Abstract
The low sensitivity inherent to both the static and magic angle spinning techniques of solid-state NMR (ssNMR) spectroscopy has thus far limited the routine application of multidimensional experiments to determine the structure of membrane proteins in lipid bilayers. Here, we demonstrate the advantage of using a recently developed class of experiments, polarization optimized experiments (POE), for both static and MAS spectroscopy to achieve higher sensitivity and substantial time-savings for 2D and 3D experiments. We used sarcolipin, a single pass membrane protein, reconstituted in oriented bicelles (for oriented ssNMR) and multilamellar vesicles (for MAS ssNMR) as a benchmark. The restraints derived by these experiments are then combined into a hybrid energy function to allow simultaneous determination of structure and topology. The resulting structural ensemble converged to a helical conformation with a backbone RMSD ∼ 0.44 Å, a tilt angle of 24° ± 1°, and an azimuthal angle of 55° ± 6°. This work represents a crucial first step toward obtaining high-resolution structures of large membrane proteins using combined multidimensional O-ssNMR and MAS-ssNMR.
Keywords: Oriented Solid-State NMR (OSS-NMR), Magic Angle Spinning Solid State NMR, Membrane Proteins, DUMAS-ssNMR, MEIOSIS, Sarcolipin, Magnetically Aligned bicelles, Hybrid Method for Membrane Protein Structure Determination
Introduction
Membrane protein function is modulated by lipid membranes (Tamm 2005; White 2009). Therefore, understanding the structure-function relationship of these proteins requires their characterization in fully hydrated lipid bilayers. Multidimensional solid-state NMR, i.e. oriented solid-state NMR (O-ssNMR) and magic angle spinning solid-state NMR (MAS-ssNMR), allows for such structural characterization in the presence of fluid lipid bilayers – the most faithful mimetic of natural membranes. O-ssNMR provides information on dipolar couplings and anisotropic chemical shifts that enable the determination of topological parameters, such as the tilt and rotation angles of transmembrane and membrane anchored domains (Veglia et al. 2012). On the other hand, MAS-ssNMR provides chemical shift information on both main chain and side chains that can be converted into torsion angles for assessing secondary structure elements. MAS-ssNMR can also be utilized to measure residual dipolar couplings in aligned bicelles (Canlas et al. 2008). In the case of fast uniaxial rotation around membrane normal, MAS-ssNMR can provide orientational restraints that can further refine the structure of small peptides (Hu et al. 2010) as well as large membrane proteins (Park et al. 2012; Das et al. 2012).
As we demonstrated for phospholamban (Traaseth et al. 2009; Verardi et al. 2011), the ab initio determination of the structural topology of a membrane protein in the absence of homology models entails the combination of all of the above information. Lack of information or even relying on only one approach to determine the structural topology of membrane proteins leads to misinterpretation of the experimental results (Traaseth et al. 2007; Shi et al. 2011b).
A major advantage of combining MAS-ssNMR and O-ssNMR data is to obtain structural and topological data using similar lipid compositions in the sample preparations, circumventing the necessity for cross-validation of the structural information obtained from different membrane mimetic systems. Nonetheless, the ssNMR experiments applied to membrane proteins are inherently insensitive. This is due in part to the need for high lipid-to-protein ratios that, while maintaining protein's functional integrity, reduce the effective protein concentrations in the sample. In addition, recurring secondary structure elements (multiple transmembrane α helices) as well as sample heterogeneity tend to give poorly resolved spectra for these proteins. As a result only a few structures of membrane proteins have been determined by ssNMR techniques (Ketchem et al. 1993; Traaseth et al. 2009; Cady et al. 2009; Sharma et al. 2010; Park et al. 2010; Verardi et al. 2011). Our work is motivated by the necessity of improving these techniques for membrane protein samples. Toward this goal, we recently designed several new pulse sequences to improve the sensitivity of both O-ssNMR and MAS-ssNMR experiments. An underlining concept for these new experiments is the optimization of nuclear spin polarization (polarization optimized experiments, POE) to augment the NMR signals or deconvolute multiple excitation pathways to obtain several 2D or 3D experiments simultaneously. The latter translates into a substantial decrease in experimental time.
For O-ssNMR, the polarization is optimized by recovering the previously discarded dipolar (Gopinath and Veglia 2009) and chemical shift coherences (Gopinath et al. 2010a). We demonstrated that these experiments improve the sensitivity of traditional rotating frame SLF experiments such as PISEMA (Wu et al. 1994), HIMSELF (Dvinskikh et al. 2006), SAMPI-4 (Nevzorov and Opella 2003) and HETCOR (Maudsley and Ernst 1977) by ∼40%, thereby cutting experimental time in half (Gopinath et al. 2010a; Gopinath et al. 2010b). Such a strategy has also been demonstrated for the original PELF experiment (Schmidt-Rohr et al. 1994), and enhances sensitivity by > 100% for highly mobile segments in proteins (Gopinath et al. 2011). The gain in sensitivity has allowed us to record comparatively insensitive constant-time experiments, such as the SECT-PISEMA (Gopinath and Veglia 2010) for increasing resolution in 2-dimensional experiments (Mote et al. 2011). In the 3D SE-PISEMAI-HETCOR experiment (Gopinath et al. 2010a), we designed a pulse scheme to recover chemical shift coherences in addition to retaining the sensitivity gain obtained by the polarization inversion. The resulting boost in sensitivity is 80-180%, which enables the implementation of 3D spectroscopy to correlate 15N chemical shifts, 15N-1H dipolar couplings, and 1H chemical shifts. The three- to seven-fold savings in time alone bring a number of membrane protein systems within reach of this technique. This experiment is used here to obtain the complete set of backbone orientation restraints for sarcolipin in oriented bicelles (Dürr et al. 2012).
In contrast to O-ssNMR, MAS-ssNMR experiments have reached a much higher level of sophistication. In the past decade, many groups have used MAS to for high-resolution structural studies of macromolecular systems, including microcrystalline proteins (Castellani et al. 2002; Wylie et al. 2011; Knight et al. 2012), fibrillar systems (Loquet et al. 2012), membrane proteins (Shi et al. 2011a; Tang et al. 2011), protein complexes (Nieuwkoop and Rienstra 2010), cell extracts (Miao et al. 2012) and whole cell (Renault et al. 2012b). Nonetheless, membrane protein structural biology still offers the biggest challenge for MAS techniques, due to dynamic and static disorder that broaden the resonances of 13C and 15N spectra. Various approaches have been devised to overcome these hurdles: faster spinning rates (Demers et al. 2011), extensive (Reif 2012) and partial deuteration (Asami et al. 2012) coupled with 1H detection (Reif et al. 2001; Zhou et al. 2012; Marchetti et al. 2012), sparse 13C labeling to reduce spectral complexity (Sperling et al. 2010) and minimize 13C-13C couplings as a source of line-broadening (Loquet et al. 2011) as well as the use of techniques such as RELOAD (Lopez et al. 2009) to gain sensitivity . One can boost sensitivity to an even higher degree by Dynamic Nuclear Polarization (DNP) (Hall et al. 1997), opening up this technique to even bigger systems (Renault et al. 2012a).
