Abstract
β-barrel proteins, or outer membrane proteins (OMPs), perform many essential functions in Gram-negative bacteria, but questions remain about the mechanism by which they are assembled into the outer membrane (OM). In Escherichia coli, β-barrels are escorted across the periplasm by chaperones, most notably SurA and Skp. However, the contributions of these two chaperones to the assembly of the OM proteome remained unclear. We used differential proteomics to determine how the elimination of Skp and SurA affects the assembly of many OMPs. We have shown that removal of Skp has no impact on the levels of the 63 identified OM proteins. However, depletion of SurA in the skp strain has a marked impact on the OM proteome, diminishing the levels of almost all β-barrel proteins. Our results are consistent with a model in which SurA plays a primary chaperone role in E. coli. Furthermore, they suggest that while no OMPs prefer the Skp chaperone pathway in wild-type cells, most can use Skp efficiently when SurA is absent. Our data, which provide a unique glimpse into the protein content of the non-viable surA skp mutant, clarify the roles of the periplasmic chaperones in E. coli.
Keywords: βeta-barrel, periplasm, proteomics, chaperone, outer membrane
1 Introduction
The outer membrane (OM) of Gram-negative bacteria is an essential organelle that functions as a permeability barrier protecting them from a wide variety of toxic molecules [1-3]. The OM is unique among biological membranes; it is an asymmetric lipid bilayer with lipopolysaccharide (LPS) in the outer leaflet and phospholipids in the inner leaflet [4]. There are two major classes of proteins associated with the OM: lipoproteins, most of which are present in the periplasm but are anchored by a lipid moiety to the inner leaflet of the OM, and integral membrane proteins which generally adopt a β-barrel conformation, and are often referred to simply as β-barrels or OMPs [2, 5-7].
Many proteins that play essential roles in OM assembly have been recently identified, such as the LptD/E complex [4, 8, 9] and the 5-member Bam complex [10], which are involved in LPS and β-barrel insertion in the OM, respectively. Despite these recent breakthroughs, we still have a relatively poor understanding of how OM components, which are synthesized in the cytoplasm or at the inner membrane, are transported across the periplasm and inserted into the OM.
OMPs, which enter the periplasm as unfolded polypeptides, need to cross this hydrophilic compartment before being inserted into the OM. It is generally considered that unfolded β-barrels interact with periplasmic chaperones that prevent their aggregation in this compartment and facilitate their delivery to the Bam assembly complex [3, 11, 12].
In agreement with this model, several proteins that assist in the transport of OMPs across the periplasm have been identified. These proteins include protein-folding catalysts such as peptidyl-prolyl cis/trans isomerases (PPIases) [13] and proteins that catalyze the formation and isomerization of protein disulfide bonds [14, 15]. Two proteins, SurA and Skp, exhibit general chaperone activity and have been shown to play a role in β-barrel transport [13]. These two chaperones operate in two different, partially redundant, pathways, as indicated by the synthetically lethal phenotype of the surA skp double mutant [16].
SurA is a PPIase that has general chaperone activity, and is known to be involved in the assembly of OMPs [17-19]. Strains lacking surA have a decreased OM density [20] and are hypersensitive to detergents and hydrophobic antibiotics [18, 19], which is indicative of OM perturbations. Recently, we used a differential proteomic approach based on label-free two-dimensional liquid chromatography tandem mass spectrometry (2D-LC-MS/MS) analysis to study the impact of surA deletion on the OM proteome [21]. We found that several OMPs, including FhuA and LptD, and the major β-barrels OmpA, OmpF and OmpX, were less abundant in strains lacking SurA. However, these studies were complicated by the fact that the lack of SurA causes massive induction of the σE stress response [18], resulting in the induction of sRNAs that decrease the synthesis of many OMPs [22-24]. FhuA and LptD were the only β-barrel proteins for which the decrease in abundance could not be attributed, at least in part, to decreased mRNA levels, which led us to identify these two proteins as substrates highly dependent on SurA for their assembly [21].
Skp is a small periplasmic protein that is homologous to the eukaryotic chaperone prefoldin and functions as a trimer [25]. Skp copurifies with numerous β-barrel proteins [26], binds denatured β-barrels such as OmpA, OmpC, OmpF and LamB with high selectivity [27] and forms stable complexes with refolded OmpA in vitro [28]. However, skp mutants have a much less severe phenotype than surA mutants. They are slightly more sensitive to hydrophobic antibiotics and detergents than wild-type strains, and although these mutants have been reported to have moderately decreased levels of the major β-barrels [27], other more recent papers demonstrate that the OMP defect in a skp mutant is minimal [20, 29]. Moreover, the loss of Skp has no effect on membrane density [20]. This could mean either that Skp plays a minor role in the transport of many OMPs, or that some minor OMPs are particularly dependent on Skp for their transport. Recent experiments in Neisseria meningitidis suggest that PorA and PorB, two major OMPs in that organism, use Skp preferentially [30].
The periplasm also contains a third protein, DegP, which plays a role in β-barrel transport and assembly. DegP forms large 12-mer and 24-mer oligomeric cages with an inner cavity that can accommodate and stabilize small folded proteins [31, 32]. DegP acts as a chaperone at low temperature, but as a protease at elevated temperature [33]. While the role of DegP as a protease is well-documented, the physiological role of its chaperone activity is less clear. Previous studies have shown that both surA degP and surA skp double mutants have a synthetically lethal phenotype, but that skp degP mutants do not [16]. Moreover, it was shown that a protease-deficient DegP protein can complement the degP surA synthetic lethality at temperatures up to 37° C [16]. This demonstrates that it is the loss of the chaperone activity of the DegP protein that is the cause of the synthetically lethal degP surA phenotype, not the loss of the protease activity. Thus, in the absence of SurA, the chaperone activity of both Skp and DegP is required to support growth [16].
Two models have been put forward to explain the functional relationship between SurA and Skp in E. coli. In the first model, SurA and Skp/DegP function in two parallel, partially redundant pathways [16]; the SurA pathway transports most β-barrel proteins across the periplasm, while the Skp/DegP pathway rescues substrates that fall off the SurA pathway [20]. The major argument in favor of this model is that depletion of SurA in a skp strain significantly decreases OM density due to the decreased delivery of OMPs to the OM, and that this depletion leads to cell death. In an alternative model, it has been suggested that Skp and SurA cooperate sequentially in β-barrel assembly: Skp interacts first with unfolded OMPs as they enter the periplasm, and SurA is involved in a later stage, assisting the folding of the β-barrel proteins when they arrive at the Bam insertion machinery [34]. We do not favor this model, as we would expect that two proteins functioning sequentially would not show the synthetically lethal phenotype that has been observed.
