Abstract
Objectives
There is growing evidence to suggest that human endogenous retroviruses (HERVs) have contributed to human evolution, being expressed in development, normal physiology and disease. A key difficulty in the scientific evaluation of this potential viral contribution is the accurate demonstration of virally expressed protein in specific human cells and tissues. In this study, we have adopted the endogenous retrovirus, ERV3, as our test model in developing a reliable high-capacity methodology for the expression of such endogenous retrovirus-coded protein.
Design
Two affinity-purified polyclonal antibodies to ERV3 Env-encoded protein were generated to detect the corresponding protein expression pattern in specific human cells, tissues and organs.
Participants
Sampling included normal tissues from 144 individuals ranging from childhood to old age. This included more than forty different tissues and organs and some 216 different cancer tissues representing the twenty commonest forms of human cancer.
Setting
The Rudbeck Laboratory, Uppsala University and Uppsala University Hospital, Uppsala, Sweden.
Main Outcome Measures
The potential expression at likely physiological level of the ERV3Env encoded protein in a wide range of human cells, tissues and organs.
Results
We found that ERV3 encoded Env protein is expressed at substantive levels in placenta, testis, adrenal gland, corpus luteum, Fallopian tubes, sebaceous glands, astrocytes, bronchial epithelium and the ducts of the salivary glands. Substantive expression was also seen in a variety of epithelial cells as well as cells known to undergo fusion in inflammation and in normal physiology, including fused macrophages, myocardium and striated muscle. This contrasted strongly with the low levels expressed in other tissues types. These findings suggest that this virus plays a significant role in human physiology and may also play a possible role in disease.
Conclusion
This technique can now be extended to the study of other HERV genomes within the human chromosomes that may have contributed to human evolution, physiology and disease.
Keywords: Endogenous retrovirus 3 (ERV3), Envelope protein, human tissues, antibody based proteomics, Tissue microarry (TMA)
Introduction
The ongoing Human Protein Atlas Project, recently reviewed by Pontén et al.,1 has adopted an antibody-based proteomic approach by combining the large-scale generation of antibodies with high-throughput immunohistochemistry to map the expression of human proteins in a multitude of tissues, cells and cancers. This analyses the relative abundance and distribution of expressed proteins in normal human tissues and cancer tissues as well as on the subcellular level in cell lines. All data including underlying high-resolution images are made freely available at the annually updated Human Protein Atlas portal, www.proteinatlas.org.2 The current version of the Human Protein Atlas includes protein profiles for over 15,000 unique proteins, corresponding to over 75% of all protein-encoding human genes.3 This same strategy can also be used to visualise the expression of endogenous retroviral gene products with a cellular resolution in both normal and diseased human tissues and, as this study exemplifies, provides an attractive approach to map the contribution of human endogenous retroviruses, or HERVs, to human tissues in normal physiology and disease.
HERVs are considered exogenous retroviruses that have inserted into the germ cell line of our primate and human ancestors. They make up roughly 9% of our human DNA. Two previous reviews in this journal presented the growing evidence that HERVs have made a significant contribution to human evolution, development and physiology, as well as playing possible roles in human diseases.4,5 A reliable methodology for the assessment of virus-derived proteins in tissues and organs is important to our understanding of the role of HERVs in human evolution, physiology and disease. Our purpose was to study the expression of a single HERV, known as ERV3, and from this to derive a novel and reliable system for the detection of HERV-encoded proteins in human cells, tissues and organs.
The prototype ERV3 was one of the first human endogenous retroviruses to be discovered. Regarded as a member of the HERV-R family, this virus has roughly 50 insertions throughout the human chromosomes. Only one of these insertions, the so-called ERV3 locus, appears to have been conserved by natural selection for an evolutionary role. In this communication, the term ERV3 will refer to this specific viral locus,6 which is situated on chromosome 7q11.21.7 Its genetic structure is shown in schematic form in Figure 1. The ERV3 locus is unique to the great apes and Old World monkeys, its insertion into the primate genome dated to roughly 30–40 millions ago. It comprises the typical retroviral ‘genes’, gag, pro, pol, env, flanked by the regulatory regions known as ‘long terminal repeats’, the 5′ and 3′ LTRs. In this locus gag, pro, pol, have been degraded by mutations, meanwhile the env gene and the relevant promoter region in the 5′ LTR have been selectively conserved. Previous studies have shown that the ERV3 env contributes to the structure and function of the placenta8 where it operates in a complex coordination with other endogenous retroviruses, including HERV-W, HERV-FRD, ERV-9 and HERV-K.9–13 Other studies have also suggested that ERV3 env sequences are expressed at RNA level in many human tissues, especially those involved in hormonal synthesis and in tissues with close contact with external surfaces.6
Figure 1.
