Abstract
During translation elongation, the compositional factors, elongation factor G (EF-G; encoded by fusA) and transfer RNA (tRNA), alternately bind to the ribosome to direct protein synthesis, in turn regulating the conformation of the ribosome. Here, we use single-molecule fluorescence with zero-mode waveguides to correlate directly ribosome conformations and compositions during multiple rounds of elongation at high factor concentrations in Escherichia coli. Our results show that EF-G-GTP continuously samples both rotational sates of the ribosome, binding with higher affinity to the rotated state. Upon successful accommodation into the rotated ribosome, the EF-G-ribosome complex evolves through several rate-limiting conformational changes and the hydrolysis of GTP, which results in a transition back to the non-rotated state, in turn driving translocation and facilitating both EF-G-GDP and E-site tRNA release. These experiments highlight the power of tracking single-molecule conformation and composition simultaneously in real-time.
Biological systems evolve temporally in composition and conformation. In translation, the ribosome processively coordinates the binding and dissociation of multiple translation factors and transfer RNAs (tRNAs) to synthesize protein encoded by a messenger RNA (mRNA). During each cycle of elongation, the bacterial ribosome selects the aminoacyl-tRNA (aa-tRNA), in a ternary complex (TC) with GTP-bound elongation factor-Tu (EF-Tu-GTP), and positions the tRNA in the A site. Upon A-site tRNA accommodation, the ribosome rapidly catalyzes peptide bond formation with the P-site tRNA1,2. Translocation moves A- and P-site tRNA-mRNA complexes to the E and P-site respectively, catalyzed by the GTPase elongation factor-G (EF-G)3. The compositional dynamics of tRNA-EF-Tu and EF-G on the ribosome, here defined as the relative timing of their arrival and departure during elongation, play a central role in directing successful protein synthesis.
Ribosomal conformational changes coordinate movements and occupancy of ligands during elongation and underlie translocation. Structural studies have revealed two global intersubunit conformational states for the 70S ribosome involved in elongation: the “unlocked”, rotated (pre-translocation) state and the “locked”, non-rotated (post-translocation) state4,5. The two conformers differ by an intersubunit rotation of 3~10° of the body of the small (30S) subunit with respect to the large (50S) subunit4,6. The ribosome starts each round of elongation in the non-rotated state. In this “locked” state, the P-site tRNA is stably bound in the classical state, preserving the reading frame of the mRNA7. Upon A-site tRNA selection and peptide bond formation, the 30S subunit rotates 3~10° counterclockwise with respect to the 50S subunit to the rotated state (pre-translocation)4,6,8. This “unlocked” state permits tRNA motions and the tRNA can fluctuate freely between the classical state and hybrid state, and possibly other intermediate hybrid states9, thus facilitating translocation of tRNA and movement of ribosome by one codon over mRNA1,10. Peptide-bond formation also triggers spontaneous fluctuations of the L1 stalk between open and closed conformations as well as spontaneous rotations in ribosome conformations8,11, which direct tRNA movement and are likely linked to the fluctuations of tRNA12. EF-G then catalyzes translocation, and the ribosome returns to the non-rotated state (post-translocation), with the L1 stalk in the open conformation12,13, allowing for E-site tRNA release.
EF-G catalyzes translocation, but the precise mechanism remains unclear. Structural studies have suggested that EF-G controls the conformational changes of the ribosome that accompany translocation14,15. Rapid-mixing kinetic approaches16–18 and single-molecule fluorescence studies12,13,19 have probed the kinetics underlying translocation and accompanying structural rearrangements. These experiments suggested that EF-G-GTP binding may bias ribosomal and tRNA conformations, and that GTP hydrolysis by EFG hydrolysis catalyzes translocation by ca. 50-fold; the GDP form of EF-G dissociates from the post-translocation ribosome. Yet current mechanisms were constructed from temporal comparisons of bulk rate constants or single-molecule studies of isolated steps. Thus, the mechanism for how ribosomal conformation and factor composition are correlated and dynamically control translocation remain unclear. To circumvent these limitations, we probed with single-molecule methods directly the interactions between EF-G, tRNA, and single translating ribosomes to correlate ligand composition to the global conformational state of the ribosome in real-time at codon resolution.