Clearly, fundamental questions about the role of these chaperone pathways in β-barrel assembly remain unanswered and addressing them will require the use of novel experimental approaches to overcome the technical difficulties inherent in working with membrane proteins. We sought to clarify the relationship between SurA, Skp, and their substrates using a differential proteomic approach based on 2D-LC-MS/MS. By monitoring protein abundance on a global level, this technique provides invaluable insight into how the loss of a chaperone affects the protein content of the cell envelope. We had previously determined the impact of surA deletion on the OM proteome [21]. Here, we determined how the elimination of Skp or of both Skp and SurA affects the OM protein content.
2 Materials and methods
Bacterial strains
Bacterial strains used in this study are described in Table S1 (Supporting Information). All strains are derivatives of the E. coli K-12 strain MC4100 [35]. All alleles were moved by P1 transduction using standard procedures [36]. The SurA depletion strain was constructed by transducing the λInCh pBAD-surA-bla construct from MB292 [20] into an MC4100 strain containing Δ(araBAD)567 from the BW25113 strain. The resulting strain was then transduced with the ΔsurA::kan Keio allele from the Keio collection [37], and the kan cassette was flipped out using the unstable plasmid pCP20 [38], generating strain JAS260. JAS261 was constructed by transducing the Δskp::kan Keio allele [37] into JAS260, and flipping out the kan cassette [38]. To achieve more rapid depletion, a lysate made from the ara+ strain NR754 [39] was transduced into both strains. JAS417 was constructed in a similar way: the ara+ mutation was transduced into an MC4100 strain containing the cured Δskp allele from the Keio collection [37, 38] and the Δ(araBAD)567 from the BW25113 strain.
Strains NR744 and NR745 were constructed by transducing the zae-502::Tn10 insertion [40] either alone or linked to a Δskp allele [27] into MC4100. To confirm the presence of the skp deletion in NR745, the resulting tetR transductants were screened by PCR using primers external to the deletion. Unless indicated, all bacterial cultures were grown under aerobic conditions at 37°C in LB media. When appropriate, media was supplemented with 0.2% L-arabinose (w/v).
Growth conditions
NR744 and NR745 cells were grown aerobically in LB medium at 37°C to an OD600 of 0.8. Overnight cultures of the surA depletion strain (JAS260) and the surA depletion skp strain (JAS261) were grown in LB medium in the presence of 0.2% L-arabinose (w/v). The overnight cultures were then washed three times with fresh LB medium and subcultured at a 1:100 dilution in LB media with or without 0.2% L-arabinose. After 3 hours of growth, the cultures were diluted 100 fold in LB with and without 0.2% L-arabinose to an OD600 of about 0.03-0.04. Cells were grown for two more hours at 37°C.
Preparation of outer membrane fraction and proteolytic digestion
Envelope proteins were extracted by using the osmotic shock procedure as described by Vertommen et al. (2009) [21] and OM proteins isolated by ultracentrifugation at 180,000 × g for 1 h at 4°C. Protein concentration was determined using the Bradford assay.
150 μg of OM proteins were then precipitated by adding trichloroacetic acid (TCA) to a final concentration of 10% v/v, followed by incubation on ice for 30 min. Samples were centrifuged at 15,000 × g for 20 min and the resulting pellets washed with 5% ice-cold TCA (v/v). The pellets were then resuspended in 100 μl of denaturing buffer (6M urea, 200 mM Tris-HCl pH 8.5, 10 mM EDTA) supplemented with 10 mM dithiothreitol (DTT) to reduce disulfide bonds. After a one hour incubation at 25°C, we added 100 μl of denaturing buffer supplemented with 100 mM iodoacetamide to titrate out the remaining DTT and alkylate all reduced cysteines. The reaction was stopped by addition of 10% TCA (v/v) and the proteins were collected by centrifugation at 15,000 × g for 20 min. The resulting pellet was successively washed with 5% TCA (v/v) and 100% ice-cold acetone, dried in a Speedvac and resuspended by sonication in 50 μl 0.1 M NH4HCO3 pH 8.0, containing 0.2% (w/v) RapiGest (Waters). The samples were then diluted two fold with 50 mM NH4HCO3 pH 8.0 containing 3 μg sequencing grade trypsin, and digested overnight at 30°C. The acido-labile detergent was then removed by acidification with HCl according to the manufacturer’s instructions and the samples were stored at −20°C.
Differential analysis of OM proteins by label-free 2D-LC-MS/MS
Peptides were loaded onto a strong cation exchange column GROM-SIL 100 SCX (100 × 2 mm, GROM, Rottenburg, Germany) equilibrated with solvent A (5% acetonitrile v/v, 0.05% v/v formic acid pH 3.5 in water) and connected to an Agilent 1100 HPLC system. Peptides were separated using a 50 min elution gradient that consisted of 0%-50% solvent B (5% acetonitrile v/v, 1 M ammonium formate adjusted to pH 3.5 with formic acid in water) at a flow rate of 200 μl/min. Absorbance was monitored at 280 nm to ensure that all samples contained similar amount of material. Fractions were collected at 2 min intervals (20 in total) and dried using a Speedvac. Peptides were resuspended in 10 μl of solvent C (5% acetonitrile v/v, 0.1% v/v TFA in water) and analyzed by LC-MS/MS as described below.
The LC-MS/MS system consisted of an LTQ XL ion trap mass spectrometer (ThermoFisher, San José, CA, USA) equipped with a microflow electrospray ionization source and interfaced to an LCPackings Ultimate Plus Dual gradient pump, Switchos column switching device, and Famos Autosampler (Dionex, Amsterdam, Netherlands). Two reverse phase peptide traps C18 Pepmap 100 Dionex (0.30 mm × 5 mm) were used in parallel with two analytical BioBasic-C18 columns from ThermoFisher (0.18 mm × 150 mm). Samples (6.5 μl) were injected and desalted on the peptide trap equilibrated with solvent C at a flow rate of 30 μl/min. After valve switching, peptides were eluted in backflush mode from the trap onto the analytical column equilibrated in solvent D (5% acetonitrile v/v, 0.1% v/v formic acid in water) and separated using a 100 min gradient from 0% to 70% solvent E (80% acetonitrile v/v, 0.1% formic acid in water) at a flow rate of 1.5 μl/min.
The mass spectrometer in positive mode was set up to acquire one full MS scan in the mass range of 400-2000 m/z, followed by five MS/MS spectra of the five most intense peaks in the mass range 400-1500 m/z. The dynamic exclusion feature was enabled to obtain MS/MS spectra on co-eluting peptides, and the exclusion time was set at 1 min.