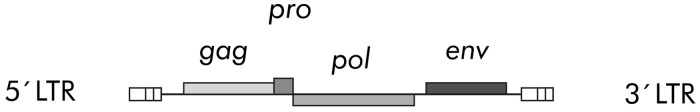
Schematic of the ERV3 locus on chromosome 7q11.21. This is a complete HERV-R retroviral genome in which the gag, pro, pol, env genetic domains have been silenced by mutation. The env domain and the viral promoter of this domain within the 5′ LTR have been conserved as a functional unit by selection.
One of the most important roles for HERVs in the human placenta is fusiogenic. This has led to the proposal that HERVs might contribute to cell fusion in terms of both physiology and pathology in a variety of cells and organs other than placenta.14,15 In addition to the fusiogenic properties, the envelope proteins of some HERVs contain a transmembrane protein (TM) with complex immunosuppressive properties. This offers the potential of a variety of different roles in the normal immune responses within different tissues, for example in the placenta, where several HERVs are being investigated for a potential role in the suppression of the maternal rejection of the fetus.16
HERV expression has been extensively linked to disease. For example, ERV3 gene expression appears to be downregulated in choriocarcinoma.17 Meanwhile increased HERV expression has been reported widely in autoimmunity, such as multiple sclerosis (MS) and disseminated lupus erythematosis (DLE).5 Approximately 20% of human cancers have been attributed to exogenous virus infection, in particular human papilloma virus, hepatitis B, Epstein Barr virus and adenoviruses, but retroviruses such as human immunodeficiency virus and human T-cell lymphotropic virus are also potential carcinogenic. HERV gene expression has been variously linked to the myeloproliferative disorders, including primary proliferative polycythaemia, essential thrombocythaemia, leukaemia, Hodgkin’s lymphoma and various cancer cell lines as well as cancers of the lung, stomach, intestine, bone marrow, bladder, prostate, cervix and breast, as well as melanomas, seminomas and teratocarcinomas.18,19 However, limitations of the prevailing methodology and a lack of understanding of the putative physiological roles of viral genes and other genetic sequences have hampered further study of such viral gene expression and its potential link to pathogenesis.
Most earlier studies have examined ERV3 gene expression at messenger RNA level through real-time PCR, qPCR and Northern blot in situ hybridization rather than the more difficult definition of protein expression within cells and tissues. This, while pioneering and useful, suffers from the limitation that the expression of viral genes at RNA level may be an unreliable guide to protein expression because of differences in mRNA turnover, stability and efficiency in translation. Thus, a reliable technique for detecting and measuring virally encoded proteins in cells, tissues and organs is an essential step in the evaluation of any putative evolutionary, physiological or pathological role for the expressed viral gene. We have attempted to solve this problem by adopting the ERV3 locus as a pilot system, generating monospecific polyclonal antibodies that are capable of reliably detecting the expression of the ERV3 Env-encoded proteins in human cells, tissues and organs.
Methods
To determine the relative (semi-quantitative) level of protein expression of ERV3 Env proteins, antibodies were used to immunohistochemically stain human tissues assembled in tissue microarray (TMA) blocks.20 Tissues were collected from archival material at the Department of Pathology, Uppsala University Hospital. Tissue cores of 1 mm diameter were sampled from paraffin blocks. Sampling was conducted as previously described in brief representative regions of normal tissues corresponding to 144 individuals, representing over 40 different normal human tissue types and in 216 different cancer tissues representing the 20 most common forms of human cancer.21 Some 45 different cancer cell lines and leukaemia patient samples assembled into cell microarrays were also used for basic protein profiling22 as well as 45 different cancer cell lines assembled into nine different arrays (2 normal tissues, 6 cancer tissues and 1 cell lines). We also examined tissues representing granulomas rich in giant cells (5 cases), tumours with giant cells (5 cases), skin with sebaceous glands (6 cases), ovary with corpus luteum (2 cases) and 11 placental specimen from different gestational time, to create a TMA with respect to tissues that would include cells involved in fusion.