All of the above experiments, however, do not take full advantage of the sparse nuclear spin polarization. To address this issue, we introduced the Dual-Acquisition MAS strategy (DUMAS) (Gopinath and Veglia 2012a). This novel strategy allows the concatenation of 2D or 3D pulse sequences into a single experiment and thus potentially combines all the aforementioned advances, boosting the capacity of any NMR spectrometer at least two-fold without the need for sample manipulation or additional hardware. At the heart of this approach is simultaneous cross-polarization from 1H to 13C and 15N, which has also been used in time-shared experiments (Linser et al. 2011; Nielsen et al. 2012), and long lived 15N longitudinal magnetization (Giraud et al. 2005) which can be stored and recalled to perform additional experiments. The DUMAS scheme enables one to acquire two experiments: the first with 100% of the sensitivity with respect to classical pulse sequences, and a second experiment generated from the 15N polarization with typically 80% of the sensitivity (Gopinath and Veglia 2012a). A well-designed combination of these experiments allows sequential assignment as well as side chain assignment in a single 3D experiment (Gopinath and Veglia 2012b). Using the initial scheme of the DUMAS experiments, it is possible to decode the coherences in multidimensional NMR experiments to recover the orphan spin operators that are discarded from the classical experiments. This approach that we called MEIOSIS (Multiple Experiments via Orphan SpIn operatorS) enables one to collect up to four multidimensional NMR experiments with a time saving greater than 50% (Gopinath and Veglia 2013). See Supplementary Tables 1-3 and Fig S5 and S6 for a detailed comparison of time savings using different sequences on SLN.
In this article, we use POE both in O-ssNMR and MAS spectroscopy to determine the structural topology of sarcolipin, a 3.8 kDa single transmembrane regulator of the sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA) (Odermatt et al. 1998; Traaseth et al. 2008). The traditional approach to solve membrane protein structure by ssNMR would involve the following experimental steps before structure calculation: a) Obtaining preliminary 2D spectra to optimize elements in the pulse sequence such as DARR mixing times, indirect evolution times as well as the determining the relative sensitivity and resolution attainable in each dimension, b) Obtaining 3D experiments based on the knowledge gained from the 2D experiments to sequentially assign the protein and c) complementing these secondary structure restraints obtained from MAS-ssNMR with orientation restraints on the backbone amide residues from O-ssNMR. We show the utility of DUMAS and MEIOSIS in combining multiple experiments to reduce experimental time, so one can simultaneously optimize mixing times, indirect evolution times as well as make decisions about which 3D experiments have the best chance of giving a complete sequential assignment. We also show the use of complementary 3D experiments simultaneously acquired to sequentially assign the protein. Finally we show the tremendous time savings that can be obtained by using the sensitivity enhanced versions of 3D O-ssNMR pulse sequences which give the complete set of orientation restraints on the protein backbone, viz. 15N and 1H anisotropic chemical shifts and 15N-1H dipolar couplings. The individual restraints (anisotropic and isotropic) obtained exclusively by these solid state NMR approaches are then combined in a manner analogous to the previously described hybrid approach (Shi et al. 2009b; Veglia et al. 2012) using an improved algorithm to converge to a structure ensemble with sub-angstrom precision.
Experimental Methods
Protein expression, purification and sample preparation
SLN was over-expressed in E. coli bacteria fused to a maltose binding protein (MBP), cleaved, and purified as described earlier (Buck et al. 2003; Veglia et al. 2010). Recombinant SLN was purified using reverse-phase HPLC. The fractions containing the protein were lyophilized and used for all solid state NMR experiments. The protein concentration was assessed using UV spectrophotometry using absorption at 280 nm (ε=10,300 M-1cm-1) as well as SDS-PAGE gels using densitometry. For the bicelle preparations, approximately 3.5mg SLN was dissolved in 6.7mg D6PC in 20mM HEPES/100mM NaCl buffer at pH 7.0 and then added to 31mg 4:1 DMPC/POPC vesicles suspension. After several freeze thaw cycles, the bicellar phases were formed and transferred to a cylindrical sample holder. For MAS preparations, 2.2mg of uniformly-[13C,15N]-SLN was reconstituted in DPC micelles and added to 15mg deuterated DMPC lipids dissolved in DPC micelles. The detergent was removed by incubating the solution with Biobeads SM2 (30:1 ratio with the detergent) for 3h at room temperature. The protein-lipid vesicles were collected as a pellet by centrifugation at 200,000xg at 4°C for 1 hour and packed in a 3.2mm MAS rotor for NMR experiments without further treatment.
NMR Spectroscopy
O-ssNMR Experiments
Experiments on bicelle preparations were carried out on a 16.85T VNMRS Spectrometer with a low-E bicelle probe built by the RF Program at NHMFL, Tallahassee, FL (Gor'kov et al. 2007). Temperature was maintained constant at 298.15 K and bicelle alignment was confirmed by a 31P spectrum. After initial optimization, the 3D-SE-PISEMAI-HETCOR experiment (Fig 1A) was performed with 80 transients for each of the 20 t1 (1H-15N-DC dimension, evolution time of 2.4ms) and 15 t2 (1H-CS dimension, evolution time of 1.13ms). A cross polarization time of 1000 μs, direct dimension acquisition time of 5ms, and a recycle delay of 3s were used in all experiments. A 50-70Hz Lorentz-to-Gauss apodization was used in the direct dimension and a 100Hz Gaussian apodization was used in the indirect dimensions. The matrix size before Fourier transformation was 8192 × 256 × 256.
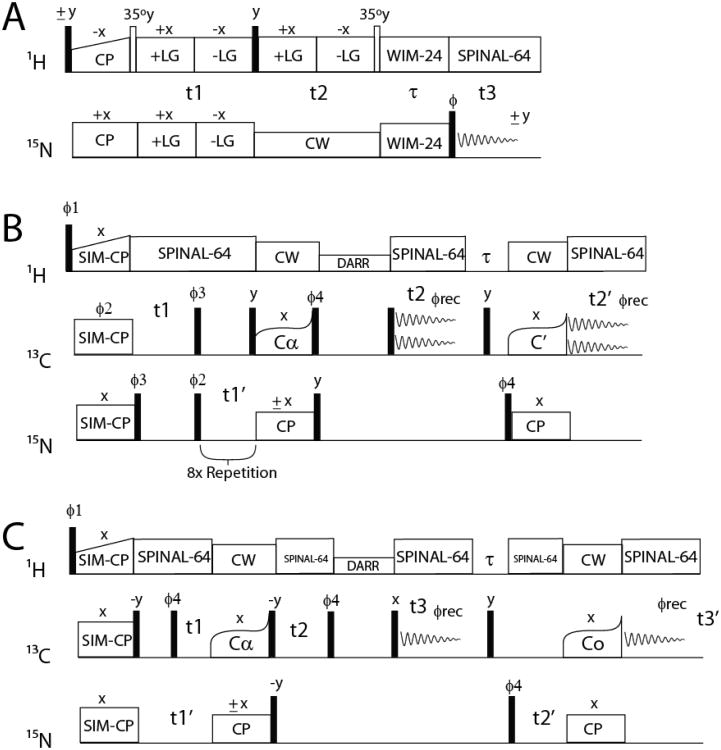
Fig. 1.
Pulse sequences O-ssNMR and MAS-ssNMR experiments.
(A) SE-PISEMAI-HETCOR, (B) 2D-MEIOSIS-(DARR/N(CA)CX/CA(N)CO/NCO) with the following phases: Φ; 1={y y y y −y −y −y −y}, Φ2={x x −x −x}2, Φ3={y −y y −y}2, Φ4={y y −y −y}, Φrec={x −x −x x −x x x −x} and (C) 3D-DUMAS(CANCO/NCACX) with the following phases: Φ1 = (y, −y)4, Φ:2 = (y, y, y, y, −y, −y, −y, −y), Φ3 = (x, x, −x, −x)2, Φrec = (x, −x, −x, x, −x, x, x, −x). For the pulse sequence in (B), the 13C carrier frequency is placed in the center of the spectral window for DARR (100ppm). SPECIFIC-CP is achieved by a phase modulated pulse on 13C that effectively changes the offset to 13Cα (64ppm) for the first transfer and to C′ (178ppm) for the second transfer.