Protein identification and quantification
Raw data collection of approximately 130,000 MS/MS spectra per 2D-LC-MS/MS experiment was followed by protein identification using Sequest. In detail, peak lists were generated using spectrum selector in Proteome Discoverer 1.2. From raw files, MS/MS spectra were exported as individuals files in .dta format with the following settings: peptide mass range: 400-3500 Da, minimal total ion intensity: 500, minimal number of fragment ions: 12, precursor mass tolerance: 1.1 Da. The resulting peak lists were searched against a target-decoy E. coli protein database (release 18.01.2007, 8690 entries comprising forward and reversed sequences) obtained from Swissprot (ftp://ftp.expasy.org/databases/complete_proteomes/fasta/) using Sequest by comparison with the theoretical spectra of all possible peptides fragments from the target-decoy database. The following parameters were used: trypsin was selected with proteolytic cleavage only after arginine and lysine but not at Arg/Pro or Lys/Pro, number of internal cleavage sites was set to 1, mass tolerance for precursor and fragment ions was 1.1 Da and 1.0 Da, respectively, considered variable modifications were +15.99 Da for oxydized methionine and +57.02 Da for carboxyamidomethylcysteine. Peptide matches were filtered using charge-state versus cross-correlation scores (Xcorr) ensuring an estimated false-positive rate below 5% calculated by target-decoy database searching. The filtered Sequest output files for each peptide were grouped according to the protein from which they were derived using the multiconsensus results tool within Proteome Discoverer. The analysis was repeated on three biological replicates for each strain. Sampling statistics such as unique peptides, spectral counts and sequence coverage were exported in Microsoft Excel spreadsheets. The spectral counts data were normalized by dividing the protein spectral count in a particular experiment by the average spectral count across all the proteins in that experiment. Relative quantification of protein abundance was estimated by calculating, after normalization, the ratio of spectral counts. Statistical significance was tested with the unpaired Student’s t-test and significance was defined as a P < 0.05 (two-tail two-sample equal variance test). The extracted peak list for all datasets were exported in Mascot Generic Format and made freely available at the Tranche Network (http//proteomecommons.org) under the following Hash code: c/huk7kU9mKvkgIJ7A7RtYb6YfvAAWe1vnnwk5jPhOlj2uwlzvxrmnuZxvi4DbpvpKFu5eocxb mC8LoK3FVyuFSldeAAAAAAAAAGuQ==
Immunoblotting
Complete depletion of SurA in either the surA depletion strain (JAS260) or the surA depletion skp strain (JAS261) was achieved as described in Growth conditions. The skp strain (JAS417) was subjected to the same treatment as the depletion strain. At 5 hours after the initial subculture, 1 mL samples were pelleted (16,000 × g, 1 min), and re-suspended at a volume equal to the OD600/7 in SDS sample buffer containing β-mercaptoethanol and boiled. 10 μL of each re-suspended sample was analyzed by SDS-PAGE. Immunoblotting was done using rabbit anti-LamB antibody (which cross-reacts with OmpA) [41] at a 1:30,000 dilution and rabbit anti-BamA antibody [42] at a 1:20,000 dilution. Donkey anti-rabbit antibody conjugated to horseradish peroxidase (GE Life Sciences) was used at a dilution of 1:8,000, with the ECL antibody detection kit (GE Life Sciences) to visualize the Western blots.
RNA extraction and RT-qPCR
Depletion of SurA was achieved as described under Growth conditions. RNA was prepared from 1 mL of a 5 mL culture 5 hours after the initial subculture using the Ambion Ribopure™-Bacteria Kit, including the optional DNase I treatment described in the manual. cDNA was made from the RNA using the Applied Biosystems High Capacity cDNA Reverse Transcription Kit using the protocol described in the manual. The Reverse Transcriptase was inactivated by the addition of 15 μL 1 N NaOH. After a 30 minute incubation at 65°C, 15 μL of 1 N HCl were added. The cDNA was purified using the Qiagen MinElute PCR Purification Kit in an elution volume of 15 μL. The purified cDNA was then diluted 1:40. The qRT-PCR was performed using Applied Biosystems 7900 HT Fast Real-Time PCR System, in 25 μL reactions using PerfeCTa™ SYBR® Green FastMix™ ROX by Quanta Biosciences, Inc. Analysis was done using Applied Biosystems SDS software, version 2.3. Cycle threshold (Ct) values were determined after automatic adjustment to the baseline. For comparative, quantitative analysis, transcript levels were normalized to the level of rpoD and the ΔCt was determined. The comparative quantitation method (ΔΔCt) was used to compare the different strains and transformed to absolute values for obtaining relative fold changes. All the assays were performed in triplicate on each of three independent RNA preparations.
3 Results
Proteomic analysis of the skp strain
We first examined the impact of skp deletion using a more comprehensive and precise approach than had been previously undertaken by comparing the OM protein content of the NR745 skp strain to that of the NR744 wild-type strain by label-free 2D-LC-MS/MS. This approach allows a global and semi-quantitative analysis of protein expression ratios. This is a reliable and powerful technique that recently allowed us to identify the substrates of the protein folding catalysts DsbA and DsbC [43] and to characterize the role of the chaperone SurA in β-barrel assembly [21]. For quantification of protein abundance, we used the number of spectral counts (SC) reported for every protein (See Supporting Information Tables S2 and S3). The number of SC for a protein is the total number of MS/MS spectra taken on peptides from this protein in a given 2D-LC-MS/MS analysis. This value is linearly correlated with the protein abundance [21, 43, 44]. It is important to note that the number of SC can be used to compare the abundance of a given protein between two different strains but not to compare the abundance of different proteins in the same biological sample [21].
In order to identify the β-barrel proteins that are affected by the loss of Skp, we prepared OM extracts from wild-type and skp strains using a protocol similar to that described in [21]. Briefly, an extract containing periplasmic and OM proteins was prepared, the OM proteins were isolated by ultracentrifugation and acid-precipitated. After solubilization with an acid-labile surfactant, the OM proteins were digested with trypsin and the pool of tryptic peptides was analyzed by 2D-LC-MS/MS. The experiments were repeated using three independent cultures for both strains. In total, 120 raw files were submitted to a database search using Sequest against a target-decoy E. coli database obtained from SwissProt. Results were filtered using charge state versus cross-correlation scores (Xcorr) and peptide probabilities to obtain a false positive rate below 5% calculated according to the formula (FPR= 2x false positives (FP)/(false positives (FP) + true positives (TP)).
An average of 786 ± 98 and 766 ± 102 proteins were identified for the wild-type and skp strain, respectively. These proteins included up to 90 OM proteins (lipoproteins and β-barrel proteins) as well as soluble proteins that contaminated the OM fraction. Importantly, 19 of the 20 proteins with the highest SC values corresponded to envelope proteins. However, only 63 OM proteins (23 β-barrel proteins and 40 lipoproteins) that could be reproducibly identified at least twice in the experiments under either condition were kept for further analysis. The SC values of those proteins are presented in the Supporting Information Table 2. We noted that the SC values of most proteins are similar in all three independent analyses of the same strain, which highlights the reproducibility of our approach. The average spectral count across all the proteins was 24.9 ± 1.0 and 22.2 ± 3.8 for the wild-type and skp strain, respectively, which denotes a similar sampling rate between samples from the two strains. We used these values for normalization (see Supporting Information Table 2). Moreover, the total number of SC reported for the OM proteins was similar for the wild-type and skp strains (see Supporting Information Table S2), which confirms that the amount of OM material used for the proteomic analysis was similar.