The set-up of protein profiling was in accordance with strategies used in the Human Protein Atlas described above.
Antibodies
Two affinity-purified polyclonal antibodies towards ERV3 Env (HPA017209 and HPA046508) were generated applying a strategy based on the bioinformatic identification of protein coding sequences with low homology between different genes23 and the use of recombinant Protein Epitope Signature Tags (PrEST) as antigens. Monospecific antibodies were obtained by affinity purification of polyclonal antibodies generated towards recombinant PrEST protein fragments. This strategy enabled large-scale production of antibodies targeting proteins encoded by the human genome, meanwhile avoiding the time-consuming screening step required for the production of monoclonal antibodies. The PrESTs were designed to contain 100–150 amino acids in a region of the protein showing low homology to other human proteins. PrEST gene fragments were cloned into an expression vector to produce recombinant PrEST proteins fused to a dual tag facilitating purification. The recombinant PrEST protein was then employed as an antigen towards which polyclonal antibodies were generated, and affinity purification using the PrEST as ligands finally produced monospecific antibodies.24 Continuous quality controls were performed throughout to ensure specificity of the antibody, including DNA sequencing of all cloned PrESTs, determination of the mass of the produced recombinant PrEST proteins with mass spectrometry and, finally, the binding specificity of the generated antibodies were verified on protein microarrays containing various PrESTs. Approved monospecific antibodies were further analysed by Western blotting, immunohistochemistry on designated TMAs, and in addition, immunofluorescence and confocal microscopy was used to determine the subcellular localisation of each protein.25 The appropriate PrESTs for ERV3 Env used as immunogen for generating the antibodies were 130 amino acids long, corresponding to aa 278–407 (HPA017209), and 141 amino acids long corresponding to aa 80–220 (HPA046508).
Immunohistochemistry
Four-micrometer sections were cut from the TMA blocks, mounted on adhesive slides and baked at 60℃ for 45 min. TMA slides were then deparaffinized in xylene, followed by hydration in graded alcohols and blocking for endogenous peroxidase in 0.3% hydrogen peroxide. For antigen retrieval, the slides were boiled in a pressure boiler (Decloaking chamber; Biocare Medical, Walnut Creek, CA, USA) for 4 min at 125℃ in Target Retrieval Solution, pH 6 (Dako, Glostrup, Denmark). Automated immunohistochemistry was performed as previously described26 using a LabVision Autostainer 480 S (Thermo Fisher Scientific, Runcorn, UK). The dilution of each of the primary antibodies in UltraAb Diluent (Thermo Fisher Scientific, Fermont, CA, USA) was optimized (1:350 for HPA017209 and 1:300 for HPA046508), and the antibodies were applied for 30 min at room temperature (RT). The slides were further incubated with the secondary reagent anti-rabbit/mouse horse radish peroxidase-conjugated UltraVision (Thermo Fischer Scientific) for 30 min at RT. Following washing steps, the slides were developed for 10 min, using diaminobenzidine as a chromogen, and counterstained with Mayer’s haematoxylin for 5 min (Sigma-Aldrich). The slides were then mounted with Pertex (Histolab AB, Gothenburg, Sweden) and scanned using the Aperio ScanScope XT for the generation of high-resolution digital images. Pathology-based annotation and scoring of immunohistochemistry outcome was performed on scanned images as previously described.27 The set-up for protein profiling was in accordance with strategies used in the Human Protein Atlas.1