MAS-ssNMR Experiments
MAS experiments on SLN in lipid vesicles were carried out on either a 14.09T or a 16.85T VNMRS Spectrometer with a 3.2mm BioMAS™ probe (Agilent Technologies) at a spinning speed of 10.0kHz and a temperature of 277.15 K. Two-dimensional MAS-ssNMR experiments were carried out by combining 13C-13C DARR, CA(N)CO, NCO and N(CA)CX experiments into a single 2D experiment using the MEIOSIS (Multiple Experiments via Orphan Spin Operators) strategy (Fig 1B) (Gopinath and Veglia 2013). Simultaneous cross-polarization from 1H to 13C and 15N was obtained using a contact time of 300μs, with the RF power of 15N and 13C channels set to 35kHz and 1H RF power optimized to achieve the n=2 side-band matching condition (59kHz). The phase of 15N spinlock during NCA transfer is altered between +x and −x and the resultant data is stored in separate files. Sum and difference of the two data sets with phases x and −x respectively gives DARR and N(CA)CX for the first acquisition, and NCO and CA(N)CO for the second acquisition. Fig S3 and Fig S4 show the dependence of the contact time for the first SPECIFIC-CP step on the distribution of polarization amongst different pathways. It was found that 3000μs was the optimal contact time, as it gave the maximum NCα transfer for both N(CA)CX and CA(N)CO experiments. As with a microcrystalline preparation of ubiquitin (Gopinath and Veglia 2013), we found that at this contact time, 54% residual polarization is left on 13C (to be used for a 13C-13C DARR experiment) and 34% residual polarization is left on 15N (to be used for a NCO experiment).
The 13C-edited experiments (DARR and CA(N)CO) were acquired with a dwell time of 32μs for 128 indirect points (t1 evolution of 4.096ms) while the 15N-edited experiments were acquired with a dwell time of 320μs for 16 indirect points (t1 evolution of 4.8ms). Acquisition time of 15ms and recycle delay of 2s was used. 64 transients were used for each t1 increment. Due to the reduced spectral width in the 15N dimension, the 15N-edited experiments can be acquired eight times along with a single acquisition of 13C-edited experiments. The number of transients for the NCO and N(CA)CX experiments thus becomes 512 (64 × 8). We acquired two independent experiments at the mixing times of 10ms and 50ms during the DARR transfer. As this transfer does not affect the NCO and CA(N)CO experiments, a further boost in sensitivity is obtained by adding these experiments, giving the total number of transients for NCO and CA(N)CO as 1024 and 128, respectively. As these are the least sensitive amongst the four concatenated experiments (Fig S3), this design allowed us to boost its sensitivity by repetition, even while optimizing the other experiments. Thus, in the end, we had a total of 6 spectra from 2 separate experiments − 13C-13C DARR with 2 different mixing times (with 64 transients each), N(CA)CX with 2 different mixing times (with 512 transients each), CA(N)CO (128 transients) and NCO (1024 transients). Note that the traditional approach would have allowed the recording of only 2 of these 6 experiments in the same time.
3D-DUMAS-ssNMR
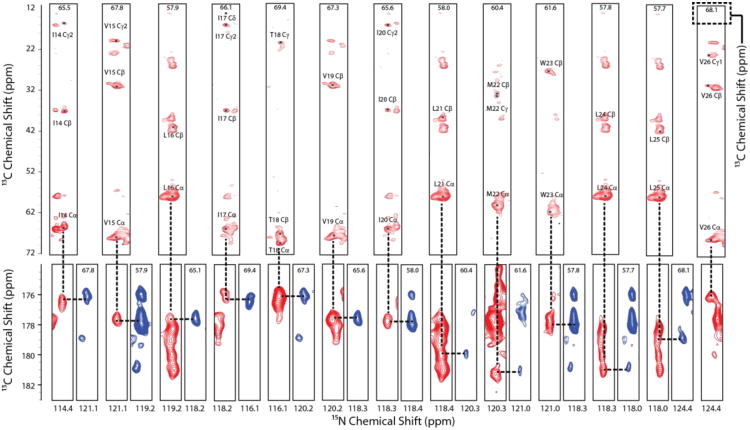
To assign the backbone and side chain atoms, we performed the 3D-DUMAS-NCACX-CANCO experiment (Fig 1C) at 277.15 K and 10.0 kHz MAS spinning. The t1 dimensions for both experiments were co-evolved with a dwell time of 150μs for the 13C-dimension (CANCO) and 300μs for the 15N-dimension (NCACX). 16 t1 points were obtained, giving a total of 2.4ms evolution for 13C and 4.8ms evolution for 15N respectively. Bidirectional cross polarization (Gopinath and Veglia 2012b) from 13Cα to 15N (CANCO) and 15N to 13Cα (NCACX) was achieved with a contact time of 3ms at RF field strength of 25 kHz (5.ωr/2) on 15N, and a 13C RF amplitude of 15 (3.ωr/2) and 35 (7.ωr/2) kHz for Cα and CO transfer respectively. 15N magnetization was stored along the z-direction with a 90° pulse after the t1 evolution periods. 13C-dimension (NCACX) was then evolved, followed by a 10ms DARR mixing period and finally detected (NCACX) with a 20ms acquisition time. After this acquisition, the 15N magnetization was brought to the transverse plane with another 90° pulse and evolved (CANCO). Magnetization was then transferred to 13CO with a 3ms contact time. The second detection of the 13C dimension (CANCO) was again carried out with an acquisition time of 20ms. The t2 dimensions were thus evolved independently for both experiments with 16 points each, giving a 15N (CANCO) evolution time of 4.8ms and 13C (NCACX) evolution time of 2.4ms. 512 transients were co-added for each t1 and t2 points. A recycle delay of 2 sec was used.
The 13C-dimension was referenced externally with adamantane at 40.48ppm and the 15N dimension was referenced indirectly from the 13C-dimension. All spectra were processed with NMRPipe (Delaglio et al. 1995) and analyzed with Sparky (Goddard and Kneller 2008). A Lorentz-to-Gauss apodization of 60-80Hz was used in the direct and indirect dimensions. The matrix size before Fourier transformation was 16384 × 512 × 512.
Structure Calculations
As the experiments were performed in lipid bicelles with the bicelle-director perpendicular to the external magnetic field, the 15N average isotropic chemical shift of 120ppm and 1H isotropic chemical shift of 8.1ppm were subtracted from each observed anisotropic chemical shift and scaled by 2.5 to obtain the input restraints to obtain the input for structure calculations. Observed dipolar couplings were also scaled by 2.5 to obtain the necessary restraints. This factor of 2.5 is a combination of the scaling of anisotropic parameters by 2 in the parallel orientation and a scaling factor of 1.25 to account for bicelle motions (Nevzorov 2011). Restraints were incorporated with an associated error of ±5ppm for 15N chemical shifts, ±1.5ppm for 1H chemical shifts and ±0.5kHz for dipolar couplings. Dihedral angle restraints from MAS-ssNMR were calculated using the isotropic chemical shifts of Cα, Cβ, C and N (amide) atoms as an input for TALOS+ (Shen et al. 2009) and implemented with an error of ±30°.
All oriented ssNMR restraints (anisotropic chemical shifts for amide 15N and amide 1H and 15N-1H dipolar couplings) and MAS-ssNMR restraints (dihedral restraints from TALOS) were combined in a single energy function along with potentials corresponding to standard peptide geometry (Echem = Ebond + Eangle + Eimproper + EvdW) (Schwieters et al. 2003). In addition, hydrogen bonds between the carbonyl oxygen and amide proton of {i} and {i+4} residues were implemented for the transmembrane segment. Orientation restraints were implemented with flat-well harmonic potentials using csaPot and rdcPot modules from XPLOR-NIH (version 2.29). These inbuilt C++ based modules allow for a faster calculation of structure than the PYTHON based modules (Bertram et al. 2000) we previously used (Shi et al. 2009b) for the structure determination. The 15N tensor was defined with the requisite parameters in XPLOR-NIH with the following traceless CSA tensor components: σ11=55.3ppm, σ22=-97.6ppm, σ33=43.3ppm, β= -17° and γ=0°. Similarly, 1H traceless tensor components were defined with σ11 = 6.3 ppm, σ22 = 1.3ppm, σ33 = -7.6ppm, β = 97° and γ = 0°(Yao et al. 2010).
