Abstract
Developing a vaccine for HIV may be aided by a complete understanding of those rare cases where some HIV-infected individuals control replication of the virus1–3. The majority of these elite controllers (ECs) express HLA-B*57 or HLA-B*273. These alleles remain by far the most robust associations with low concentrations of plasma virus4,5, yet the mechanism of control in these individuals is not entirely clear. Here we vaccinated Indian rhesus macaques that express Mamu-B*08, an animal model for HLA-B*27-mediated elite control6, with three Mamu-B*08-restricted CD8+ T cell epitopes and demonstrate that these vaccinated animals controlled replication of the highly pathogenic SIVmac239 clonal virus. High frequencies of CD8+ T cells against these Vif and Nef epitopes in the blood, lymph nodes and colon, were associated with viral control. Moreover, the frequency of the Nef RL10-specific response correlated significantly with reduced acute phase viremia. Finally, two of the eight vaccinees lost control of viral replication in the chronic phase, concomitant with escape in all three targeted epitopes, further implicating these three CD8+ T cell responses in control of viral replication. Our findings indicate that narrowly targeted vaccine-induced virus-specific CD8+ T cell responses can control replication of the AIDS virus.
A vaccine is desperately needed to curb the global HIV pandemic. Estimates project that for every HIV-infected individual initiating antiretroviral treatment, more than two individuals are newly infected7. Vaccines have historically been chosen based on their ability to induce responses that mimic successful immune responses to human pathogens, yet correlates of successful immune responses against HIV remain an enigma. Elite control of chronic phase viral replication is the best example of an effective immune response against the virus1. Understanding why ECs control viral replication may enable the design of an effective vaccine for HIV.
Recently, we have discovered similarities between an animal model of elite control and the same phenomenon in humans8. The Indian rhesus macaque MHC class I molecule Mamu-B*08 and the human HLA class I molecule HLA-B*27 bind many of the same peptides6. Despite being divergent at 28 amino acid positions, these molecules share similar peptide binding motifs, including an identical position 2 arginine primary anchor6. Both MHC class I molecules also exhibit considerable overlap in preferred binding residues at the other dominant position 1 and position 9 residues6. Remarkably, 50% of Mamu-B*08+ animals show some measure of control of the highly pathogenic SIVmac239 virus6,9. It is important to note, however, that several key differences exist between HLA-B27-mediated elite control and the Mamu-B*08+ animal model. Humans are infected with a variety of different viruses and it is widely thought that CD8+ T cells directed against conserved epitopes in Gag play an important role in viral control1. By contrast the animal model of elite control has been developed using macaques infected with the SIVmac239 clone. Furthermore, Mamu-B*08+ positive macaques do not usually recognize Gag-derived epitopes10. Finally, Indian rhesus macaques infected with SIVmac239 have average chronic phase viral loads of more than 106 vRNA copies/mL whereas humans normally have mean plasma viral concentrations of 30,000 vRNA/mL in the chronic phase. Due to the high replicative capacity of SIVmac239, the majority of drug- or vaccine-naïve SIVmac239-infected monkeys die from AIDS-defining illnesses by 2 years post-infection11.
Three Mamu-B*08-restricted CD8+ T cell responses make up more than half of the T cell response against the virus in Mamu-B*08+ ECs10,12; these CD8+ T cells recognize Vif RL8 (Vif amino acids 172–179), Vif RL9 (Vif amino acids 123–131) and Nef RL10 (Nef amino acids 137–146). CD8+ T cells directed against these three epitopes select for escape mutations preferentially in Mamu-B*08+ macaques that do not become ECs13, suggesting that these three immunodominant T cell responses are important for the development of elite control.
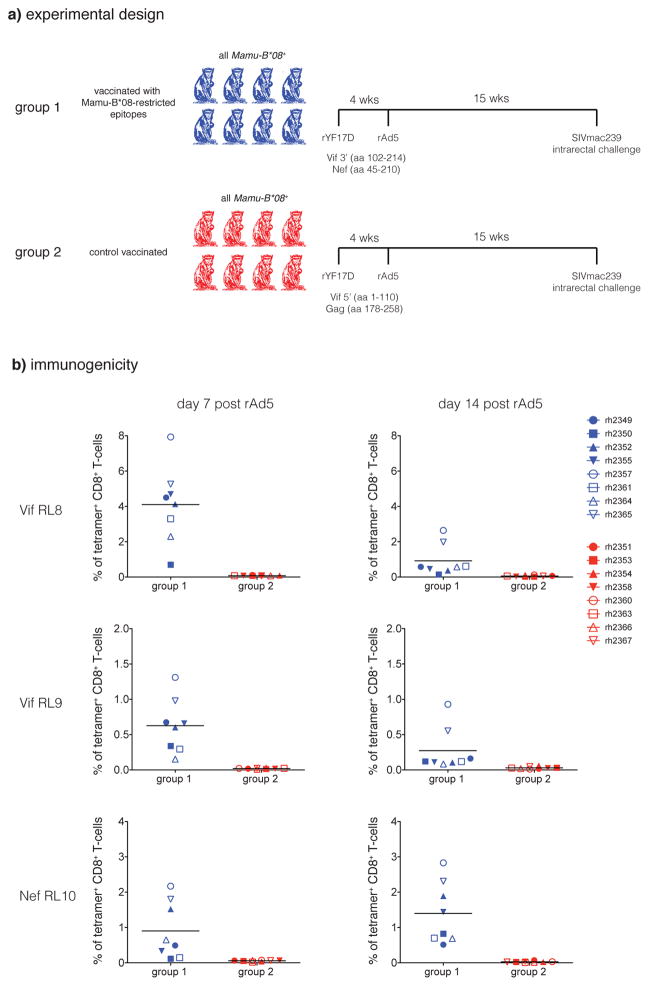
To test the hypothesis that antigen-specific CD8+ T cells are responsible for the development of elite control, we vaccinated eight Mamu-B*08+ macaques (Group 1, Fig. 1A) with two small regions of the SIV proteome that include the three immunodominant T cell epitopes bound by Mamu-B*08: Vif RL8, Vif RL9, and Nef RL10. We used a recombinant yellow fever 17D (rYF17D) prime and boosted with recombinant adenovirus serotype 5 (rAd5). As controls, we vaccinated another eight Mamu-B*08+ macaques (Group 2, Fig. 1A) with two small regions of the SIV proteome that do not encode any known Mamu-B*08 epitopes12. The Group 1 vaccinees mounted high frequency CD8+ T cell responses against the three Mamu-B*08-bound epitopes post-vaccination, whereas the Group 2 animals did not make CD8+ T cell responses against these three epitopes (Fig. 1B). Group 1 and Group 2 animals exhibited similar SIV-specific total T cell and CD4+ T cell response frequencies against the vaccine inserts post-vaccination, as demonstrated by IFN-γ ELISPOT assay (Suppl. Fig. 1A and B).