Results
Detection of ERV3 env-encoded proteins in human tissues
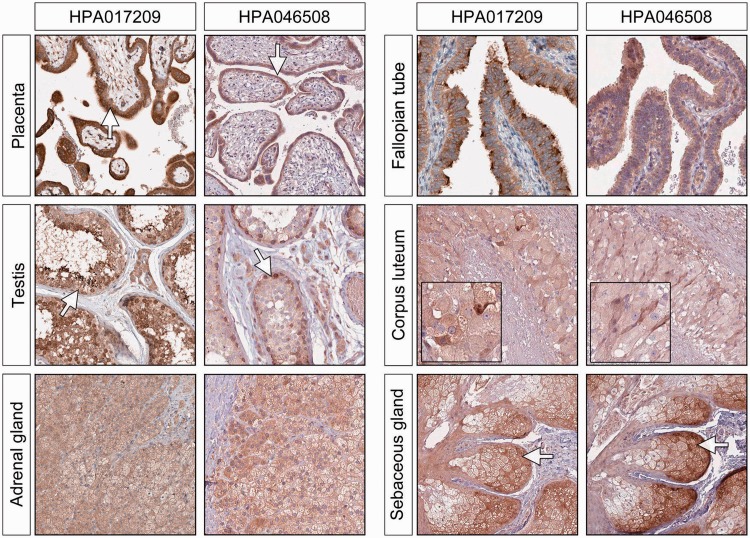
We found that ERV3 Env proteins are expressed at variable but usually low levels in the cytoplasm of cells in the majority of normal adult tissues. This may imply some minor cross-reactivity. When we compared the outcome of two different antibodies targeting different, non-overlapping epitopes of the ERV3 Env protein, we found high levels of expression of the protein in the cells of placenta, testis, adrenal glands, Fallopian tubes, corpus luteum and sebaceous glands (Figure 2). This level of protein expression would suggest a significant physiological role for the viral protein in these tissues. In placenta, ERV3 Env is highly expressed in the cyto/syncytiotrophoblastic layer, with only low levels of expression in the underlying villous stroma. A similar staining pattern was found in 11 placentas sampled from week 11 through the whole gestational period up to full term, though we acknowledge that minor variation of expression in the different samples might not be seen with this method. In testis, ERV3 Env expression was found in all the germ cell layers of the semeniferous ducts. The highest level of expression was found in more immature spermatogonia, but the specific cell type could not be identified. In the adrenal gland, high level of expression was detected in all three layers of the cortex. No differences in intensity were detected in any of the layers. The previous observation of high levels of ERV3 mRNA expression in sebaceous glands was confirmed in all samples.
Figure 2.
ERV3 Env protein expression in organs known to be highly hormonally dependent. In the placenta, the highest expression is seen in the cyto/syncytotrophoblastic cells, with only a weak staining in the stroma. In the testis, there is significant staining of immature cells at different intensity. In the Fallopian tube, there is dense staining of the tips of the cells, which is particularly marked with the antibody HPA017209. The large luteinised cells in ovarian corpus luteum, and all of the cells in the adrenal cortex are shown to be moderately to strongly positive.
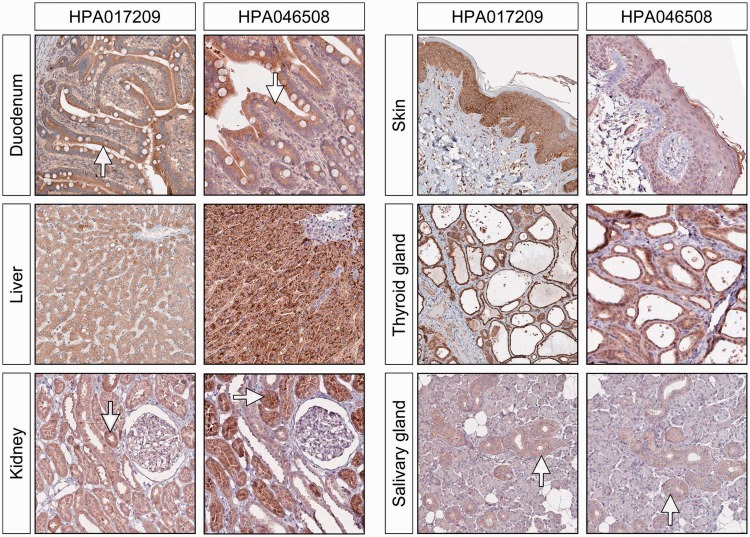
Moderate to high levels of ERV3 Env protein expression was also found in epithelial cells from the gut (duodenum), liver, kidney and thyroid gland, as well as in several more differentiated epithelia, including skin (squamous), urinary and glandular. We also observed moderate to high level expression of ERV3 Env protein in bronchial epithelium and in the ducts of the salivary glands. The majority of these are illustrated in Figure 3. When we extended our testing to malignant tissues, several epithelial tumours displayed what appeared to be significant positivity, although we saw no obvious pattern that could be associated with a specific group morphology. For example, certain scattered cells within gliomas showed strongly positive for Env protein expression, a finding that may indicate some cellular heterogeneity.