The hybrid energy function (EO-ssnmr, EMas-ssnmr and Echem) was minimized using a simulated annealing protocol in the torsion angle space. Starting from an extended conformation of SLN, we carried out simulated annealing after an initial equilibration step at 6000K for 10ps. The temperature was decreased in steps of 5 K with a 0.5 ps time step to reach a final temperature of 10 K. A knowledge based Ez-potential (Senes et al. 2007) was implemented as this stage to insert SLN into a virtual membrane bilayer. The structure was then equilibrated at 10 K for 20 ps and subjected to Powell Minimization in Cartesian space. Of 200 structures generated, 10 lowest energy structures were selected for simulated annealing in the Cartesian space to relax local geometries. 1H-CSA restraints were included at this stage. After a 10ps equilibration at 300K, the temperature was reduced to 5K in steps of 2.5K, with 0.5ps simulation at each step. Ez-potential was again implemented and a final equilibration at 5K for 20ps followed by Powell minimization was carried out to give the final structure. One hundred structures were generated and the ten lowest energy conformers were selected for further analysis. The convergence of the lowest energy structures was determined by aligning the projections of each of the structures on the XY plane (Shi et al. 2009b). The RMSD of each conformer from the average structure was calculated without further manipulation of the coordinates in order to maintain the topology calculated from anisotropic NMR parameters. The tilt angle (with respect to the bilayer normal) and rotation angle (with respect to the Cα atom of residue R6) were calculated by aligning a model 20-residue alpha helix with the residues 14-28. Quality of the final structure ensemble was validated by ProCheck (Laskowski et al. 1996) and MolProbity (Davis et al. 2007), as well as the convergence of R-values for individual orientation restraints.
Results
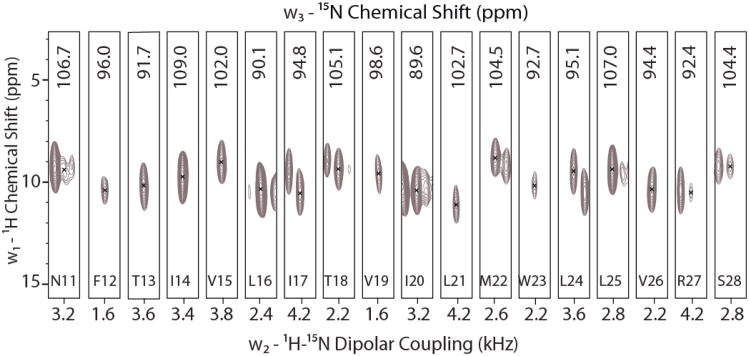
Magnetically aligned bicelle samples give resolved spectra with a sensitivity and resolution higher than mechanically aligned samples. The latter combined with the ease of sample preparation, better hydration, pH control and sample stability makes bicelles an excellent membrane mimetic for O-ssNMR experiments (Veglia et al. 2012; Dürr et al. 2012). This allows highly sensitive samples to be prepared for 3-dimensional spectroscopy. Fig S1 shows a typical 1D spectrum after initial optimization of CP conditions. For SLN, these preparations give 2D SLF spectra with remarkable resolution and a distinct PISA-wheel pattern (Page and Cross 2008; Mote et al. 2011). All these improvements make it possible to acquire 3D experiments with the sensitivity enhancement pulse sequences. Figure 2A shows strip plots from the 3D-SE-PISEMAI-HETCOR experiment. All of the resonances belonging to the transmembrane domain of SLN are present and well resolved, with characteristic oscillatory patterns (Mesleh et al. 2002; Mascioni and Veglia 2003) evident for the chemical shifts and the dipolar couplings. We have previously assigned the 2D SE-PISEMA spectrum for SLN in these lipids using a 3D-SE-PISEMA-PDSD experiment that allowed for a sequential assignment using 15N-15N correlations established by proton driven spin diffusion (Mote et al. 2011). This allowed a straightforward transfer of the assignment to the resonances observed in the 3D SE-PISEMAI-HETCOR experiment. The high quality of the spectrum, with linewidths for 15N-CS of ∼150Hz, 1H-CS of ∼800-1000 Hz and the 15N-1H DC of ∼100Hz allowed us to obtain the set of orientation restraints that completely describe the backbone topology.
Fig. 2.
Strip plot of the 3D-SE-PISEMAI-HETCOR spectrum. All resonances from N11 to S28 are completely resolved and assigned based on our previous assignment for the 2D spectrum. Average signal is 55σ (range of 24σ-124σ). Spectrum is plotted starting at 18σ with contours progressing by factor of 1.2.
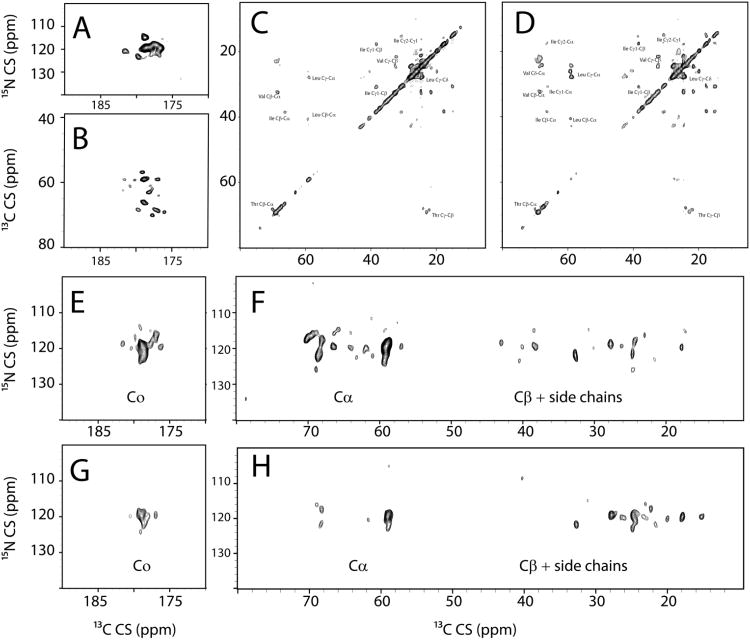
For a rapid screen to determine the relative dispersion attainable in each dimension, as well as optimizing mixing times for the DARR element (Takegoshi et al. 2001) in the pulse sequence, we used the 2D-MEIOSIS experiment. Figure 3 shows a variant of this experiment, i.e. MEIOSIS-(DARR-CA(N)CO-NCO-N(CA)CX), in which the four 2D spectra are obtained simultaneously within a single experiment. Although several peaks are resolved and residue type assignments are easily obtained from these experiments, they give only an ambiguous assignment. The comparatively low resolution in the 2D spectra is a direct result of fast T2 relaxation, which is also observed for other small membrane embedded proteins (Su et al. 2011), and underscores the importance of 3D experiments to assign these proteins. In the N(CA)CX obtained at 10ms mixing time, it is possible to identify several intense correlations, indicating that the mixing period is sufficient to transfer the polarization from the backbone to the side chains. At a 10ms mixing time, the N(CA)CX spectrum shows several Cβ and C' resonances. At a 50ms mixing time, the Cβ and possibly the other side chain peaks increase in intensity at the expense of both C' and Cα. Our assignment strategy relies on obtaining information on residue type using statistically predicted chemical shifts and connecting these residues via a ‘backbone-walk’. Based on this screening, we concluded that the DUMAS-CANCO/NCACX pulse sequence with a 10ms mixing time during the DARR transfer in NCACX is optimal for assigning SLN, as it would give information on the residue type in the NCACX spectrum and also allow a simultaneous main-chain walk via the common carbonyl resonance of the {i} and {i+1} spin systems. A mixing time of 10ms was deemed optimal as longer mixing times decreased the intensity of the C' resonances. Therefore, we acquired two 3D experiments simultaneously using the 3D-DUMAS-CANCO/NCACX pulse sequence, with a 10ms mixing time for the NCACX spectrum during 13C-13C DARR transfer. After assignment of a majority of residue types in the transmembrane segment based on the statistical Cα and Cβ chemical shifts obtained from the 2D and the 3D spectra, we sequentially assigned the transmembrane segment from residue E7 to V26 by the classical ‘main chain walk’, aligning the carbonyl chemical shifts for {i} and {i+1} residues. This assignment was aided by relatively high sequence variability in the SLN transmembrane domain, which has threonine, methionine and tryptophan residues in addition to the hydrophobic leucine, isoleucine and valine residues frequently found in TM domains. The leucine region of the spectrum (∼56ppm) suffers from severe overlap of resonance. However, with resolution in the carbonyl region of the spectrum, we were also able to assign these residues. Residues N4-R6 which are known to undergo conformational dynamics, were not assigned, possibly due to inefficient cross-polarization for these residues. The terminal residues M1-I3 and Y31 were assigned in a refocused-INEPT experiment at 298.15K (Supplementary Fig 2).