Figure 1.
Experimental design. A) Mamu-B*08+ macaques were vaccinated with an identical rYF17D/rAd5 regimen. Group 1 received two SIVmac239 constructs: Vif 3′ (amino acids 102–214; includes the Mamu-B*08-restricted Vif123–131RL9 and Vif172–179RL8 epitopes) and Nef (amino acids 45–210; includes the Mamu-B*08-restricted Nef137–146RL10 epitope). Group 2 received two SIVmac239 constructs: Gag (amino acids 178–258) and Vif 5′ (amino acids 1–110), which do not contain any known Mamu-B*08-restricted CD8+ T cell epitopes10. Animals were challenged with SIVmac239 intrarectally 15 weeks after the rAd5 boost. B) Frequencies of tetramer+ CD8+ T-cells in PBMC at days 7 and 14 following the rAd5 boost. Lines represent mean frequency.
Fifteen weeks after the final vaccine boost, we challenged all 16 animals intrarectally with a high dose of SIVmac239 (10,000 TCID50). Four of eight Group 1 animals and six of eight Group 2 animals were infected after this first challenge. The remaining uninfected macaques were re-challenged with a second high dose of SIVmac239 (10,000 TCID50) three weeks after the initial challenge. All Group 2 control-vaccinated animals were infected after this second challenge. A single Group 1 vaccinee, rh2355, resisted four high-dose SIVmac239 challenges and was only infected after the fifth challenge.
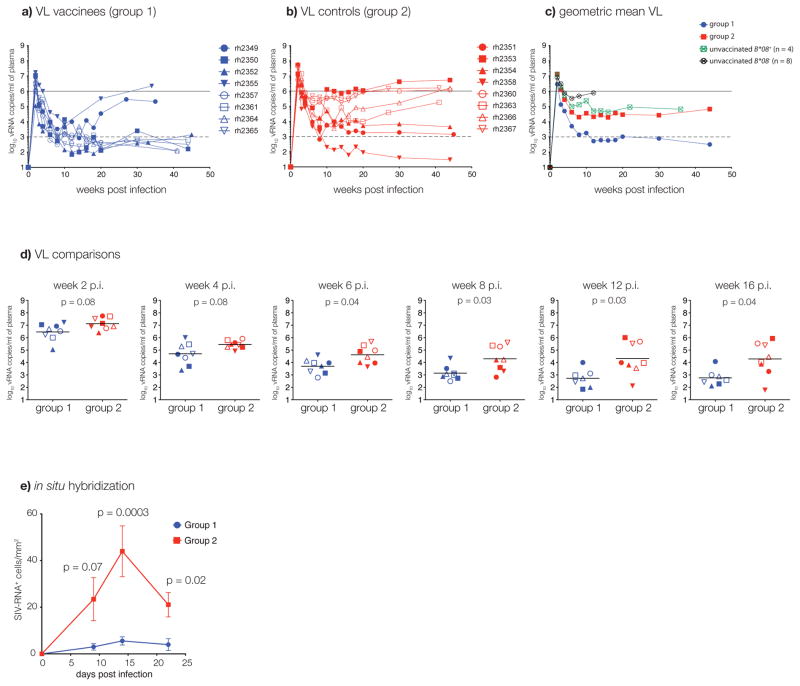
Surprisingly, all eight infected Group 1 animals, vaccinated with the three immunodominant Mamu-B*08-restricted T cell epitopes, controlled viral replication during acute infection when compared with the Group 2 animals (Fig. 2A–D). Remarkably, six of the seven Group 1 vaccinated Mamu-B*08+ animals that were infected after one or two challenges became ECs with post acute phase viral loads of less than 1,000 vRNA copies/mL (Fig. 2A). The seventh animal (rh2349) controlled viral replication to less than 10,000 vRNA copies/mL. The Group 1 animal (rh2355) that required five high-dose challenges for infection controlled viral replication to less than 10,000 vRNA copies/mL by 10 weeks post-infection. By contrast, viral replication in the Group 2 animals was indistinguishable from viral replication in four concurrently challenged unvaccinated Mamu-B*08+ controls (Fig. 2C). We also measured viral replication during acute infection in lymph node biopsies using in situ hybridization14,15. Group 1 animals demonstrated significantly less viral replication in lymph node tissue at time-points near peak plasma viral load when compared with Group 2 animals (Fig. 2E).