Figure 3.
ERV3 Env protein expression in epithelial cells in the gut (duodenum), liver, skin and thyroid gland. There is good concordance between the two antibodies although the intensity of the staining can vary. The tubular cells of the kidney are moderately stained, but we see very little staining in the glomerular cells. In the salivary glands, the staining is mainly seen in the epithelial cells of the ducts.
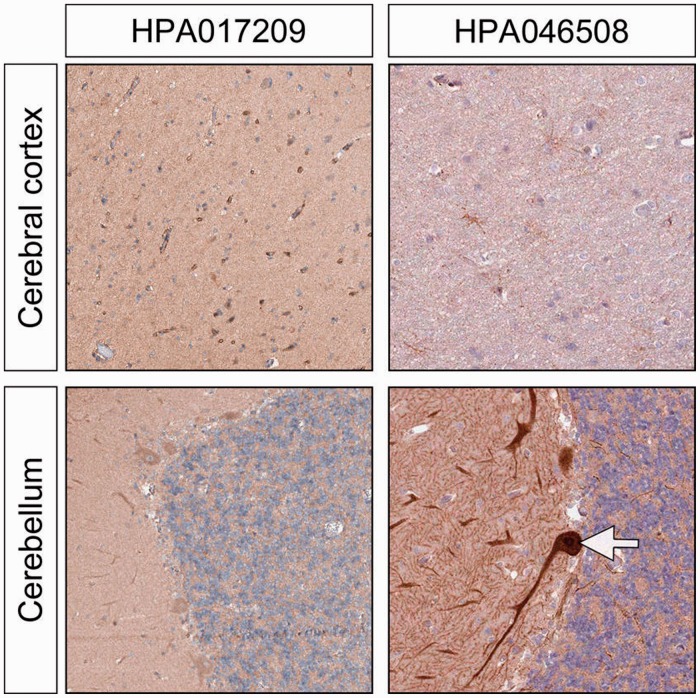
In the normal brain, there was no clear pattern of ERV3 Env expression in neural tissues, but scattered specific cells, probably astrocytes, displayed a moderate to high level of expression. It is intriguing that astrocytes, which are glial cells involved in a wide spectrum of functions, including local inflammatory and immunological processes, also show significant expression of syncytin-1, the protein encoded by the env gene of a different endogenous retrovirus known as HERV-W, where it may also be associated with the neurotoxicity associated with MS. In the cerebellum, we obtained a somewhat contradictory result with the Purkinje cells staining strongly positive with only one of the antibodies, HPA046508. We cannot as yet explain this curious finding. These results are illustrated in Figure 4.
Figure 4.
ERV3 Env protein expression in brain. There is striking intensity of staining in the Purkinje cells of the cerebellum, but this is only seen with one of the antibodies (HPA046508). There was also weak to moderate staining in astrocytes and also a faint staining in the surrounding neuropil.