Fig. 3.
Spectra obtained with MEIOSIS-(DARR/CA(N)CO/N(CA)CX/NCO) pulse sequence (A) NCO Correlation, (B) CA(N)CO Correlation. (C) 13C-13C DARR Correlation with 10ms mixing time, (D) 13C-13C DARR Correlation with 10ms mixing time, (E,F) N(CA)CX correlations with 10ms mixing time and (G,H) N(CA)CX correlation with 50ms mixing time. (C) and (D) are plotted starting at 7σ and contours progressing by a factor of 1.1. All other spectra are plotted starting at 6σ with contours progressing by a factor of 1.1.
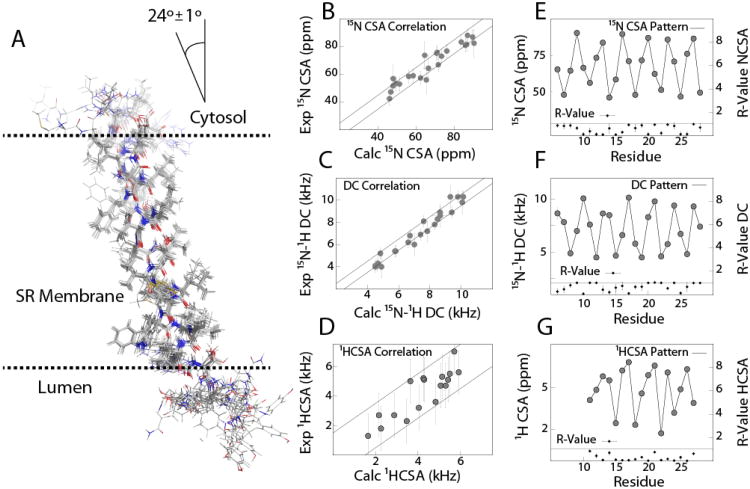
Figure 5A shows the ensembles of the SLN conformers obtained. The high precision of the ensemble (backbone, incl. HN + Cβ RMSD ∼0.44 Å) is also reflected in low energies for all structural restraints. The global as well as the individual R-values for all orientation restraints are converged below 1 (Figure 5 E-G), indicating a good agreement between the calculated structures and the experimental O-ssNMR data. Table 1 summarizes the structural statistics for these calculations. The hybrid NMR ensemble shows disordered N- and C- termini in agreement with the solution NMR ensemble obtained in DPC micelles (Mascioni et al. 2002; Buffy et al. 2006a). In fact, experiments utilizing cross polarization both in bicelles and lipid bilayers are unable to detect the resonances corresponding to residues 1-3 and 31. These were assigned using the refocused-INEPT experiment at 298K (Fig S3). The resonances corresponding to residues 29-31 detected by SE-PISEMA experiments in aligned bicelles show dipolar couplings that are averaged by the substantial motions in these segments and hence were not included in the final structure calculations. For the most structured residues (Residues 6-28), hybrid NMR ensemble shows a higher precision than the corresponding solution NMR ensemble (PDB Code: 1JDM). The topology is converged to a well-defined minimum in the conformational space. The tilt angle for SLN with respect to the bilayer normal was found to be 24±1° and the rotation angle was 57±5°. The indole ring of W23 is oriented with its plane parallel to the bilayer normal, with the N-H bond vector oriented in the direction of the C-terminal bilayer interface, possibly making contact with the phosphate head groups. Along with W23, the conserved residues R6, located near the cytosolic N-terminus, and R27, near the lumenal C-terminus, anchor SLN to the lipid membrane, modulating the tilt and rotation angles which seem to be only minimally affected in the different membrane mimetic systems (Mote et al. 2011). The NMR structural ensembles determined by the hybrid method are in good agreement with molecular dynamics simulations (Shi et al. 2009a) which show a similar orientation of the indole side chain of W23, and a previous oriented solid state NMR study (Buffy et al. 2006b) carried out mechanically aligned DOPC/DOPE bilayers, which shows a similar the tilt angle of ∼23° with respect to the bilayer.
Fig. 5.
(A) Structure ensemble obtained for SLN from the hybrid MAS/O-ssNMR structure calculations (10 lowest energy structures) (B-D) Correlation of experimental O-ssNMR restraints with the back-calculated restraints (E-F) Oscillation patterns for all orientation restraints and the R-values of back-calculated restraints
Table 1. Summary of Structure Ensemble Statistics (10 structures).
| RMSD from MAS NMR Restraints | |
| NH-C(O) hydrogen bond, Å | 0.052 ± 0.004 |
| Dihedral Angles (TALOS) | 0.000 ± 0.000 |
| PISEMA R-factors | |
| 15N CSA | 0.5 ± 0.4 |
| 15N-1H DC | 0.7 ± 0.3 |
| 1H CSA | 0.3 ± 0.3 |
| RMSD from Idealized Covalent Geometry | |
| Bond, Å | 0.004 ± 0.000 |
| Angle, ° | 0.60 ± 0.01 |
| Improper angle, ° | 0.13 ± 0.01 |
| Measure of Structure Quality, % | |
| Most Favored Region | 99 ± 1 |
| Additionally Allowed Region | 1 ± 1 |
| Generously Allowed Region | 0 |
| Disallowed Region MolProbity® Score | 098 ± 2 percentile |
| Precision of Structures (RMSD for atoms NH+HN+C'+0+Cα+Cβ), Å | |
| Domain Ib+II (Res 6-28) | 0.43 |
| Domain Ib (Res 6-13) | 0.44 |
| Domain II (Res 14-28) | 0.40 |
| Protein Topology | |
| Tilt Angle (Res 6-28),° | 24 ± 1 |
| Rotation Angle (from R6-Cα), ° | 55 ± 5 |
| Depth of Insertion (R6-Cα), Å | 16.1 ± 0.2 |
Discussion
While multidimensional MAS-ssNMR techniques are mature for structure determination of large systems, their application to membrane proteins is limited by the sensitivity and resolution that can be attained in these systems. Obtaining complete resolution is often not possible without 3D experiments, which take a long time due to the poor sensitivity of these systems. The DUMAS strategy allowed us to substantially reduce experimental time as compared with traditional MAS-ssNMR pulse sequences.
The initial experiments with MEIOSIS allowed the determination of parameters, such as the appropriate mixing time, that were important to set up the 3-dimensional experiments for assignments. These initial experiments also allowed us to determine the best possible experimental approach to assign the protein based on relative resolution attainable in each dimension. The DUMAS-CANCO/NCACX experiment allowed us to assign a majority of the protein sequentially as the experiment combines the complementary pulse sequence into a single experiment. Moreover, the simultaneous acquisition of both the experiments allowed the data to be fairly compared without the need for concern regarding sample stability or correction factors.