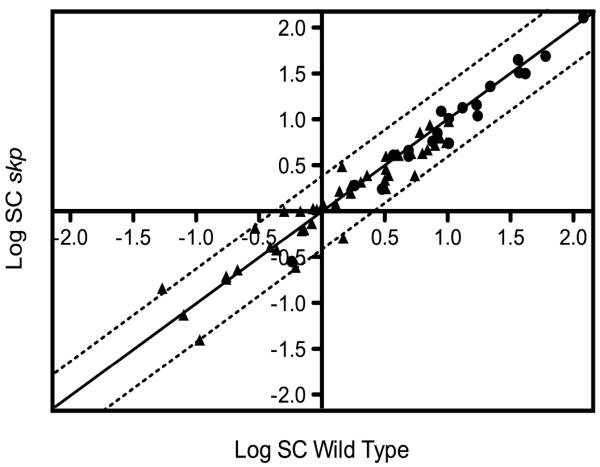
To identify β-barrel proteins that are substrates of Skp, we searched for proteins that were significantly less abundant in the OM of the skp strain. We consider that the abundance of a protein is changed if it is decreased or increased by at least 2-fold. To test the significance of the data, we used the unpaired Student’s t test and defined significance as a P < 0.05 (2-tail 2-sample equal variance test). We did not identify a single OMP that was significantly affected by the deletion of skp. As we might have expected, given the dependence of lipoproteins on the LolA pathway rather than the SurA or Skp/DegP pathways [7], only one lipoprotein (LptE) out of the 40 identified was present at decreased levels and none were present at increased levels. However, the decrease in LptE levels is barely within our defined criteria for significance described above, and we were unable to confirm this decrease by Western blot analysis. Thus, the skp mutant has an envelope proteome that is indistinguishable from that of the wild type, as reflected by the linear distribution observed when the SC values from the skp strain are plotted against those from the wild type (Figure 1). We confirmed the results of the 2D-LC-MS/MS analysis for a few selected proteins such as LamB, OmpA and BamA using Western blot analysis. Because OMPs that do not reach the OM are degraded in the periplasm, the levels of these proteins in whole-cell extracts are indicative of levels in the OM. We confirm that the levels of those proteins remain largely unchanged in a skp strain (JAS417) (Figure 4).
Figure 1. skp deletion has no effect on the OM proteome.
Two-dimensional logarithmic plot for the mean SC values of the identified lipoproteins and β-barrel proteins. Spots on the diagonal and between the two dotted lines correspond to proteins that are not significantly affected by the loss of Skp (they differ in their SC values by a factor of less than 2). Closed circles (●) correspond to β-barrel proteins and closed triangles (▴) to lipoproteins. Only one spot corresponding to the LptE lipoprotein (above the upper dotted line) is decreased by more than twofold in the skp strain (NR745).
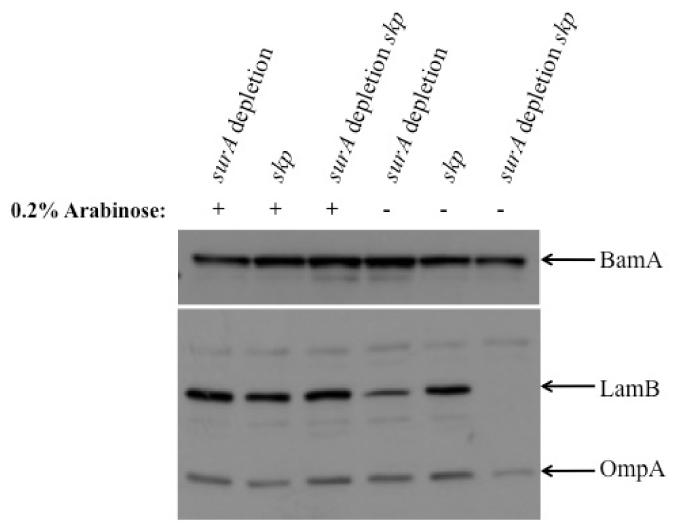
Figure 4. Western blot analysis confirms the proteomic results.
Depletion of SurA in JAS260 (surA depletion strain) leads to increased levels of BamA (compare lane 4 to lane 1), as reported in our previous proteomic analysis of that strain [21]. When SurA is depleted in the surA depletion skp strain (JAS261), BamA levels appear only slightly decreased (compare lane 6 to lane 3), in good agreement with our 2D-LC-MS/MS analysis where they are not changed. The levels of LamB and OmpA are greatly decreased when the surA depletion skp strain (JAS261) is grown under non-permissive conditions (compare lane 6 to lane 3), as observed in the 2D-LC-MS/MS experiments. The levels of all proteins examined remained largely unchanged in the skp strain (JAS417) (lanes 2 and 5).
Proteomic analysis of the surA depletion skp strain
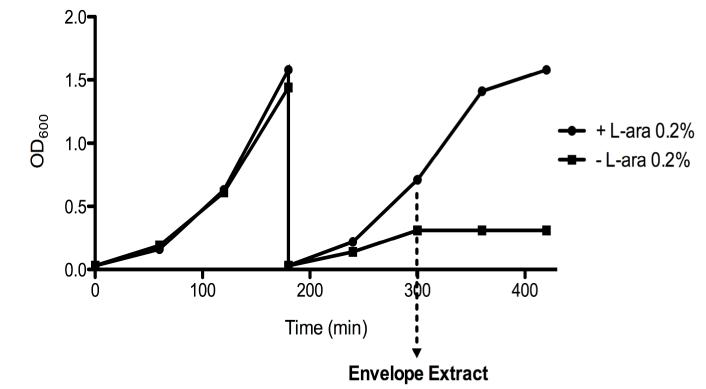
We further investigated Skp’s role, as well as its functional relationship to SurA, by determining how the elimination of both SurA and Skp impacts the OM proteome. As explained above, surA skp mutants have a synthetically lethal phenotype [16]. In order to circumvent the lethality of the double mutant, we used a depletion strain. This strain is a double surA skp mutant in which a copy of the surA gene under the control of the araBAD promoter is inserted into the chromosome at the λ attachment site (strain JAS261). When cultured in the presence of L-arabinose, this strain is viable because SurA is produced. However, subculture of the strain in the absence of L-arabinose leads to the progressive depletion of SurA, growth of the strain slows and then ceases (Figure 2). Thus, this strain offers the unique opportunity to look at the physiological consequences of the loss of both SurA and Skp.
Figure 2. Bacteria cease growing upon SurA depletion in the skp strain.
After 3 hours of growth at 37°C, cells were diluted 100-fold in a medium with or without 0.2% of L-arabinose. After two more hours, envelope proteins were extracted by an osmotic shock procedure. Growth of the surA depletion skp strain requires L-arabinose for viability. In the presence of L-arabinose (●), the depletion strain exhibits normal growth. In the absence of L-arabinose (■), cells cease growing. The data of a representative experiment are shown.