Cells undergoing fusion
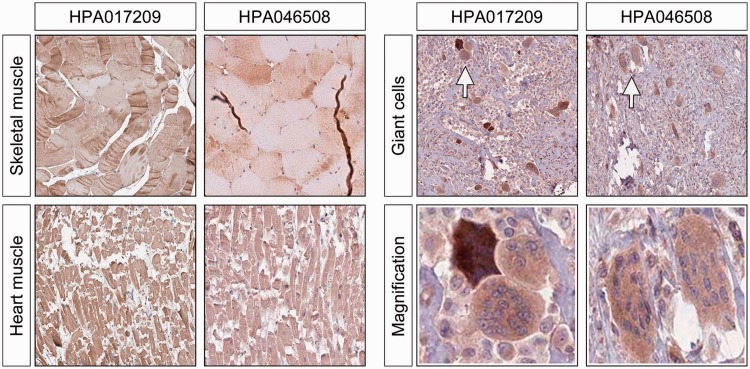
We also investigated tissue cells known to undergo fusion and form giant cells as a response to injury or other pathological stimuli, including giant cells (fused macrophages). These cells were consistently positive for ERV3 Env protein expression (Figure 5), as were cells in benign tumours with prominent giant cells. We also saw a similar consistent pattern of ERV3 Env protein expression in myocardial muscle cells and in the striated cells of normal muscle, both tissues in which fusion of cells may be a feature.
Figure 5.
ERV3 Env protein expression in tissues that undergo fusion, including myocardium, skeletal muscle and giant cells. A high intensity of staining was obtained in myocytes from both heart and striated muscle fibres, as well as giant cells in histiocytomas/benign giant cell tumours. The latter is also highlighted under higher magnification.
Discussion
This is the most comprehensive study to date of the protein expression of the ERV3 HERV locus in the human genome. Although we cannot claim to titrate absolute quantitative levels of Env protein expression in specific tissues, with few exceptions this methodology resulted in a strongly supportive overlapping pattern of heavily concentrated staining involving the sibling antibodies in all of the tissues and organs studied. Our findings also fit the in situ and qPCR experiments of Andersson et al.6 reported earlier and have been further corroborated by deep sequencing data that will be published elsewhere.
There is growing evidence that retroviral colonisation with subsequent holobiontic genomic evolution has played an important role in the evolution of the mammals, primates and humans.28 Preliminary research in our laboratories suggests that the ERV3 findings are likely to extrapolate to some of the other viral loci within the human chromosomes. Future research might include the potential for interaction of viral proteins with host physiological and pathological pathways, especially those involved in cellular differentiation, inflammation and immunity.
For example, since HERVs, including ERV3, have been strongly associated with cell fusion in placenta, it has been suggested that HERV Env proteins could be involved in physiological and pathological situations in other tissues that involve cell fusion. Thus, it may be relevant that we found strong staining for ERV env protein in fused macrophages (giant cells), including osteoclasts, as well as other multinucleated macrophages. Other HERVs have also been suspected of involvement in these processes, suggesting that several different fusiogenic proteins may work together in a complex coordination, which might also allow for some degree of redundancy. It might also be worthwhile to apply the present methodology to metastatic malignancy, where others have proposed that HERVs might play a role in fusing tumour cells with endothelium.
This study also confirmed that ERV3 env gene is highly expressed in tissues involved in steroid synthesis and reproduction. Some colleagues have proposed that some HERVs could be involved in protection against superinfection by a putative exogenous retrovirus (so-called intragenomic vaccination) in analogy with the FV1 restriction, a process that would be facilitated by the expression of protective viral sequences in cells exposed to the surrounding environment. Strong viral expression in sebaceous glands undergoing holocrine secretion could lead to the export of an Env protein to the cell surface. We have confirmed a high level of ERV3 Env expression in sebaceous glands, particularly so in cells undergoing holocrine secretion. Moreover, our findings of significant expression of ERV Env protein in skin is supported by the work of Otsuka in transgenic rats,29 where they found that the expression of HERV-R (ERV3) is linked to both the development and differentiation of squamous cells.
HERV expression has been associated with a wide range of cancers as well as most forms of autoimmune diseases, in particular multiple sclerosis and disseminated lupus erythematosis. These findings have also been confounded by a lack of understanding of the role of the relevant viruses in normal genetic and physiological function. Our study, and its potential extrapolation to the many other HERVs that have colonised the human genome, may assist such understanding and suggest new avenues of investigation.