Figure 2.
In vivo viral replication. A) Plasma VLs for Group 1 animals. The dashed line (103 vRNA copies/mL) represents the threshold that defines elite control in Indian rhesus macaques9. B) Plasma VLs for Group 2 animals. C) Geometric mean plasma VLs for Groups 1, 2, and unvaccinated Mamu-B*08+ (n = 4) or Mamu-B*08− (n = 8) control macaques. D) Plasma VL comparisons between Groups 1 and 2. Lines represent geometric means. E) Viral replication measured using in situ hybridization14,15 in lymph node biopsy specimens. The mean value and the standard error of the mean are shown.
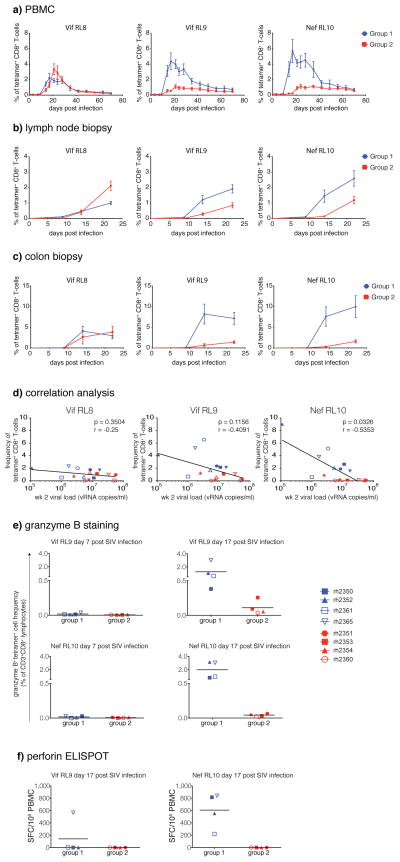
To evaluate potential correlates of viral protection in the Group 1 vaccinees, we characterized the kinetics of T cell responses after SIV infection using peptides and Mamu-B*08 tetramers. We first measured SIV-specific T cell responses post-infection in both Group 1 and Group 2 vaccinated animals with a panel of synthetic peptides spanning both the vaccine inserts and the entire SIV proteome using IFN-γ ELISPOT (Suppl. Fig. 1A and B). Interestingly, PBMC responses against the entire proteome were equivalent in the two groups after infection. At d14 and d17 post-infection, however, higher frequency insert-specific (total T cell and CD4+ T cell) and proteome-specific (CD4+ T cell) responses were observed in the Group 1 animals, perhaps reflecting preservation of the CD4+ T cell responses due to reduced peak viral replication in this group. These preserved antigen-specific CD4+ T cell responses may have facilitated the control of chronic phase viral replication in the Group 1 animals. We also found that Group 1 vaccinated macaques exhibited earlier and higher magnitude Vif RL9- and Nef RL10-specific responses in the peripheral blood when compared with Group 2 vaccinated macaques (Fig. 3A). This pattern was also detected in both lymph node biopsy specimens (Fig. 3B) and colon biopsy tissue (Fig. 3C). Interestingly, T cell responses directed against the Vif RL8 epitope were similar between the Group 1 and Group 2 macaques (Fig. 3A–C). Taken together, these results suggest that early, high frequency Vif RL9- and Nef RL10-specific responses in the blood and in key immunological tissues were directly responsible for viral control in Group 1 Mamu-B*08+ macaques.
Figure 3.
CD8+ T-cell responses in vaccinated animals following SIVmac239 infection. Peptide/Mamu-B*08 tetramer staining in A) PBMC, B) lymph nodes, and C) colon biopsies. Error bars represent standard error of the mean. D) Correlations between the frequency of tetramer+ cells and VL at week 2 post infection. E) Granzyme B production by SIV-specific CD8+ T-cells in Group 1 and Group 2 vaccinees at days 7 and 17 post-infection. F) Perforin production by SIV-specific CD8+ T-cells in Group 1 and Group 2 vaccinees at day 17 post-infection. Assay results are shown as sport forming cells (SFC) per 106 PBMC.
Remarkably, we found a significant correlation between high frequency Nef RL10-specific responses and decreased day 14 peak viral loads (Fig. 3D) in our cohort of infected macaques. Furthermore, these vaccine-induced Nef RL10-specific CD8+ T cells expressed both granzyme B and perforin at d17 post-infection, at a time when Nef RL10-specfic CD8+ T cells showed little or no expression of these important markers of cytotoxicity in the Group 2 animals (Fig. 3E and F and Suppl. Fig. 2A). Additionally, the frequency of Nef RL10- (but not Vif RL8- or Vif RL9-) specific CD8+ T cell responses has been recently shown to correlate with control of chronic phase viral replication in SIVmac239-infected, unvaccinated Mamu-B*08+ macaques (D. Douek, pers. com.). These results suggest that early, high frequency cytotoxic Nef RL10-specfic responses in the blood and in key immunological tissues were directly responsible for viral control in Group 1 Mamu-B*08+ macaques. This is the first described correlate of viral control in the Mamu-B*08 model of elite control.