RESULTS
Correlating ribosome conformation with tRNA translocation
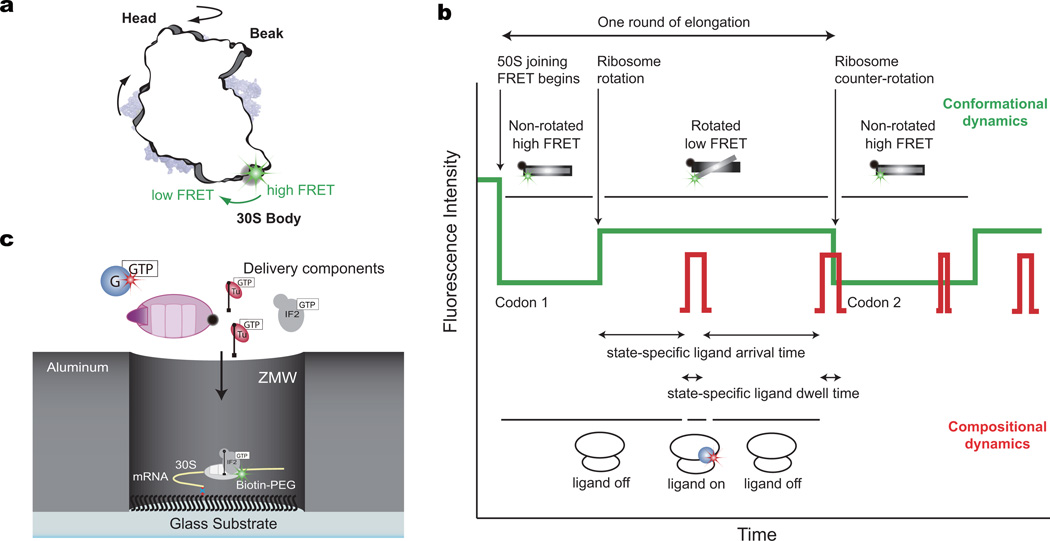
To monitor the rotational state of the ribosome in real time, we employed Förster resonant energy transfer (FRET) between the small (30S) and large (50S) subunits. The 30S subunit was site-specifically labeled with Cy3B on helix 44, and a nonfluorescent quencher, BHQ-2, was placed on helix 101 of 50S subunit7,20,21. The labeling sites are distant from all active sites and dynamic regions of the ribosome (far from head and platform domains of 30S), as well as EF-G binding site and tRNA A, P, and E sites (not within FRET distance), while reporting on the rotational state of the 30S body domain7,22 and excluding complications of other rotational movements during elongation (Fig. 1a). Substituting the traditional FRET acceptor, Cy5, with BHQ-2 allowed the use of Cy5 to label other translation components for correlation studies, as validated previously23; here tRNAs or EF-G were labeled with Cy5, without perturbing function (Supplementary Fig. 1). The Cy3B intensity reports on the conformational state of the ribosome, while Cy5 pulses indicate arrival, occupancy, and departure of ribosomal ligands (Fig. 1b). To track tRNA and EF-G dynamics on translating ribosomes at near-physiological concentrations of fluorescent factors (0.1 – 1 µM), we used zero-mode waveguides (ZMWs) to detect hundreds of individual ribosomes with 30 frames per second (fps) (~30 ms exposure time) time resolution24–26 (Fig. 1c). We use ZMWs here to correlate directly EF-G and tRNA arrival and departure dynamics with the conformational states of the ribosome during continuous translation elongation at near-physiological concentrations.
Figure 1. Correlating conformation and composition with nonfluorescent FRET acceptor and ZMW.
a, Position of the Cy3B FRET probe on the body domain of the 30S subunit with respect to the nonfluorescent FRET acceptor, BHQ, on the 50S subunit, reporting on the rotational state of the 30S subunit. The tRNA (in A, P, and E sites) and EF-G are not within FRET distance with the Cy3B and BHQ dye on the ribosome.
b, Expected sequence of fluorescence signals using Cy3B and BHQ FRET to correlate conformational and compositional dynamics. Cy3B intensity (green) reports on the conformation of the ribosome, while Cy5 pulses (red) report on ligand occupancy on the ribosome. This allows the correlation of conformation and composition to extract state-specific ligand dynamics.
c, Experimental setup. Pre-initiation complex with Cy3B-30S, fMet-tRNAfMet and IF2-GTP are immobilized on the bottom of the ZMW well through a biotinylated mRNA. The reaction was started by delivering IF2-GTP, BHQ-50S, Lys-tRNALys ternary complex, and either Cy5 labeled EF-G-GTP or Phe-tRNAPhe ternary complex to the surface.