The profile of protein expression in different human cells, tissue and organs is critical to the understanding of human biology. In particular, we need to know how a large fraction of our genes encode for cell-type specific proteins, house-keeping proteins or proteins differentially expressed across different cell types. In this study, we showed by immunohistochemistry that the ERV3 protein was detected in a multitude of human tissues, but at relatively different intensity levels. This is in accord with a previous study using 5934 antibodies to target 4842 proteins, which suggested that cellular phenotype and function is not primarily determined by the expression of cell-type specific proteins, i.e. proteins expressed in only one cell type, but rather on a tight regulation of protein levels expressed by a large proportion of the genome,30 and that in fact, only a handful of proteins appear to be exclusively expressed by a limited number of cell types. Based on this finding, coupled with the present study, one can categorise ERV3 into the dominant group of quite abundantly expressed proteins whose relative levels vary across cell types and tissues. This would infer that ERV3 actively participates in normal cellular function and human biology and that this may extrapolate to HERVs in general.
Declarations
Competing interests
None declared
Funding
Grant number KAW 0143 (Human Protein Atlas Project)
Guarantor
EL
Ethical approval
Ethical approval for this study was obtained from the local ethics committees at Uppsala University (number: 03-618)
Contributorship
CF analyzed the Immunostainings and prepared a draft; CA analyzed immunostainings and participating in the preparation of the manuscript; P-HE supervised the immunostainings, prepared figures and wrote parts of the article; FP designed the epitopes used for immunisation and preparation of the tissue microarrays; WWZ participated in the writing the manuscript; EL designed the study, chose the tissues for immunohistochemistry and wrote parts of the article including discussions and FPR designed and wrote parts of the article
Acknowledgements
This work was supported by the Lion Foundation for Cancer Research Uppsala and by the Knut and Alice Wallenberg Foundation
Provenance
Not commissioned; peer-reviewed by Martin Pall and Michael Syvanen
References
- 1.Pontén F, Schwenk JM, Asplund A, Edqvist PH. The Human Protein Atlas as a proteomic resource for biomarker discovery. J Intern Med 2011; 270: 428–46 [DOI] [PubMed] [Google Scholar]
- 2.Berglund L, Björling E, Oksvold P, et al. A gene-centric human protein atlas for expression profiles based on antibodies. Mol Cell Proteom 2008; 7: 2019–27 [DOI] [PubMed] [Google Scholar]
- 3.Uhlen M, Oksvold P, Fagerberg L, et al. Towards a knowledge-based human protein atlas. Nature Biotechnol 2010; 28: 1248–50 [DOI] [PubMed] [Google Scholar]
- 4.Ryan FP. An alternative approach to medical genetics based on modern evolutionary biology. Part 2: retroviral symbiosis. J R Soc Med 2009; 102: 324–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryan FP. An alternative approach to medical genetics based on modern evolutionary biology. Part 3: HERVs in diseases. J R Soc Med 2009; 102: 415–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersson A-C, Yun Z, Sperber GO, Larsson E, Blomberg J. ERV3 and related sequences in humans: structure and RNA expression. J Virol 2005; 79: 9270–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim HS, Yi JM, Hirai H, et al. Human Endogenous Retrovirus (HERV)-R family in primates: chromosomal location, gene expression, and evolution. Gene 2006; 370: 34–42 [DOI] [PubMed] [Google Scholar]
- 8.Kato N, Pfeifer-Ohlsson S, Kato SM, et al. Tissue-specific expression of human provirus ERV3 mRNA in human placenta: two of the three ERV3 mRNAs contain human cellular sequences. J Virol 1987; 61: 2182–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mi S, Lee X, Li X-P, Veldman GM, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 2000; 403: 785–9 [DOI] [PubMed] [Google Scholar]