After controlling viral replication during the acute phase, virus replication was observed in two of the Group 1 vaccinees during the chronic phase (rh2349 and rh2355). Viral sequencing revealed that despite the presence of wild type virus in one of these two animals at 6 weeks post-infection, all three targeted epitopes had escaped by the time the virus had started to replicate in the chronic phase (Table 1 and Suppl. Fig. 3A and B). By contrast, the majority of these three epitopes were intact in virus from two of the ECs in Group 1 at 48 weeks post-infection (Table 1 and Suppl. Fig. 3B). This finding further implicates CD8+ T cells against these three epitopes in control of viral replication and suggests that the virus cannot replicate in the face of these CD8+ T cell responses. The canonical escape mutants that arose in these animals did not seem to affect viral fitness since the virus replicated to high levels in these two “breakthrough” animals. Furthermore, we have previously engineered mutant viruses bearing several of the escape mutants observed in these animals and these mutations cause abrogation of CD8+ T cell recognition and they do not appear to cause significant fitness loses in vitro and in vivo10.
Table 1.
Viral sequence analysis.
| Acute phase | |||
|---|---|---|---|
| Group 1 animals | Vif RL8 | Vif RL9 | Nef RL10 |
| rh2349 (wk 6) | nd | nd | nd |
| rh2355 (wk 6) | 100% | 100% | 100% |
| rh2350 (wk 6) | 99% | 27% | nd |
| rh2352 (wk 6) | 97% | 47% | nd |
| rh2357 (wk 6) | 87% | 75% | 97% |
| rh2361 (wk 6) | 100% | 100% | 100% |
| rh2364 (wk 6) | 59% | 37% | 92% |
| rh2365 (wk 6) | nd | nd | nd |
| Group 2 animals | Vif RL8 | Vif RL9 | Nef RL10 |
|---|---|---|---|
| rh2351 (wk 6) | 22% | 33% | 97% |
| rh2353 (wk 6) | 38% | 40% | 95% |
| rh2354 (wk 6) | 92% | 92% | 100% |
| rh2358 (wk 6) | 100% | 100% | 99% |
| rh2360 (wk 6) | 50% | 64% | 99% |
| rh2363 (wk 6) | 94% | 99% | 95% |
| rh2366 (wk 6) | 100% | 69% | 99% |
| rh2367 (wk 6) | 97% | 78% | 100% |
| Chronic phase | |||
|---|---|---|---|
| Group 1 animals | Vif RL8 | Vif RL9 | Nef RL10 |
| rh2349 (wk 35) | 4% | 4% | 7% |
| rh2355 (wk 37) | 0% | 0% | 0% |
| rh2350 (wk 48) | nd | nd | 91% |
| rh2352 (wk 48) | 97% | 17% | 69% |
| rh2357 (wk 45) | nd | nd | 99% |
| rh2361 (wk 48) | 99% | 68% | 99% |
| rh2364 (wk 45) | nd | nd | nd |
| rh2365 (wk 48) | nd | nd | nd |
454 sequencing of the three Mamu-B*08-restricted Vif RL8, Vif RL9, and Nef RL10 CD8+ T-cell epitopes. Vif and Nef were amplified independently and PCR amplicons sequenced by 454. Sequence analysis was carried out using RC454 and V-phaser as previously described21,22. Percentages indicate frequencies of wild type epitope sequence reads for both the acute and chronic phases of infection. Breakthrough animals in Group 1 are identified in yellow. nd = not determined.
With this report, we demonstrate that vaccine-induced Vif and Nef-specific CD8+ T cells can control replication of a highly pathogenic AIDS virus in an animal model of MHC class I-associated elite control. Further analysis of the Vif RL9- and Nef RL10-specific CD8+ T cell responses in Mamu-B*08+ macaques could shed light on the general principles of an effective AIDS virus-specific CD8+ T cell response. It is possible that vaccines that generate high frequency efficacious effector CD8+ T cells against only a few epitopes in the mucosae might intercept the virus during this crucial acute phase and result in long-term virological control. These types of effective immune responses might be inducible in the setting of an appropriate vaccination regimen16–19. Therefore, understanding why these particular T cell responses control viral replication when most other T cell responses do not may enable the design of an effective approach to HIV vaccination.
Online-only Methods
Animals
The sixteen Mamu-B*08+ Indian rhesus macaques in Groups 1 and 2 were purchased from the Oregon National Primate Research Center. Among the four unvaccinated Mamu-B*08+ macaques displayed in figure 2C, one animal came from Pfizer (Kalamazoo, MI), one came from the Harlow Primate lab at the University of Wisconsin-Madison, and the two remaining ones were raised in-house at the Wisconsin National Primate Research Center (WNPRC). All eight unvaccinated, Mamu-B*08− animals shown in figure 2C were also raised in-house at WNPRC. All animals were housed and studies were conducted at WNPRC. Animals were cared for according to the regulations and guidelines of the University of Wisconsin Institutional Animal Care and Use Committee, Animal Welfare Assurance No. A3368-01. Full details of the study were approved (UW-Madison Animal Care and Use Protocol No. G00639) by the University of Wisconsin Institutional Animal Care and Use Committee in accordance with the recommendations of the Weatherall report. Macaques in Groups 1 and 2 (n = 16) were males, with a mean age of 7.95 years (range: 7.1–9.2). The four unvaccinated, Mamu-B*08+ control animals were also males and their mean age was 12.4 years (range: 7.5–18.3). Among the eight unvaccinated, Mamu-B*08− control animals, six were males and two were females; their mean age was 9.7 years (range: 7–9). Once infected, animals were singly housed to prevent cross contamination of SIV infection and spread of opportunistic infections. Animals were closely monitored daily for pain or discomfort and treated accordingly by a veterinarian to ameliorate any suffering. Following progression to AIDS or at the end of the study, animals with high viral loads were humanely euthanized. Animals that controlled viral replication during this study have been maintained for continuing study as a part of the elite controller resource at the Wisconsin National Primate Research Center. Group 1 vaccinee rh2349 was euthanized at 35 weeks post-infection with 20% loss of its body weight. At necropsy, this animal had right-sided dilated cardiomyopathy and cardiomyocyte necrosis, with minimal inflammation. This may be related to SIV infection but it is not a typical lesion. All animals were MHC class I typed to verify the presence of the Mamu-B*08 allele according to a previously reported protocol9.
