi: 10.1021/bi00172a004. [DOI] [PubMed] [Google Scholar]
- Parker B. O., Marinus M. G. Repair of DNA heteroduplexes containing small heterologous sequences in Escherichia coli. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1730–1734. doi: 10.1073/pnas.89.5.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan M., Lu C., Woodgate R., O'Donnell M., Goodman M. F., Echols H. Activity of the purified mutagenesis proteins UmuC, UmuD', and RecA in replicative bypass of an abasic DNA lesion by DNA polymerase III. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10777–10781. doi: 10.1073/pnas.89.22.10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy M. K., Weitzel S. E., von Hippel P. H. Assembly of a functional replication complex without ATP hydrolysis: a direct interaction of bacteriophage T4 gp45 with T4 DNA polymerase. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3211–3215. doi: 10.1073/pnas.90.8.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp W. D., Wilde C. E., 3rd, Reno D. L., Howard-Flanders P. Exchanges between DNA strands in ultraviolet-irradiated Escherichia coli. J Mol Biol. 1971 Oct 14;61(1):25–44. doi: 10.1016/0022-2836(71)90204-x. [DOI] [PubMed] [Google Scholar]
- Schmid S. E., Daune M. P., Fuchs R. P. Repair and mutagenesis of plasmid DNA modified by ultraviolet irradiation or N-acetoxy-N-2-acetylaminofluorene. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4133–4137. doi: 10.1073/pnas.79.13.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick S. G. Misrepair of overlapping daughter strand gaps as a possible mechanism for UV induced mutagenesis in UVR strains of Escherichia coli: a general model for induced mutagenesis by misrepair (SOS repair) of closely spaced DNA lesions. Mutat Res. 1976 Dec;41(2-3):185–200. doi: 10.1016/0027-5107(76)90091-9. [DOI] [PubMed] [Google Scholar]
- Sommer S., Bailone A., Devoret R. The appearance of the UmuD'C protein complex in Escherichia coli switches repair from homologous recombination to SOS mutagenesis. Mol Microbiol. 1993 Dec;10(5):963–971. doi: 10.1111/j.1365-2958.1993.tb00968.x. [DOI] [PubMed] [Google Scholar]
- Steinborn G. Uvm mutants of Escherichia coli K12 deficient in UV mutagenesis. I. Isolation of uvm mutants and their phenotypical characterization in DNA repair and mutagenesis. Mol Gen Genet. 1978 Sep 20;165(1):87–93. doi: 10.1007/BF00270380. [DOI] [PubMed] [Google Scholar]
- Veaute X., Fuchs R. P. Greater susceptibility to mutations in lagging strand of DNA replication in Escherichia coli than in leading strand. Science. 1993 Jul 30;261(5121):598–600. doi: 10.1126/science.8342022. [DOI] [PubMed] [Google Scholar]
- Witkin E. M. Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol Rev. 1976 Dec;40(4):869–907. doi: 10.1128/br.40.4.869-907.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgate R., Rajagopalan M., Lu C., Echols H. UmuC mutagenesis protein of Escherichia coli: purification and interaction with UmuD and UmuD'. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7301–7305. doi: 10.1073/pnas.86.19.7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgate R., Sedgwick S. G. Mutagenesis induced by bacterial UmuDC proteins and their plasmid homologues. Mol Microbiol. 1992 Aug;6(16):2213–2218. doi: 10.1111/j.1365-2958.1992.tb01397.x. [DOI] [PubMed] [Google Scholar]