Compared to MAS-ssNMR, multidimensional O-ssNMR spectroscopy is still in its infancy, in spite of the fact that the number of membrane protein structures being solved by this technique is greater. Sensitivity gains obtained from the 3D-SE-PISEMAI-HETCOR are a substantial leap forward in obtaining complete resolution for these studies. It must be noted here that the resolution of the structure depends primarily on the O-ssNMR data, as the secondary structure obtained from MAS-ssNMR experiments contains no information on the topological parameters of tilt and rotation angle. As such, the O-ssNMR experiments described herein are indispensable in order to characterize the structure of membrane proteins precisely. TALOS based dihedral angle restraints obtained from MAS-ssNMR are a perfect complement to these orientation restraints from O-ssNMR and help convergence of the structure ensemble in spite of inherent degeneracy in the observed values of dipolar couplings and anisotropic chemical shifts.
In the past these MAS-ssNMR and O-ssNMR techniques were thought to be mutually exclusive and most of the researchers used either MAS or O-ssNMR to obtain de novo structure determination. Recent developments of structural biology emphasize the use of multiple techniques to substantiate biological and structural data (Traaseth et al. 2007; Shi et al. 2011b). In this spirit, we combined MAS-ssNMR and O-ssNMR restraints into a unique refinement protocol. The idea to combine these techniques is similar to solution NMR, where distance and torsion angle restraints are energy minimized together with residual dipolar couplings to obtain high-resolution structures and reciprocal orientations of protein domains. While some concerns can be raised regarding the compatibility of solution NMR restraints in micelles, which can also serve as complements to O-ssNMR restraints, the agreement between MAS and O-ssNMR data stems from the similarity between the two membrane mimicking environments.
Conclusions
We show here that the complete set of high resolution restraints for both MAS-ssNMR and O-ssNMR can be conveniently obtained for membrane proteins using POE for both oriented and MAS-ssNMR spectroscopy. The combination of structural and topological restraints from these techniques into a hybrid NMR calculation approach makes it possible to determine the high resolution structure and topology of membrane proteins. Although we anticipate that POE experiments will have the biggest impact on data acquisition of membrane proteins, they are universally applicable to other biomacromolecular systems.
Supplementary Material
Fig. 4.
Strip plots from the 3D-DUMAS-NCACX (red)/CANCO (blue) experiment. NCACX spectrum is plotted starting at 12σ (Cα-Cβ region) and 8.5σ (C region) and contours progressing by a factor of 1.1. Average signal per residue for NCACX is 28σ (Cα-Cβ region, range 12σ-63σ) and 15σ (carbonyl region, range 9σ to 25σ). CANCO spectrum is plotted starting at 6σ and contours progressing by a factor 1.1. The average signal for per residue is 10σ (range 7σ to 20σ). The average linewidth in each spectrum is ∼1.1 ppm for 13Cα (indirect dimension), 1.0ppm 13C (direct dimension) and 2.5ppm for 15N (indirect dimension)
Acknowledgments
We thank Dr. Martin Gustavsson and Dr. Vitaly Vostrikov for helpful discussions. This work is supported by the National Institute of Health (GM64742 to G.V.). The experiments were carried out at the University Of Minnesota NMR Center.
Abbreviations
- SLN
Sarcolipin
- SERCA
sarcoplasmic reticulum Ca2+-ATPase
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphocholine
- D6PC
1,2-dihexanoyl-sn-glycero-3-phosphocholine
- POPC
1-palmitoyl, 2-oleyl-sn-glycero-3-phosphocholine
- PISA
Polarity Index Slant Angle
- DUMAS
Dual Acquisition Magic Angle Spinning
- MEIOSIS
Multiple Experiments via Orphan Spin Operators
References
- Asami S, Szekely K, Schanda P, Meier BH, Reif B. Optimal degree of protonation for 1H detection of aliphatic sites in randomly deuterated proteins as a function of the MAS frequency. J Biomol NMR. 2012;54:155–168. doi: 10.1007/s10858-012-9659-9. [DOI] [PubMed] [Google Scholar]
- Bertram R, Quine JR, Chapman MS, Cross TA. Atomic refinement using orientational restraints from solid-state NMR. J Magn Reson. 2000;147:9–16. doi: 10.1006/jmre.2000.2193. [DOI] [PubMed] [Google Scholar]
- Buck B, Zamoon J, Kirby TL, DeSilva TM, Karim C, Thomas D, Veglia G. Overexpression, purification, and characterization of recombinant Ca-ATPase regulators for high-resolution solution and solid-state NMR studies. Protein Expression Purif. 2003;30:253–261. doi: 10.1016/s1046-5928(03)00127-x. [DOI] [PubMed] [Google Scholar]
- Buffy JJ, Buck-Koehntop BA, Porcelli F, Traaseth NJ, Thomas DD, Veglia G. Defining the intramembrane binding mechanism of sarcolipin to calcium ATPase using solution NMR spectroscopy. J Mol Biol. 2006a;358:420–429. doi: 10.1016/j.jmb.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Buffy JJ, Traaseth NJ, Mascioni A, Gor'kov PL, Chekmenev EY, Brey WW, Veglia G. Two-dimensional solid-state NMR reveals two topologies of sarcolipin in oriented lipid bilayers. Biochemistry. 2006b;45:10939–10946. doi: 10.1021/bi060728d. [DOI] [PubMed] [Google Scholar]
- Cady SD, Mishanina TV, Hong M. Structure of amantadine-bound M2 transmembrane peptide of influenza A in lipid bilayers from magic-angle-spinning solid-state NMR: the role of Ser31 in amantadine binding. J Mol Biol. 2009;385:1127–1141. doi: 10.1016/j.jmb.2008.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canlas CG, Ma D, Tang P, Xu Y. Residual Dipolar Coupling Measurements of Transmembrane Proteins Using Aligned Low-q Bicelles and High-Resolution Magic Angle Spinning NMR Spectroscopy. J Am Chem Soc. 2008;130:13294–13300. doi: 10.1021/ja802578z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellani F, Rossum B, van Diehl A, Schubert M, Rehbein K, Oschkinat H. Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy. Nature. 2002;420:98–102. doi: 10.1038/nature01070. [DOI] [PubMed] [Google Scholar]
- Das BB, Nothnagel HJ, Lu GJ, Son WS, Tian Y, Marassi FM, Opella SJ. Structure determination of a membrane protein in proteoliposomes. J Am Chem Soc. 2012;134:2047–2056. doi: 10.1021/ja209464f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Demers JP, Chevelkov V, Lange A. Progress in correlation spectroscopy at ultra-fast magic-angle spinning: basic building blocks and complex experiments for the study of protein structure and dynamics. Solid State Nucl Magn Reson. 2011;40:101–113. doi: 10.1016/j.ssnmr.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Dürr UHN, Gildenberg M, Ramamoorthy A. The magic of bicelles lights up membrane protein structure. Chem Rev. 2012;112:6054–6074. doi: 10.1021/cr300061w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvinskikh SV, Yamamoto K, Ramamoorthy A. Heteronuclear isotropic mixing separated local field NMR spectroscopy. J Chem Phys. 2006;125:034507–034519. doi: 10.1063/1.2212939. [DOI] [PubMed] [Google Scholar]
- Giraud N, Blackledge M, Goldman M, Böckmann A, Lesage A, Penin F, Emsley L. Quantitative analysis of backbone dynamics in a crystalline protein from nitrogen-15 spin-lattice relaxation. J Am Chem Soc. 2005;127:18190–18201. doi: 10.1021/ja055182h. [DOI] [PubMed] [Google Scholar]
- Goddard TG, Kneller DG. SPARKY. Vol. 3. University of California; San Francisco: 2008. p. 114. [Google Scholar]