- 10.Blond J-L, Lavillette D, Cheynet V, et al. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol 2000; 74: 3321–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blaise S, de Parseval N, Bénit L, Heidmann T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci USA 2003; 100: 13013–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kudaka W, Oda T, Jinno Y, Yoshimi N, Aoki Y. Cellular localization of placenta-specific human endogenous retrovirus (HERV) transcripts and their possible implication in pregnancy-induced hypertension. Placenta 2008; 29: 282–9 [DOI] [PubMed] [Google Scholar]
- 13.Kämmerer U, Germeyer A, Stengel S, Kapp M, Denner J. Human endogenous retrovirus K (HERV-K) is expressed in villous and extravillous cytotrophoblast cells of the human placenta. J Reprod Immunol 2011; 91: 1–8 [DOI] [PubMed] [Google Scholar]
- 14.Bjerregaard B, Holck S, Christensen IJ, Larsson LI. Syncytin is involved in breast cancer-endothelial cell fusions. Cell Mol Life Sci 2006; 63: 1906–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Søe K, Andersen TL, Hobolt-Pedersen AS, Bjerregaard B, Larsson LI, Delaissé JM. Involvement of human endogenous retroviral syncytin-1 in osteoclast fusion. Bone 2011; 48: 837–46 [DOI] [PubMed] [Google Scholar]
- 16.Mangeney M, Renard M, Schlecht-Louf G, et al. Placental syncytins: genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc Natl Acad Sci USA 2007; 104: 20534–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato N, Larsson E, Cohen M. Absence of expression of a human endogenous retrovirus is correlated with choriocarcinoma. Intern J Cancer 1988; 41: 380–5 [DOI] [PubMed] [Google Scholar]
- 18.Ryan FP. An alternative approach to medical genetics based on modern evolutionary biology. Part 4: HERVs in cancer. J R Soc Med 2009; 102: 474–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruprecht K, Mayer J, Sauter M, Roemer K, Mueller-Lantzsch N. Endogenous retroviruses and cancer. Cell Mol Life Sci 2008; 65: 3366–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kononen J, Bubendorf L, Kallionimeni A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nature Med 1998; 4: 844–7 [DOI] [PubMed] [Google Scholar]
- 21.Kampf C, Andersson A-C, Wester K, Björling E, Uhlen M, Pontén F. Antibody-based tissue profiling as a tool for clinical proteomics. Clin Proteom 2004; 1: 285–300 [Google Scholar]
- 22.Andersson AC, Strömberg S, Backvall H, et al. Analysis of protein expression in cell microarrays: a tool for antibody-based proteomics. J Histochem Cytochem 2006; 54: 1413–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berglund L, Björling E, Jonasson K, et al. A whole-genome bioinformatics approach to selection of antigens for systematic antibody generation. Proteomics 2008; 8: 2832–39 [DOI] [PubMed] [Google Scholar]
- 24.Nilsson P, Paavilainen L, Larsson K, et al. Towards a human proteome atlas: high-throughput generation of mono-specific antibodies for tissue profiling. Proteomics 2005; 5: 4327–37 [DOI] [PubMed] [Google Scholar]
- 25.Barbe L, Lundberg E, Oksvold P, et al. Toward a confocal subcellular atlas of the human proteome. Mol Cell Proteom 2008; 7: 499–508 [DOI] [PubMed] [Google Scholar]
- 26.Paavilainen L, Wernérus H, Nilsson P, et al. Evaluation of mono-specific antibodies: a comparison study with commercial analogues using immunohistochemistry on tissue microarrays. Appl Immunohistochem Mol Morphol 2008; 16: 493–502 [DOI] [PubMed] [Google Scholar]
- 27.Björling E, Lindskog C, Oksvold P, et al. A web-based tool for in silico biomarker discovery based on tissue-specific protein profiles in normal and cancer tissues. Mol Cell Proteom 2008; 7: 825–44 [DOI] [PubMed] [Google Scholar]
- 28.Villarreal LP, Ryan FP. Viruses in host evolution: general principles and future extrapolations. Curr Top Virol 2011; 9: 79–90 [Google Scholar]
- 29.Otsuka N, Miyatake Y, Ishizu A, et al. Expression of human endogenous retrovirus-R gene links to differentiation of squamous cells. AIDS Res Hum Retroviruses 2006; 22: 1148–51 [DOI] [PubMed] [Google Scholar]
- 30.Pontén F, Gry M, Fagerberg L, et al. A global view of protein expression in human cells, tissues, and organs. Mol Syst Biol 2009; 5: 337–337 [DOI] [PMC free article] [PubMed] [Google Scholar]