The surA depletion skp strain was first grown overnight with L-arabinose, and diluted 1:100 in the presence or absence of L-arabinose. After 3 hours, the cells were diluted 100-fold in fresh medium (with or without L-arabinose). After two additional hours of growth, corresponding to the time at which the cells cultured in the non-permissive condition cease growing (Figure 2), envelope extracts were prepared and the OM proteins were analyzed by 2D-LC-MS/MS as explained above. This experiment was repeated on three independent cultures.
An average of 710 ± 183 and 1158 ± 461 proteins were identified when the surA depletion skp strain was grown under permissive and non-permissive conditions, respectively. Most of the additional proteins that were identified in the sample prepared under non-permissive conditions correspond to cytoplasmic proteins that were released during the osmotic shock procedure, probably because of the decreased envelope integrity of the surA skp double mutant. Here also, we analyzed only the 51 OM proteins (20 β-barrel proteins and 31 lipoproteins) that could be reproducibly identified at least twice in the experiments under either condition. The SC values of those proteins are presented in the Supporting Information Table S3. The average spectral count across all the proteins (9.6 ± 2.8 and 5.7± 1.5 when the strain was grown with or without L-arabinose, respectively) was not statistically different (p=0.09), indicating that the sampling rate is similar between samples prepared under the two conditions. We used these values for normalization (see Supporting Information Table S3).
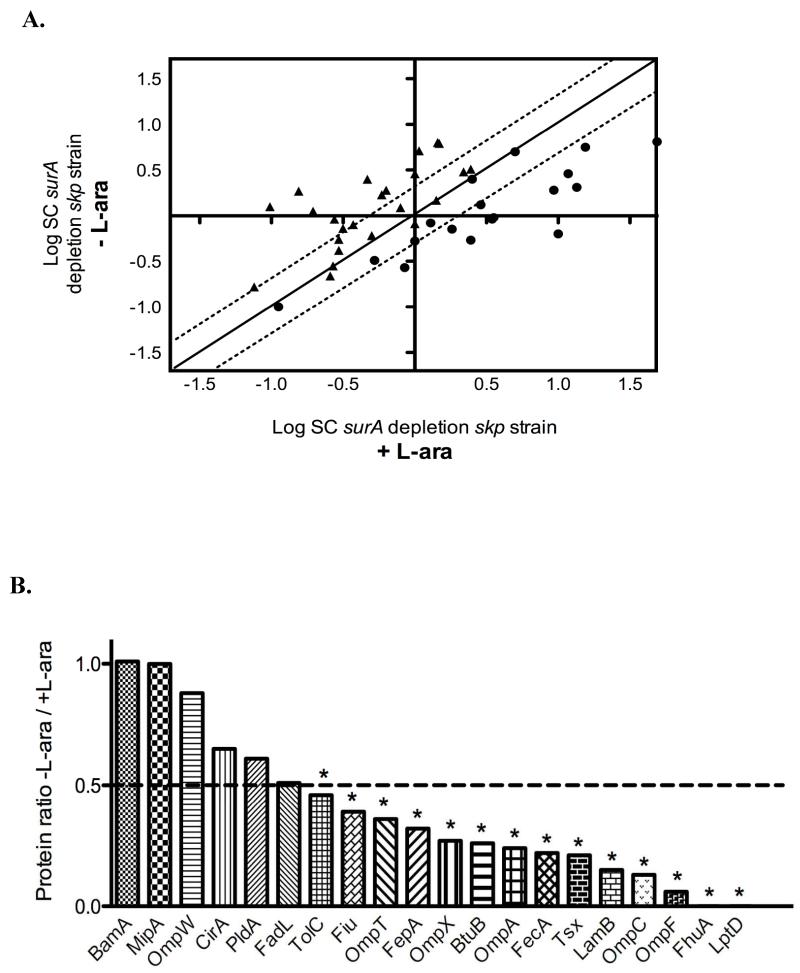
Depleting SurA in a skp strain has a marked impact on the OM proteome and affects the abundance of both β-barrel proteins and lipoproteins. This is reflected by the dispersed distribution of the SC values when those reported for proteins from the surA depletion skp mutant grown with L-arabinose are plotted against those from the same strain grown in the absence of L-arabinose (Figure 3A).
Figure 3. The loss of SurA and Skp has a severe impact on the OM proteome.
A. Two-dimensional logarithmic plot for the mean SC values of the identified lipoproteins and β-barrel proteins from the surA depletion skp strain grown in the presence or absence of L-arabinose. Spots on the diagonal and between the two dotted lines correspond to proteins that are not significantly affected by the depletion of SurA (they differ in their SC values by a factor of less than 2). Spots that are above the upper dotted line or below the lower dotted line correspond to proteins whose abundance is increased or decreased by more than two fold in the surA depletion skp strain, respectively. Closed circles (●) correspond to β-barrel proteins and closed triangles (▴) to lipoproteins. B. Ratio of OM proteins calculated by considering that SC values obtained for proteins from the surA depletion skp strain grown with L-arabinose are equal to 1. SurA depletion in the surA depletion skp strain leads to a significant decrease (more than two fold) in the abundance of 14 β-barrel proteins. The OM proteins for which the decrease is statistically significant (p< 0.05) are indicated by an asterisk.
We found that SurA depletion in the skp strain leads to a significant decrease in the abundance of 14 β-barrel proteins (representing about 70% of the identified β-barrels) (Figure 3B and Table 1). These proteins include the abundant proteins OmpA, OmpF and LamB, the unusual β-barrel protein TolC, as well as FhuA and LptD. The latter two proteins, which were identified previously as SurA-dependent substrates [21], were the most affected by the depletion of SurA. An additional protein, FadL, is also decreased but fails the t-test. The proteomic data were confirmed for a few selected proteins, such as LamB and OmpA, using Western blot analysis (Figure 4).
Table 1.
Outer membrane proteins identified by 2-D LC-MS/MS in the surA depletion skp strain
| β-barrel proteins | Ratioa) | Lipoproteins | Ratioa) |
|---|---|---|---|
| BamA | 1.01 | YbjP | 13.07c) |
| MipA | 1.00 | YajI | 11.94c) |
| OmpW | 0.88 | LptE | 5.78c) |
| CirA | 0.65 | YedD | 5.33c) |
| PldA | 0.61 | BamD | 4.78c) |
| FadL | 0.51 | BamC | 4.31c) |
| TolC | 0.46 | YajG | 4.17c) |
| Fiu | 0.39 | OsmE | 3.35 |
| OmpT | 0.36 | LolB | 3.04c) |
| FepA | 0.32 | BamB | 2.90c) |
| OmpX | 0.27 | YdcL | 2.89c) |
| BtuB | 0.26 | MltC | 2.26 |
| OmpA | 0.24 | NlpI | 2.17 |
| FecA | 0.22 | CutF | 2.13 |
| Tsx | 0.21 | NlpD | 1.89 |
| LamB | 0.15 | MltA | 1.56 |
| OmpC | 0.13 | MltB | 1.43 |
| OmpF | 0.06 | Pal | 1.38 |
| FhuA | 0.00 | SlyB | 1.31 |
| LptD | 0.00 | VacJ | 1.20 |
| YggN | 1.04 | ||
| Lpp | 1.04 | ||
| YgeR | 0.83 | ||
| RlpA | 0.82 | ||
| YeaY | 0.00 | ||
| YncD | 0.00 | ||
| YddW | 0.00 | ||
| YgdI | b) | ||
| YbaY | b) | ||
| YgdR | b) | ||
| YifL | b) |
The ratios were calculated by considering that the SC values obtained for proteins from the depletion strain grown with L-arabinose are equal to 1.