- Gopinath T, Mote KR, Veglia G. Proton evolved local field solid-state nuclear magnetic resonance using Hadamard encoding: theory and application to membrane proteins. J Chem Phys. 2011;135:074503. doi: 10.1063/1.3622604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath T, Traaseth NJ, Mote K, Veglia G. Sensitivity enhanced heteronuclear correlation spectroscopy in multidimensional solid-state NMR of oriented systems via chemical shift coherences. J Am Chem Soc. 2010a;132:5357–5363. doi: 10.1021/ja905991s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath T, Veglia G. Sensitivity enhancement in static solid-state NMR experiments via single- and multiple-quantum dipolar coherences. J Am Chem Soc. 2009;131:5754–5756. doi: 10.1021/ja900096d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath T, Veglia G. Improved Resolution in Dipolar NMR Spectra Using Constant Time Evolution PISEMA Experiment. Chem Phys Lett. 2010;494:104–110. doi: 10.1016/j.cplett.2010.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath T, Veglia G. Dual acquisition magic-angle spinning solid-state NMR-spectroscopy: simultaneous acquisition of multidimensional spectra of biomacromolecules. Angew Chem, Int Ed. 2012a;51:2731–2735. doi: 10.1002/anie.201108132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath T, Veglia G. 3D DUMAS: simultaneous acquisition of three-dimensional magic angle spinning solid-state NMR experiments of proteins. J Magn Reson. 2012b;220:79–84. doi: 10.1016/j.jmr.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath T, Veglia G. Orphan spin operators enable the acquisition of multiple 2D and 3D magic angle spinning solid-state NMR spectra. J Chem Phys. 2013;138:184201. doi: 10.1063/1.4803126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath T, Verardi R, Traaseth NJ, Veglia G. Sensitivity enhancement of separated local field experiments: application to membrane proteins. J Phys Chem B. 2010b;114:5089–5095. doi: 10.1021/jp909778a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gor'kov PL, Chekmenev EY, Li C, Cotten M, Buffy JJ, Traaseth NJ, Veglia G, Brey WW. Using low-E resonators to reduce RF heating in biological samples for static solid-state NMR up to 900 MHz. J Magn Reson. 2007;185:77–93. doi: 10.1016/j.jmr.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Hall DA, Maus DC, Gerfen GJ, Inati SJ, Becerra LR, Dahlquist FW, Griffin RG. Polarization-enhanced NMR spectroscopy of biomolecules in frozen solution. Science. 1997;276:930–932. doi: 10.1126/science.276.5314.930. [DOI] [PubMed] [Google Scholar]
- Hu F, Luo W, Hong M. Mechanisms of proton conduction and gating in influenza M2 proton channels from solid-state NMR. Science. 2010;330:505–508. doi: 10.1126/science.1191714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketchem RR, Hu W, Cross TA. High-resolution conformation of gramicidin A in a lipid bilayer by solid-state NMR. Science. 1993;261:1457–1460. doi: 10.1126/science.7690158. [DOI] [PubMed] [Google Scholar]
- Knight MJ, Pell AJ, Bertini I, Felli IC, Gonnelli L, Pierattelli R, Herrmann T, Emsley L, Pintacuda G. Structure and backbone dynamics of a microcrystalline metalloprotein by solid-state NMR. Proc Natl Acad Sci U S A. 2012;109:11095–11100. doi: 10.1073/pnas.1204515109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Linser R, Bardiaux B, Higman V, Fink U, Reif B. Structure calculation from unambiguous long-range amide and methyl 1H-1H distance restraints for a microcrystalline protein with MAS solid-state NMR spectroscopy. J Am Chem Soc. 2011;133:5905–5912. doi: 10.1021/ja110222h. [DOI] [PubMed] [Google Scholar]
- Lopez JJ, Kaiser C, Asami S, Glaubitz C. Higher sensitivity through selective (13)C excitation in solid-state NMR spectroscopy. J Am Chem Soc. 2009;131:15970–15971. doi: 10.1021/ja904963n. [DOI] [PubMed] [Google Scholar]
- Loquet A, Lv G, Giller K, Becker S, Lange A. 13C spin dilution for simplified and complete solid-state NMR resonance assignment of insoluble biological assemblies. J Am Chem Soc. 2011;133:4722–4725. doi: 10.1021/ja200066s. [DOI] [PubMed] [Google Scholar]
- Loquet A, Sgourakis NG, Gupta R, Giller K, Riedel D, Goosmann C, Griesinger C, et al. Atomic model of the type III secretion system needle. Nature. 2012;486:276–279. doi: 10.1038/nature11079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti A, Jehle S, Felletti M, Knight MJ, Wang Y, Xu ZQ, Park AY, et al. Backbone assignment of fully protonated solid proteins by (1)H detection and ultrafast magic-angle-spinning NMR spectroscopy. Angew Chem, Int Ed. 2012;51:10756–10759. doi: 10.1002/anie.201203124. [DOI] [PubMed] [Google Scholar]
- Mascioni A, Karim C, Barany G, Thomas DD, Veglia G. Structure and orientation of sarcolipin in lipid environments. Biochemistry. 2002;41:475–482. doi: 10.1021/bi011243m. [DOI] [PubMed] [Google Scholar]
- Mascioni A, Veglia G. Theorotical Analysis of Residual Dipolar Couplings in Regular Secondary Structure of Proteins. 2003;125:12520–12526. doi: 10.1021/ja0354824. [DOI] [PubMed] [Google Scholar]
- Maudsley AA, Ernst RR. Indirect detection of magnetic resonance by heteronuclear two-dimensional spectroscopy. Chem Phys Lett. 1977;50:368–372. [Google Scholar]
- Mesleh MF, Veglia G, DeSilva TM, Marassi FM, Opella SJ. Dipolar waves as NMR maps of protein structure. J Am Chem Soc. 2002;124:4206–4207. doi: 10.1021/ja0178665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Qin H, Fu R, Sharma M, Can TV, Hung I, Luca S, Gor'kov PL, Brey WW, Cross TA. M2 Proton Channel Structural Validation from Full-Length Protein Samples in Synthetic Bilayers and E. coli Membranes. Angew Chem, Int Ed. 2012;51:8383–8386. doi: 10.1002/anie.201204666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mote KR, Gopinath T, Traaseth NJ, Kitchen J, Gor'kov PL, Brey WW, Veglia G. Multidimensional oriented solid-state NMR experiments enable the sequential assignment of uniformly 15N labeled integral membrane proteins in magnetically aligned lipid bilayers. J Biomol NMR. 2011;51:339–346. doi: 10.1007/s10858-011-9571-8. [DOI] [PubMed] [Google Scholar]
- Nevzorov AA. Orientational and motional narrowing of solid-state NMR lineshapes of uniaxially aligned membrane proteins. J Phys Chem B. 2011;115:15406–15414. doi: 10.1021/jp2092847. [DOI] [PubMed] [Google Scholar]
- Nevzorov AA, Opella SJ. A “magic sandwich” pulse sequence with reduced offset dependence for high-resolution separated local field spectroscopy. J Magn Reson. 2003;164:182–186. doi: 10.1016/s1090-7807(03)00240-4. [DOI] [PubMed] [Google Scholar]
- Nielsen AB, Székely K, Gath J, Ernst M, Nielsen NC, Meier BH. Simultaneous acquisition of PAR and PAIN spectra. Journal of biomolecular NMR. 2012;52:283–288. doi: 10.1007/s10858-012-9616-7. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop AJ, Rienstra CM. Supramolecular protein structure determination by site-specific long-range intermolecular solid state NMR spectroscopy. J Am Chem Soc. 2010;132:7570–7571. doi: 10.1021/ja100992y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odermatt A, Becker S, Khanna VK, Kurzydlowski K, Leisner E, Pette D, MacLennan DH. Sarcolipin regulates the activity of SERCA1, the fast-twitch skeletal muscle sarcoplasmic reticulum Ca2+-ATPase. J Biol Chem. 1998;273:12360–12369. doi: 10.1074/jbc.273.20.12360. [DOI] [PubMed] [Google Scholar]