Vaccination
Vaccinated animals received constructs as diagramed in Figure 1A. Animals receiving the three immunodominant Mamu-B*08-restricted epitopes were vaccinated with two separate recombinant yellow fever 17D (rYF17D) constructs. Recombinant YF17D viruses were created with a single insert between the viral proteins E and NS1, as previously described23,24. The first was engineered to contain SIVmac239 Vif amino acids 102–214 (including the Vif123–131RL9 and Vif172–179RL8 epitopes), and the second contained SIVmac239 Nef amino acids 45–210 (including the Nef137–146RL10 epitope). Animals were vaccinated subcutaneously with 2.0 × 105 PFU of each of these constructs in separate locations on each forearm. The animals were then boosted intramuscularly in each thigh 4 weeks later with two separate doses of 1011 particles of recombinant adenovirus serotype 5 (rAd5) vectors (Viraquest, North Liberty, IA, USA) each containing SIVmac239 Vif and Nef inserts identical to those in the recombinant YF17D. Control vaccinated animals underwent the same vaccination regimen, except recombinant YF17D and recombinant adenoviral vectors included portions of SIVmac239 without known Mamu-B*08 epitopes10 – SIVmac239 Vif amino acids 1–110 and SIVmac239 Gag amino acids 175–255.
SIV infection and viral load measurement
Animals in Groups 1, 2, and the unvaccinated Mamu-B*08+ controls were challenged intrarectally with 10,000 TCID50 (8.15 × 107 vRNA copies) of SIVmac239 fifteen weeks after vaccination with rAd5. Animals that remained uninfected after the initial inoculation were challenged a second time 3 weeks later. Group 1 vaccinee rh2355 resisted a total of 4 challenges. Each of these intrarectal inoculations occurred approximately 3 weeks after the previous challenge. The eight unvaccinated, Mamu-B*08− animals described in figure 2C were used as experimental controls as part of two SIV vaccine trials conducted in our laboratory (Martins et al., Wilson et al., manuscripts in preparation). They were challenged intrarectally every week with the same stock of SIVmac239 described above, albeit at doses ranging from 800 to 50,000 TCID50 (6.52 × 106 to 4.07 × 108 vRNA copies, respectively).
Clonal SIVmac239 was generated by transfection of Vero cells with plasmid DNA encoding proviral sequences. Activated PBMC from 4 SIV-naïve rhesus macaques were then added to the culture 1 day later and removed from the Vero cells into flasks 3 days after the initial transfection. Virus was amplified in the activated PBMC and cell-free supernatant was collected two days after peak syncytium formation. Viral loads were measured from EDTA anti-coagulated plasma using a previously described protocol20. Viral RNA was dissolved in 30 μL molecular biology grade water with 1 mM DTT and 1 U/μL RNAseOUT (Invitrogen, Carlsbad, CA, USA). Samples were run in duplicate. Each reaction contained 10 μL RNA and 20 μL of a reverse transcriptase (RT) master mix so that the final reactions contained 50 mM Tris-Cl, pH 8.3 and 50 mM KCl (1x PCR II Buffer; Applied Biosystems, Foster City, CA, USA), 0.05% gelatin (Sigma, St Louis, MO, USA), 0.02% Tween 20 (Sigma), 5 mM MgCl2, 0.5 mM of each dNTP, 150 ng random hexamer primers (Promega, Madison, WI, USA), 20 U RNAseOUT, and 20 U Superscript II reverse transcriptase (Invitrogen). Viral RNA was converted to cDNA using the following conditions: 25°C for 15 minutes, 42°C for 40 minutes, 85°C for 10 minutes, 25°C for 30 minutes, and 5°C until the reaction was removed from the thermocycler. At the second 25°C stage, or later, the reactions were unsealed and 20 μL of a cocktail containing primers, probe and thermostable polymerase were added so that the final reactions contained 1X PCR II Buffer, 0.03% gelatin, 0.012% Tween 20, 4.5 mM MgCl2, 600 nM of each primer – forward primer (SGAG21), 5′-GTCTGCGTCATPTGGTGCATTC-3′; reverse primer (SGAG22), 5′-CACTAGKTGTCTCTGCACTATPTGTTTTG-3′, 100 nM probe (pSGAG23), 5′-(FAM) CTTCPTCAGTKTGTTTCACTTTCTCTTCTGCG-(BHQTM1)-3′, 50 nM passive reference dye, 5′-(FAM)TTTTTTTTTT(ROX)(C3-blocked)-3′, and 1.25 U Taq-Gold polymerase (Applied Biosystems). P and K are non-standard bases (Glen Research, Sterling, VA, USA) that minimize the impact of potential sequence mismatches at positions of described heterogeneity in SIV isolates (Los Alamos Sequence Database), FAM is the reporter fluorochrome 6-carboxyfluoroscein, BHQTM1 is Black Hole QuencherTM 1 (Biosearch Technologies, Novato, CA, USA), and ROX is 5-carboxyrhodamine. The final reactions were then run on an ABI 7700 Sequence Detection System (Applied Biosystems). The run conditions were 95°C for 10 minutes, followed by 45 cycles of 95°C for 15 seconds and 60°C for 1 minute. Copy numbers for test samples were determined using an RNA standard curve run at the same time. Ten viral copy equivalents per mL of plasma is the limit of reliable quantification for this assay.