Lipoproteins that were only detected when the strain was grown in the absence of L-arabinose.
Lipoproteins for which the ratio is statistically significant.
Interestingly, whereas most β-barrel proteins were affected by the loss of SurA and Skp, the levels of a subset of OM proteins did not appear to be significantly decreased. The most striking example is the essential β-barrel protein BamA, which appears to be present at approximately wild-type levels. This was also confirmed by Western blot analysis (Figure 4). The other β-barrel proteins that appear less affected by the simultaneous absence of SurA and Skp include the colicin IA receptor protein CirA, the phospholipase PldA, the MltA-interacting protein MipA and OmpW.
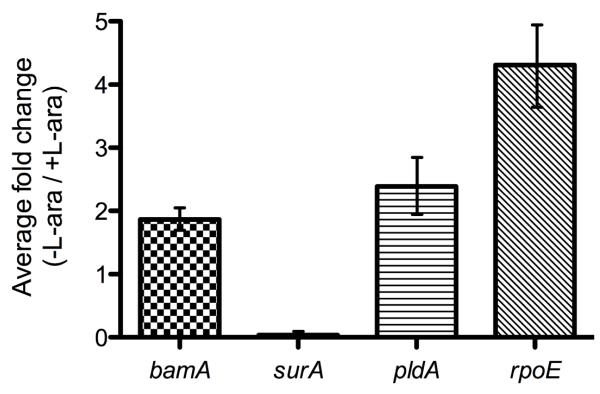
To further examine the contributions of transcription and assembly to the levels of select members of this group, we compared the levels of BamA and PldA mRNA by RT-qPCR when the surA depletion skp strain is grown with or without L-arabinose. As shown in Figure 5, we observed that growth under non-permissive conditions leads to the induction of bamA and pldA transcription by 1.9 and 2.4-fold, respectively. As we do not observe this same increase in the levels of BamA and PldA in the OM, it suggests that SurA depletion in the skp strain does affect the assembly of these proteins, neutralizing their increased synthesis.
Figure 5. Synthesis of bamA and pldA increases upon SurA depletion in the skp strain and masks the assembly defect.
The mRNA levels of BamA and PldA are increased (about 1.9 and 2.4-fold, respectively). Depletion of SurA also induces the rpoE gene, responsible for the induction of the σE response (about 4.3-fold induction).
Upon analysis of the OM lipoproteins, we found that the abundance of 14 lipoproteins is significantly increased (Table 1) when the cells are grown under non-permissive conditions. These proteins include the Bam complex lipoproteins BamD (formerly known as YfiO) and BamC (formerly known as NlpB) and the LptD partner LptE, which all belong to the σE regulon [24]. This increase in levels, together with the increased synthesis of bamA, suggested that SurA depletion in the skp strain activates the σE response, which was verified by RT-qPCR (Figure 5). However, no lipoproteins are significantly decreased upon SurA depletion in the skp strain, which is consistent with their assembly depending on the dedicated chaperone LolA [7].
4 Discussion
The roles of SurA and Skp in β-barrel assembly have been studied using bacterial genetics and biochemistry. However, it has never been possible to fully characterize the OMP assembly defect in strains lacking these chaperones. Instead, previous studies have examined only a few model proteins. Clearly, there is a need for more comprehensive approaches in order to determine how the absence of SurA and Skp affects the global OM protein content and not only the few β-barrel proteins that constitute the bulk mass of OMPs.
In our analysis of the OM proteome of the skp mutant, we found no β-barrel proteins that were present at significantly decreased levels in the OM when compared to the wild-type strain. This indicates that SurA alone is sufficient to transport most β-barrel proteins across the periplasm. Perhaps surprisingly, given the strong dependence of some OM proteins (such as LptD) on the SurA pathway [21], no β-barrel proteins seem to strongly depend on the Skp/DegP pathway for their assembly, suggesting that there are no OMPs that use the skp pathway preferentially.
This new data is striking when compared to recent analyses of Skp’s involvement in OMP biogenesis in other species of bacteria. In Neisseria meningitidis, levels of the OMPs PorA and PorB are decreased in a skp mutant, but not in a surA mutant [30]. It was also reported that there is no synthetic lethality associated with the surA skp strain in that bacterial species [30]. In Shigella flexneri, Skp is crucial for the assembly of the autotransporter IcsA. Remarkably, however, it is the N-terminal passenger domain of this protein that requires Skp, not the C-terminal β-barrel domain [45, 46]. Conversely, in E. coli at 25°, it has recently been shown that Skp interacts with the β-barrel domain of the autotransporter EspP [47]. It is clear that even homologous chaperones in different species function quite differently.
The new data also reveal that many OMPs in E. coli show no pathway preference. CirA, PldA and others were previously shown to be unaffected in a surA mutant [21] and are now shown to be unaffected in the skp mutant. However, in the absence of both chaperone pathways, the levels of almost every OMP we were able to analyze were decreased in the OM. Clearly, some proteins can use either pathway available to them, but require at least one of the two for efficient transport.
The ability of the Skp/DegP backup pathway to transport OMPs in the absence of the main SurA pathway may be additionally aided by the decrease in levels of the mRNAs for some of the most abundant β-barrels, such as OmpA and LamB, mediated by the σE stress response [24]. The reduced synthesis of these β-barrel proteins decreases the number of OMPs that the Skp/DegP backup pathway must chaperone, and so this pathway is sufficient, even in the absence of SurA. In fact, the levels of some OMPs are even higher in the OM of the surA strain [21], which is likely due to the combination of an increase in the synthesis of these proteins and the decreased load placed on the remaining chaperones and the Bam assembly complex, as has been suggested previously [21, 48, 49].
Although most OMPs are affected by the loss of both Skp and SurA, for a small number of proteins including BamA and PldA, there is no apparent decrease in OM abundance in the surA depletion skp strain. These proteins could depend on a third, unknown, chaperone pathway for their assembly, or they could be slower to deplete from the OM once SurA and Skp are gone. Alternatively, the unchanged levels of these proteins could result from the combination of two opposing processes: an increased synthesis of the protein and a decreased assembly in the cell envelope. Upon examining the synthesis of BamA and PldA, we found that their mRNA levels were indeed increased in the surA depletion skp mutant. Taking this increase in mRNA levels into account, we find that the seemingly unaffected PldA levels represent an assembly defect of about 75%. It is possible that all of the proteins with levels that seem relatively unaffected in the surA depletion skp strain follow this same pattern: an increase in synthesis masks their assembly defect.