- Page RC, Cross TA. Transmembrane helix uniformity examined by spectral mapping of torsion angles. Structure. 2008;16:787–797. doi: 10.1016/j.str.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Das BB, Casagrande F, Tian Y, Nothnagel HJ, Chu M, Kiefer H, et al. Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature. 2012;491:779–783. doi: 10.1038/nature11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Marassi FM, Black D, Opella SJ. Structure and dynamics of the membrane-bound form of Pf1 coat protein: implications of structural rearrangement for virus assembly. Biophys J. 2010;99:1465–1474. doi: 10.1016/j.bpj.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif B. Ultra-high resolution in MAS solid-state NMR of perdeuterated proteins: implications for structure and dynamics. J Magn Reson. 2012;216:1–12. doi: 10.1016/j.jmr.2011.12.017. [DOI] [PubMed] [Google Scholar]
- Reif B, Jaroniec CP, Rienstra CM, Hohwy M, Griffin RG. 1H-1H MAS correlation spectroscopy and distance measurements in a deuterated peptide. J Magn Reson. 2001;151:320–327. doi: 10.1006/jmre.2001.2354. [DOI] [PubMed] [Google Scholar]
- Renault M, Pawsey S, Bos MP, Koers EJ, Nand D, Tommassen-van Boxtel R, Rosay M, Tommassen J, Maas WE, Baldus M. Solid-state NMR spectroscopy on cellular preparations enhanced by dynamic nuclear polarization. Angew Chem, Int Ed. 2012a;51:2998–3001. doi: 10.1002/anie.201105984. [DOI] [PubMed] [Google Scholar]
- Renault M, Tommassen-van Boxtel R, Bos MP, Post JA, Tommassen J, Baldus M. Cellular solid-state nuclear magnetic resonance spectroscopy. Proc Natl Acad Sci U S A. 2012b;109:4863–4868. doi: 10.1073/pnas.1116478109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Rohr K, Nanz D, Emsley L, Pines A. NMR Measurement of Resolved Heteronuclear Dipole Couplings in Liquid Crystals and Lipids. J Phys Chem. 1994;98:6668–6670. [Google Scholar]
- Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Senes A, Chadi DC, Law PB, Walters RF, Nanda V, Degrado WF. E(z), a depth-dependent potential for assessing the energies of insertion of amino acid side-chains into membranes: derivation and applications to determining the orientation of transmembrane and interfacial helices. J Mol Biol. 2007;366:436–448. doi: 10.1016/j.jmb.2006.09.020. [DOI] [PubMed] [Google Scholar]
- Sharma M, Yi M, Dong H, Qin H, Peterson E, Busath DD, Zhou HX, Cross TA. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science. 2010;330:509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Cembran A, Gao J, Veglia G. Tilt and azimuthal angles of a transmembrane peptide: a comparison between molecular dynamics calculations and solid-state NMR data of sarcolipin in lipid membranes. Biophys J. 2009a;96:3648–3662. doi: 10.1016/j.bpj.2009.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Kawamura I, Jung KH, Brown LS, Ladizhansky V. Conformation of a seven-helical transmembrane photosensor in the lipid environment. Angew Chem, Int Ed. 2011a;50:1302–1305. doi: 10.1002/anie.201004422. [DOI] [PubMed] [Google Scholar]
- Shi L, Traaseth NJ, Verardi R, Cembran A, Gao J, Veglia G. A refinement protocol to determine structure, topology, and depth of insertion of membrane proteins using hybrid solution and solid-state NMR restraints. J Biomol NMR. 2009b;44:195–205. doi: 10.1007/s10858-009-9328-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Traaseth NJ, Verardi R, Gustavsson M, Gao J, Veglia G. Paramagnetic-based NMR restraints lift residual dipolar coupling degeneracy in multidomain detergent-solubilized membrane proteins. J Am Chem Soc. 2011b;133:2232–2241. doi: 10.1021/ja109080t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling LJ, Berthold DA, Sasser TL, Jeisy-Scott V, Rienstra CM. Assignment strategies for large proteins by magic-angle spinning NMR: the 21-kDa disulfide-bond-forming enzyme DsbA. J Mol Biol. 2010;399:268–282. doi: 10.1016/j.jmb.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Waring AJ, Ruchala P, Hong M. Structures of beta-hairpin antimicrobial protegrin peptides in lipopolysaccharide membranes: mechanism of gram selectivity obtained from solid-state nuclear magnetic resonance. Biochemistry. 2011;50:2072–2083. doi: 10.1021/bi101975v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takegoshi K, Nakamura S, Terao T. Dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett. 2001;344:631–637. doi: 10.1063/1.2364503. [DOI] [PubMed] [Google Scholar]
- Tamm LK. Protein Lipid Interactions: From Membrane Domains to Cellular Networks. Wiley-VCH Verlag GmbH & Co. KGaA; Weinheim, Germany: 2005. [Google Scholar]
- Tang M, Sperling LJ, Berthold DA, Schwieters CD, Nesbitt AE, Nieuwkoop AJ, Gennis RB, Rienstra CM. High-resolution membrane protein structure by joint calculations with solid-state NMR and X-ray experimental data. J Biomol NMR. 2011;51:227–233. doi: 10.1007/s10858-011-9565-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traaseth NJ, Ha KN, Verardi R, Shi L, Buffy JJ, Masterson LR, Veglia G. Structural and dynamic basis of phospholamban and sarcolipin inhibition of Ca(2+)-ATPase. Biochemistry. 2008;47:3–13. doi: 10.1021/bi701668v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traaseth NJ, Shi L, Verardi R, Mullen DG, Barany G, Veglia G. Structure and topology of monomeric phospholamban in lipid membranes determined by a hybrid solution and solid-state NMR approach. Proc Natl Acad Sci U S A. 2009;106:10165–10170. doi: 10.1073/pnas.0904290106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traaseth NJ, Verardi R, Torgersen KD, Karim CB, Thomas DD, Veglia G. Spectroscopic validation of the pentameric structure of phospholamban. Proc Natl Acad Sci U S A. 2007;104:14676–14681. doi: 10.1073/pnas.0701016104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veglia G, Ha KN, Shi L, Verardi R, Traaseth NJ. What can we learn from a small regulatory membrane protein? Methods Mol Biol. 2010;654:303–319. doi: 10.1007/978-1-60761-762-4_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veglia G, Traaseth NJ, Shi L, Verardi R, Gopinath T, Gustavsson M. The Hybrid Solution/Solid-State NMR Method for Membrane Protein Structure Determination. In: Egelman EH, editor. Comprehensive Biophysics. Elsevier; Amsterdam: 2012. pp. 182–198. [Google Scholar]
- Verardi R, Shi L, Traaseth NJ, Walsh N, Veglia G. Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method. Proc Natl Acad Sci U S A. 2011;108:9101–9106. doi: 10.1073/pnas.1016535108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SH. Biophysical dissection of membrane proteins. Nature. 2009;459:344–346. doi: 10.1038/nature08142. [DOI] [PubMed] [Google Scholar]
- Wu CH, Ramamoorthy A, Opella SJ. High-Resolution Heteronuclear Dipolar Solid-State NMR Spectroscopy. J Magn Reson A. 1994;109:270–272. [Google Scholar]
- Wylie BJ, Sperling LJ, Nieuwkoop AJ, Franks WT, Oldfield E, Rienstra CM. Ultrahigh resolution protein structures using NMR chemical shift tensors. Proc Natl Acad Sci U S A. 2011;108:16974–16979. doi: 10.1073/pnas.1103728108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L, Grishaev A, Cornilescu G, Bax A. The impact of hydrogen bonding on amide 1H chemical shift anisotropy studied by cross-correlated relaxation and liquid crystal NMR spectroscopy. J Am Chem Soc. 2010;132:10866–10875. doi: 10.1021/ja103629e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou DH, Nieuwkoop AJ, Berthold DA, Comellas G, Sperling LJ, Tang M, Shah GJ, Brea EJ, Lemkau LR, Rienstra CM. Solid-state NMR analysis of membrane proteins and protein aggregates by proton detected spectroscopy. J Biomol NMR. 2012;54:291–305. doi: 10.1007/s10858-012-9672-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.