Colon and lymph node biopsies
Lymph node and colon biopsies were obtained from all animals with positive viral loads on days 9, 14, and 21 post SIVmac239 infection. Animals were anesthetized and a single inguinal or axillary lymph node was obtained from a separate biopsy site each day by a veterinarian using aseptic technique. Lymph node biopsy specimens were sieved through a 100-micrometer screen to obtain a single cell suspension and any remaining red blood cells were removed by hypotonic lysis. During the same anesthetic event, colon biopsy samples (approximately 10 separate 2 × 2 × 2 mm pieces of tissue) were collected by the veterinarian using a pinch biopsy device and a fiber optic endoscope. Colon biopsy samples were washed with R10 (RPMI 1640 media supplemented with 10% FBS, 1% antibiotic/antimycotic and 1% L-Glutamine) and then re-suspended in pre-digestion buffer (HBSS supplemented with 5% FBS, 1% antibiotic/antimycotic, 5 mM EDTA and 1 mM DTT) and incubated in an orbital shaker at 50 RPM, 37°C for 30 minutes. Samples were then washed once with R10 and re-suspended in collagenase medium (R10 with 15 μg/mL type II collagenase from Clostridium histolyticum [Sigma-Aldrich, St. Louis, MO, USA]) and incubated at 50 RPM, 37°C for 30 minutes. The supernatant was collected and lamina propria lymphocytes (LPL) contained within the supernatant were washed three times with R10. The collagenase digestion was repeated two more times and collected, washed LPL were pooled in R10 and layered over a 40%–90% percoll gradient before being centrifuged for 30 minutes at 450 RCF. Purified LPL were collected from the interface between the 40% and 90% percoll layers and washed once with R10 before being counted and stained for flow cytometry.
In situ hybridization
In situ hybridization was performed as previously described14,15. Briefly, 5-mm sections were cut from 4% PFA fixed lymph node tissues. After deparaffinization, pretreatment to permeabilize the tissues and blocking of non-specific binding, the sections were hybridized to 35S-labeled SIV RNA antisense or sense (as a negative control) riboprobes covering SIV sequences at the 5′, middle and 3′ end of the genome. After overnight hybridization at 45°C, the sections were washed, digested with ribonucleases, coated with nuclear track emulsion, exposed, developed and counterstained with H&E. Viral load was quantified as SIV RNA+ cells per mm2 of tissue.
Tetramer staining and flow cytometry
For tetramer staining, we used MHC class I tetramers produced by the Wisconsin National Primate Research Center Tetramer Core conjugated to either phycoerythrin (PE) or allophycocyanin (APC). Prepared PBMC, lymph node cells or colon biopsy cells suspended in R10 were centrifuged for 5 minutes at 530 × g in 1.2 mL cluster tubes and excess media was aspirated leaving all samples in 100–200 uL of R10. Approximately 50,000–100,000 colon biopsy cells or 500,000–1,000,000 PBMC/lymph node cells were stained with tetramer for 90 minutes at 37°C. Following the incubation, surface staining antibodies were added and samples were incubated at room temperature for 30 minutes. Cells were then washed twice with FACS buffer (1x PBS with 10% FBS) and fixed with 1% paraformaldehyde in 1x PBS. Cells were run on a BD-LSRII instrument (BD, San Jose, CA, USA) and analysis was performed using FlowJo software (version 9.3.1, Tree Star, Ashland, OR, USA). We used the following antibodies from BD Biosciences during this study: anti-CD3 Alexa Fluor 700 (clone SP34-2) and anti-CD8 Pacific Blue (RPA-T8).
For the MHC class I tetramer/Granzyme B combined staining, cryopreserved PBMC were thawed at 37°C and washed twice in R10 media prior to staining. Approximately 2.0 × 105–106 PBMC were stained with 5 μL of PE-conjugated tetramer in a volume of 100–200 μL of R10 media for 90 minutes at 37°C. Cells were then stained with 2 μl anti-CD3 Alexa Fluor 700 (clone SP34-2; BD Biosciences) and 1 μL anti-CD8 Pacific Blue (clone RPA-T8; BD Biosciences) for 30 min at room temperature. Subsequent washes and fixation with 1% paraformaldehyde were performed as described above. Fixed cells were washed twice with FACS buffer, and excess liquid was aspirated to leave samples in approximately 75 μL of FACS buffer. Cells were then permeabilized by adding 100 μL Medium B (Life Technologies, Carlsbad, CA, USA), and simultaneously stained for Granzyme B with 1 μL anti-GzmB allophycocyanin (clone GB12; Life Technologies, Carlsbad, CA, USA) for 30 min at room temperature. Cells were then washed twice with FACS buffer, and fixed in 1% paraformaldehyde. Cells were run on a SORP BD-LSRII (BD, San Jose, CA, USA) and analysis was performed using FlowJo software (version 9.4.2, Tree Star, Ashland, OR, USA). Due to sample limitations, only rh2350, rh2352, rh2361, and rh2365 from Group 1 and rh2351, rh2353, rh2354, and rh2360 from Group 2 were included in this analysis.