It should be noted, however, that BamA appears to be an exceptional case. Taking the increase in mRNA levels into account, we find that BamA levels could represent an assembly defect of 50%. Clearly, even in the absence of both chaperones, a significant portion of it is able to reach the OM. As has been suggested by Bennion et al. [50], BamA’s five Polypeptide-Transport-Associated (POTRA) [51] domains may play a role in preventing its aggregation in the periplasm. Thus, more work is required to clarify the chaperone requirement in the assembly pathway for BamA.
Consistent with the idea that the Skp/DegP pathway does not play a central role in β-barrel assembly when SurA is present, we did not observe the same increase in the levels of σE-regulated OM lipoproteins in the skp strain that was observed in a surA strain [21] or in the surA depletion skp strain. This suggests that the σE response is not strongly induced, and consistent with earlier reports, that the OM is not greatly perturbed in the absence of Skp [29]. However, levels of many σE-regulated OM lipoproteins are increased in the surA depletion skp strain [24]. Because OM lipoproteins are transported by the Lol pathway, independently of the β-barrel chaperones, an increase in transcription due to increased σE activity can be directly observed as an increase in levels at the OM (Figure 5). It is clear that even when SurA is depleted quickly, the OM assembly defect is severe enough to induce the σE response when Skp is also absent.
In the absence of SurA, the Skp/DegP pathway becomes essential for the transport of OMPs. In the absence of both pathways, levels of all the β-barrels that we could detect are decreased. Apparently, when the SurA and Skp/DegP pathways are compromised simultaneously, there is no chaperone pathway available to transport β-barrels to the OM. The global impact of the loss of both of these pathways means that we could obtain no evidence supporting the presence of an additional chaperone pathway in E. coli. There are a number of other periplasmic protein folding factors, including FkpA, PpiA, and Spy [1, 52]. These proteins may play a role in OMP assembly. However, these proteins by themselves cannot support the assembly of any OMP we could identify.
Supplementary Material
Acknowledgements
We thank Geneviève Connerotte and Steve Calberson for technical assistance and Pauline Leverrier for critical reading of the manuscript. We also thank Bonnie Bassler, Ammar Zafar, and Julia van Kessel for use of their qRT-PCR machine and their assistance with the procedure and data analysis, and Natividad Ruiz for strains (NR744 & 745) and helpful discussion. DV is Collaborateur Logistique and JFC is Chercheur Qualifié of the FRS-FNRS. KD is a research fellow of the FRIA. This work was supported by the Interuniversity Attraction Pole Programme - Belgian Science Policy to JFC (network P6/05), and by grants from the FRS-FNRS to JFC. JS is supported by the Genetics and Molecular Biology Training Grant from the NIH GM07388 and T.J.S. is supported by NIGMS grant 34821.
Abbreviations
- OM
Outer Membrane
- OMP
Outer Membrane Protein (β-barrel)
- LPS
Lipopolysaccharides
- PPIase
Peptidyl-Prolyl cis-trans Isomerase
- LB
Luria Bertani
- 2D-LC-MS/MS
Two-Dimensional Liquid Chromatography Tandem Mass Spectrometry
- EDTA
Ethylenediaminetetraacetic Acid
- DTT
Dithiothreitol
- TCA
Trichloroacetic Acid
- TFA
Trifluoroacetic Acid
- SC
Spectral Count
- IM
Inner Membrane
- POTRA
Polypeptide-Transport-Associated
Footnotes
Conflict of interest statement
The authors have declared no conflict of interest.
References
- [1].Leverrier P, Vertommen D, Collet JF. Contribution of proteomics toward solving the fascinating mysteries of the biogenesis of the envelope of Escherichia coli. Proteomics. 2009;9:2432–43. doi: 10.1002/pmic.200900461. [DOI] [PubMed] [Google Scholar]
- [2].Bos MP, Robert V, Tommassen J. Biogenesis of the gram-negative bacterial outer membrane. Annu Rev Microbiol. 2007;61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. [DOI] [PubMed] [Google Scholar]
- [3].Knowles TJ, Scott-Tucker A, Overduin M, Henderson IR. Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat Rev Microbiol. 2009;7:206–214. doi: 10.1038/nrmicro2069. [DOI] [PubMed] [Google Scholar]
- [4].Ruiz N, Kahne D, Silhavy TJ. Transport of lipopolysaccharide across the cell envelope: the long road of discovery. Nat Rev Microbiol. 2009;7:677–683. doi: 10.1038/nrmicro2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tokuda H. Biogenesis of outer membranes in Gram-negative bacteria. Biosci Biotechnol Biochem. 2009;73:465–473. doi: 10.1271/bbb.80778. [DOI] [PubMed] [Google Scholar]
- [7].Tokuda H, Matsuyama S. Sorting of lipoproteins to the outer membrane in E. coli. Biochim Biophys Acta. 2004;1694 IN1-9. [PubMed] [Google Scholar]
- [8].Wu T, McCandlish AC, Gronenberg LS, Chng SS, et al. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A. 2006;103:11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ruiz N, Falcone B, Kahne D, Silhavy TJ. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell. 2005;121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- [10].Wu T, Malinverni J, Ruiz N, Kim S, et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- [11].Hagan CL, Kim S, Kahne D. Reconstitution of outer membrane protein assembly from purified components. Science. 2010;328:890–892. doi: 10.1126/science.1188919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ruiz N, Kahne D, Silhavy TJ. Advances in understanding bacterial outer-membrane biogenesis. Nat Rev Microbiol. 2006;4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- [13].Duguay AR, Silhavy TJ. Quality control in the bacterial periplasm. Biochim Biophys Acta. 2004;1694:121–134. doi: 10.1016/j.bbamcr.2004.04.012. [DOI] [PubMed] [Google Scholar]
- [14].Depuydt M, Messens J, Collet JF. How Proteins Form Disulfide Bonds. Antioxid Redox Signal. 2011;15:49–66. doi: 10.1089/ars.2010.3575. [DOI] [PubMed] [Google Scholar]
- [15].Messens J, Collet JF. Pathways of disulfide bond formation in Escherichia coli. Int J Biochem Cell Biol. 2006;38:1050–1062. doi: 10.1016/j.biocel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- [16].Rizzitello AE, Harper JR, Silhavy TJ. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J Bacteriol. 2001;183:6794–6800. doi: 10.1128/JB.183.23.6794-6800.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ureta AR, Endres RG, Wingreen NS, Silhavy TJ. Kinetic analysis of the assembly of the outer membrane protein LamB in Escherichia coli mutants each lacking a secretion or targeting factor in a different cellular compartment. J Bacteriol. 2007;189:446–454. doi: 10.1128/JB.01103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rouviere PE, Gross CA. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev. 1996;10:3170–3182. doi: 10.1101/gad.10.24.3170. [DOI] [PubMed] [Google Scholar]