Interferon-γ and perforin ELISPOT
Enzyme-linked immunosorbent spot (ELISPOT) assays were performed on either PBMC or PBMC depleted of CD8+ cells at the indicated time points. CD8 depletion was performed on PBMC prior to ELISPOT in some assays using the Miltenyi Biotec non-human primate CD8 MicroBead kit and LS columns according to the manufacturer’s protocols (Miltenyi Biotec, Bergisch Gladbach, Germany). CD8 depletion was determined to be >99% in every assay performed. All interferon-γ and perforin ELISPOT assays (ELISpotPLUS -ALP) were performed with 100,000 cells per well, in duplicate, according to the manufacturer’s instructions (MABTECH, Stockholm, Sweeden). For all interferon-γ assays, the indicated viral amino acids were tested as pools of 15mer peptides which overlap by 11 amino acids (NIH AIDS Research and Reference Reagent Program, Germantown, MD, USA), and each peptide was tested at a final concentration of 1 μM. For the perforin assays, only the minimal peptides were tested, each at a final concentration of 10μM. Each individual animal on each separate plate included at least two separate positive control wells (5 μg/mL concanavalin A) and two or more negative control wells. Wells were imaged and spots were enumerated with an AID ELISPOT reader (AID, Strassberg, Germany). Positive responses were determined as follows: test wells were run with two replicates while positive and negative control wells were run in replicates of 2, 4 or 6 depending on the assay. Responses containing < 50 spot-forming cells (SFC) per 106 cells were considered negative and not tested statistically. Positive responses were determined using a one-tailed t-test and alpha = 0.05, where the null hypothesis (H0): background level = treatment level. If determined to be positive statistically, the reported values equal the average of the test wells minus the average of all negative control wells. For the Perforin ELISPOT in figure 3F, we only included rh2350, rh2352, rh2361, and rh2365 from Group 1 and rh2351, rh2353, rh2354, and rh2360 from Group 2 in the analysis due to sample limitations.
Amplicon-Based 454 sequencing of Vif and Nef Epitopes
One to three mLs of plasma was thawed on ice and centrifuged at 20,817 × g for 1.5 hours at 4°C after which the pellet was re-suspended in 140 μl of the supernatant. The QIAamp Viral RNA Mini Kit (Qiagen) was used to isolate viral RNA per manufacturer protocol. Viral RNA was eluted in 60 μl of Buffer AVE, aliquoted, and stored at −80°C for future RT-PCR.
An amplicon-based 454 sequencing approach was used to analyze the vif and nef epitopes as previously described13. Briefly, primers were synthesized with Roche 454 amplicon (Lib-A) adapter sequences, multiplex identifier tags (MID) 1 though 18, and sequence-specific regions (Vif Forward 5′-GAAAAAGGGTGGCTCAGT-3′ Vif Reverse 5′-AGGTGGTTTACCGCCTCTCT-3′; and Nef Forward 5′-ACTGGAAGGGATTTATTAC-3′ Nef Reverse 5′-GAGTTTCCTTCTTGTCAGCC-3′), which allowed multiplexing of up to 16 animal/amplicon combinations per sequencing run.
A single-step RT-PCR was carried out for each of the unique amplicon/animal/MID sequence combinations using SuperScript III One-Step RT-PCR System with Platinum Taq High Fidelity (Invitrogen, Carlsbad, CA, USA). Each 25 μL reaction contained 12.5 μL of 2x reaction mix, an additional 0.3mM MgSO4, 1 μL of enzyme mix, 0.2 μM each of the sequence specific, adaptor/MID-tagged forward and reverse primers, and up to 10 μL of template RNA. Cycling parameters for the Vif RT-PCR were as follows: 50°C for 30 minutes, 94°C for 2 minutes followed by 40 cycles of 94°C for 15 seconds, 50°C for 30 seconds and 68°C for 30 seconds, 68°C for 5 minutes, hold at 10°C. The Nef RT-PCR parameters were identical with the exception of an annealing temperature of 54°C. Amplicons were visualized on a 1% agarose gel and purified using the Purelink Quick Gel Extraction Kit (Invitrogen, Carlsbad, CA, USA). RT-PCR products were quantified using a Promega Quantiflor-ST fluorometer (Promega, Madison, WI, USA) and analyzed for quality using an Agilent 2100 bioanalyzer with high sensitivity DNA chips (Santa Clara, CA, USA).
For each sequencing run, up to 16 animal/amplicon samples were pooled in equimolar ratios and 20 million molecules of pooled sample were added to 10 million DNA capture beads for a final ratio of 2.0 DNA molecules per bead. Emulsion PCR, enrichment, breaking and DNA sequencing were all performed according to the GS Junior FLX Titanium Series manuals for Lib-A (Roche, Indianapolis, IN, USA). Sequencing and run processing were performed on a GS Junior 454 sequencing instrument (Roche, Indianapolis, IN, USA).
We utilized the ReadClean 454 (RC454) and V-Phaser algorithms as previously described21 to call variants from the 454 datasets. Briefly, RC454 was used to align reads to SIVmac239 and reads were corrected for sequencing related artifacts such as InDels resulting from overcalls and undercalls in homopoymeric regions and Carry forward and Incomplete Extension (CAFIE) errors. Furthermore, RC454 optimizes read alignments using coding frame information. The V-Phaser algorithm was then used to distinguish an observed variant as a true variant from an amplification or sequencing artifact22.
Supplementary Material
Acknowledgments
The authors would like to thank Mario Stevenson, Matt Reynolds, Nick Maness and Jonah Sacha for helpful discussions and suggestions. We also thank David Evans, Lisa Heyer and Zachary R. Bergman for facilitating the experiments. This work was funded in part by NIH Grants R37 AI052056, RO1 AI076114, RR015371, and FAPERJ, INCTV, CNPq, MCT and FIOCRUZ.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions. DIW, TMA and PAM conceived of and designed the experiments. PAM, AJE, MAM, KAP, AS, ADG, ATB, SMP, LD, KLW, JRF, SC, MP, ATH and JDL performed the experiments. PAM, MAM, NAW, ER, MP, ATH, JDL, DCT, TMA and DIW compiled and analyzed the data. YK and DBA oversaw statistical analysis of the data and aided in interpretation. MGV, MCB and RG designed and manufactured the rYF17D vectors used in these studies. PAM, ATH, JDL, TMA and DIW wrote the manuscript.
Reprints and permissions information is available at www.nature.com/reprints. DBA has, anticipates, or has had financial interests with the Frontiers Foundation; Vivus, Inc: Kraft Foods; University of Wisconsin; University of Arizona; University of Miami; Paul, Weiss, Wharton & Garrison LLP; and Sage Publications.
All other authors declare no competing financial interests.
References
- 1.Deeks SG, Walker BD. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity. 2007;27:406–416. doi: 10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 2.Goulder PJ, Watkins DI. HIV and SIV CTL escape: implications for vaccine design. Nat Rev Immunol. 2004;4:630–640. doi: 10.1038/nri1417. [DOI] [PubMed] [Google Scholar]
- 3.Migueles SA, Connors M. Long-term nonprogressive disease among untreated HIV-infected individuals: clinical implications of understanding immune control of HIV. JAMA. 2010;304:194–201. doi: 10.1001/jama.2010.925. [DOI] [PubMed] [Google Scholar]
- 4.Fellay J, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pereyra F, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loffredo JT, et al. Two MHC class I molecules associated with elite control of immunodeficiency virus replication, Mamu-B*08 and HLA-B*2705, bind peptides with sequence similarity. J Immunol. 2009;182:7763–7775. doi: 10.4049/jimmunol.0900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnston MI, Fauci AS. An HIV vaccine--challenges and prospects. N Engl J Med. 2008;359:888–890. doi: 10.1056/NEJMp0806162. [DOI] [PubMed] [Google Scholar]
- 8.Mudd PA, Watkins DI. Understanding animal models of elite control: windows on effective immune responses against immunodeficiency viruses. Curr Opin HIV AIDS. 2011;6:197–201. doi: 10.1097/COH.0b013e3283453e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loffredo JT, et al. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J Virol. 2007;81:8827–8832. doi: 10.1128/JVI.00895-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valentine LE, et al. Infection with “escaped” virus variants impairs control of simian immunodeficiency virus SIVmac239 replication in Mamu-B*08-positive macaques. J Virol. 2009;83:11514–11527. doi: 10.1128/JVI.01298-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang Q, Hirsch VM. Rapid disease progression to AIDS due to Simian immunodeficiency virus infection of macaques: host and viral factors. Adv Pharmacol. 2008;56:369–398. doi: 10.1016/S1054-3589(07)56012-6. [DOI] [PubMed] [Google Scholar]
- 12.Loffredo JT, et al. Patterns of CD8+ immunodominance may influence the ability of Mamu-B*08-positive macaques to naturally control simian immunodeficiency virus SIVmac239 replication. J Virol. 2008;82:1723–1738. doi: 10.1128/JVI.02084-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mudd PA, et al. Escape from CD8(+) T cell responses in Mamu-B*00801(+) macaques differentiates progressors from elite controllers. J Immunol. 2012;188:3364–3370. doi: 10.4049/jimmunol.1102470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q, et al. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature. 2005;434:1148–1152. doi: 10.1038/nature03513. [DOI] [PubMed] [Google Scholar]
- 15.Li Q, et al. Glycerol monolaurate prevents mucosal SIV transmission. Nature. 2009;458:1034–1038. doi: 10.1038/nature07831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen SG, et al. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med. 2009;15:293–299. doi: 10.1038/nm.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen SG, et al. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature. 2011;473:523–527. doi: 10.1038/nature10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawada M, et al. Gag-specific cytotoxic T-lymphocyte-based control of primary simian immunodeficiency virus replication in a vaccine trial. J Virol. 2008;82:10199–10206. doi: 10.1128/JVI.01103-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsukamoto T, et al. Impact of cytotoxic-T-lymphocyte memory induction without virus-specific CD4+ T-Cell help on control of a simian immunodeficiency virus challenge in rhesus macaques. J Virol. 2009;83:9339–9346. doi: 10.1128/JVI.01120-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cline AN, Bess JW, Piatak MJ, Lifson JD. Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J Med Primatol. 2005;34:303–312. doi: 10.1111/j.1600-0684.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- 21.Henn MR, et al. Whole genome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathog. 2012;8:e1002529. doi: 10.1371/journal.ppat.1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macalalad AR, et al. Highly sensitive and specific detection of rare variants in mixed viral populations from massively parallel sequence data. PLoS Comput Biol. 2012;8:e1002417. doi: 10.1371/journal.pcbi.1002417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonaldo MC, et al. Recombinant yellow fever vaccine virus 17D expressing simian immunodeficiency virus SIVmac239 gag induces SIV-specific CD8+ T-cell responses in rhesus macaques. J Virol. 2010;84:3699–3706. doi: 10.1128/JVI.02255-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonaldo MC, et al. Construction and characterization of recombinant flaviviruses bearing insertions between E and NS1 genes. Virol J. 2007;4:115. doi: 10.1186/1743-422X-4-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.