- [19].Lazar SW, Kolter R. SurA assists the folding of Escherichia coli outer membrane proteins. J Bacteriol. 1996;178:1770–1773. doi: 10.1128/jb.178.6.1770-1773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sklar JG, Wu T, Kahne D, Silhavy TJ. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007;21:2473–2484. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Vertommen D, Ruiz N, Leverrier P, Silhavy TJ, Collet JF. Characterization of the role of the Escherichia coli periplasmic chaperone SurA using differential proteomics. Proteomics. 2009;9:2432–2443. doi: 10.1002/pmic.200800794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gogol EB, Rhodius VA, Papenfort K, Vogel J, Gross CA. Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc Natl Acad Sci U S A. 2011;108:12875–12880. doi: 10.1073/pnas.1109379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Thompson KM, Rhodius VA, Gottesman S. SigmaE regulates and is regulated by a small RNA in Escherichia coli. J Bacteriol. 2007;189:4243–4256. doi: 10.1128/JB.00020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol. 2006;4:e2. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Korndorfer IP, Dommel MK, Skerra A. Structure of the periplasmic chaperone Skp suggests functional similarity with cytosolic chaperones despite differing architecture. Nat Struct Mol Biol. 2004;11:1015–1020. doi: 10.1038/nsmb828. [DOI] [PubMed] [Google Scholar]
- [26].Jarchow S, Luck C, Gorg A, Skerra A. Identification of potential substrate proteins for the periplasmic Escherichia coli chaperone Skp. Proteomics. 2008;8:4987–4994. doi: 10.1002/pmic.200800288. [DOI] [PubMed] [Google Scholar]
- [27].Chen R, Henning U. A periplasmic protein (Skp) of Escherichia coli selectively binds a class of outer membrane proteins. Mol Microbiol. 1996;19:1287–1294. doi: 10.1111/j.1365-2958.1996.tb02473.x. [DOI] [PubMed] [Google Scholar]
- [28].Bulieris PV, Behrens S, Holst O, Kleinschmidt JH. Folding and insertion of the outer membrane protein OmpA is assisted by the chaperone Skp and by lipopolysaccharide. J Biol Chem. 2003;278:9092–9099. doi: 10.1074/jbc.M211177200. [DOI] [PubMed] [Google Scholar]
- [29].Schafer U, Beck K, Muller M. Skp, a molecular chaperone of gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J Biol Chem. 1999;274:24567–24574. doi: 10.1074/jbc.274.35.24567. [DOI] [PubMed] [Google Scholar]
- [30].Volokhina EB, Grijpstra J, Stork M, Schilders I, et al. Role of the periplasmic chaperones Skp, SurA, and DegQ in outer membrane protein biogenesis in Neisseria meningitidis. J Bacteriol. 2011;193:1612–1621. doi: 10.1128/JB.00532-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Krojer T, Sawa J, Schafer E, Saibil HR, et al. Structural basis for the regulated protease and chaperone function of DegP. Nature. 2008;453:885–890. doi: 10.1038/nature07004. [DOI] [PubMed] [Google Scholar]
- [32].Jiang J, Zhang X, Chen Y, Wu Y, et al. Activation of DegP chaperone-protease via formation of large cage-like oligomers upon binding to substrate proteins. Proc Natl Acad Sci U S A. 2008;105:11939–11944. doi: 10.1073/pnas.0805464105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Spiess C, Beil A, Ehrmann M. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell. 1999;97:339–347. doi: 10.1016/s0092-8674(00)80743-6. [DOI] [PubMed] [Google Scholar]
- [34].Tommassen J. Assembly of outer-membrane proteins in bacteria and mitochondria. Microbiology. 2010;156:2587–2596. doi: 10.1099/mic.0.042689-0. [DOI] [PubMed] [Google Scholar]
- [35].Casadaban MJ. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- [36].Silhavy TJ, Berman ML, Enquist LW. In: Laboratory CSH, editor. New York: 1984. [Google Scholar]
- [37].Baba T, Ara T, Hasegawa M, Takai Y, et al. Construction of Escherichia coli K-12 inframe, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006–0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- [39].Button JE, Silhavy TJ, Ruiz N. A suppressor of cell death caused by the loss of sigmaE downregulates extracytoplasmic stress responses and outer membrane vesicle production in Escherichia coli. J Bacteriol. 2007;180:6408–11. doi: 10.1128/JB.01534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nichols BP, Shafiq O, Meiners V. Sequence analysis of Tn10 insertion sites in a collection of Escherichia coli strains used for genetic mapping and strain construction. J Bacteriol. 1998;189:1523–1530. doi: 10.1128/jb.180.23.6408-6411.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Misra R, Peterson A, Ferenci T, Silhavy TJ. A genetic approach for analyzing the pathway of LamB assembly into the outer membrane of Escherichia coli. J Biol Chem. 1991;266:13592–13597. [PubMed] [Google Scholar]
- [42].Aoki SK, Malinverni JC, Jacoby K, Thomas B, et al. Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol Microbiol. 2008;70:323–340. doi: 10.1111/j.1365-2958.2008.06404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Vertommen D, Depuydt M, Pan J, Leverrier P, et al. The disulphide isomerase DsbC cooperates with the oxidase DsbA in a DsbD-independent manner. Mol Microbiol. 2008;67:336–349. doi: 10.1111/j.1365-2958.2007.06030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- [45].Wagner JK, Heindl JE, Gray AN, Jain S, Goldberg MB. Contribution of the periplasmic chaperone Skp to efficient presentation of the autotransporter IcsA on the surface of Shigella flexneri. J Bacteriol. 2009;191:815–821. doi: 10.1128/JB.00989-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Purdy GE, Fisher CR, Payne SM. IcsA surface presentation in Shigella flexneri requires the periplasmic chaperones DegP, Skp, and SurA. J Bacteriol. 2007;189:5566–5573. doi: 10.1128/JB.00483-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ieva R, Tian P, Peterson JH, Bernstein HD. Sequential and spatially restricted interactions of assembly factors with an autotransporter beta domain. Proc Natl Acad Sci U S A. 2011;108:383–91. doi: 10.1073/pnas.1103827108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Charlson ES, Werner JN, Misra R. Differential effects of yfgL mutation on Escherichia coli outer membrane proteins and lipopolysaccharide. J Bacteriol. 2006;188:7186–7194. doi: 10.1128/JB.00571-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Werner J, Misra R. YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol Microbiol. 2005;57:1450–1459. doi: 10.1111/j.1365-2958.2005.04775.x. [DOI] [PubMed] [Google Scholar]
- [50].Bennion D, Charlson ES, Coon E, Misra R. Dissection of beta-barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutants of Escherichia coli. Mol Microbiol. 2010;77:1153–1171. doi: 10.1111/j.1365-2958.2010.07280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kim S, Malinverni JC, Sliz P, Silhavy TJ, et al. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- [52].Quan S, Koldewey P, Tapley T, Kirsch N, et al. Genetic selection designed to stabilize proteins uncovers a chaperone called Spy. Nat. Struct. Mol. Biol. 2011;18:262–269. doi: 10.1038/nsmb.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.