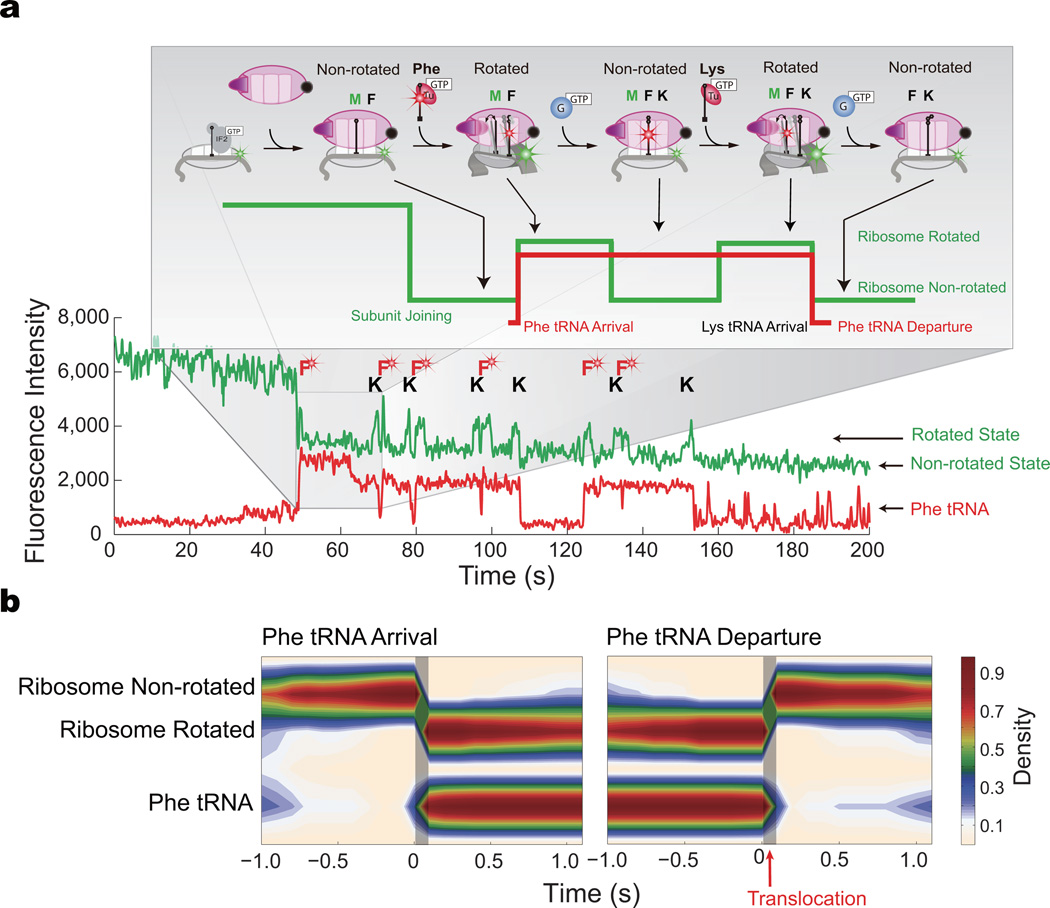
To confirm the relationship between the intersubunit rotation FRET signal and translocation of tRNA, we first correlated tRNA arrival and departure dynamics with ribosome conformation. We delivered 200 nM EF-G-GTP, 200 nM BHQ-50S, 1 µM IF2-GTP, 200 nM Phe-(Cy5)tRNAPhe-ET-Tu-GTP ternary complex, and 200 nM Lys-tRNALys-EF-Tu-GTP ternary complex to pre-initiation complex with Cy3B-30S immobilized through a biotinylated mRNA (with a sequence of fMet followed by six alternating Phe and Lys codons, Online Methods) on the bottom of the ZMW wells. Reagent delivery results in IF2-guided 70S assembly during initiation and establishment of FRET between the two ribosomal subunits: upon subunit joining27, the green (Cy3B) intensity drops, which is followed by alternating low-high-low intensities (Fig. 2a). Each alternating cycle corresponds to the ribosome translating a single codon, with the two intensity states consistent with the two rotational states of the ribosome: the low intensity state (high FRET) defining the non-rotated (locked) ribosome conformation and the high intensity state (low FRET) the rotated (unlocked) conformation, as confirmed previously7,23. The arrival of Phe-(Cy5)tRNAPhe ternary complex, indicated by the appearance of the red pulse, is correlated with the ribosome rotating at the corresponding Phe codons specified by the mRNA sequence. The red pulse persists as the ribosome translocates the (Cy5)tRNA to the P site and the next Lys-tRNALys ternary complex arrives to the A site. The next round of translocation, correlated with the ribosome counter-rotation, drives the (Cy5)tRNAPhe to the E-site, from which it rapidly departs, as indicated by the disappearance of the red pulse.
Figure 2. Ribosome conformation drives translocation and regulates tRNA dynamics.
a, Representative trace of Cy3B and BHQ labeled ribosome elongating with Phe-(Cy5)tRNAPhe. Delivery of reagents results in 50S subunit joining and the arrival of FRET between Cy3B and BHQ, followed by multiple cycles of low-high-low green intensities, each reporting on ribosome rotating and counter-rotating during one round of elongation. The arrival and departure of Phe-(Cy5)tRNAPhe ternary complex are superimposed as red pulses, allowing correlation of tRNA arrival and departure with ribosome conformation. Brief sampling pulses of Phe-(Cy5)tRNAPhe ternary complex are observed after arrival at the stop codon, as characterized by Uemura et al25.
b, Post-synchronization of the (Cy5)tRNAPhe to ribosome rotating and counter-rotating shows that tRNA arrival is correlated with ribosome rotation and that tRNA departure is correlated with ribosome counter-rotation, as emphasized by the shaded areas. Ribosome conformational counter-rotation thus underlies translocation and E-site tRNA release. The number of molecules analyzed is n = 141.
Here, “rotation” is defined as the transition between non-rotated and rotated states, and “counter-rotation” is the transition back. Observation of individual translation complexes enables the arbitrary post-synchronization of both the FRET and red fluorescence data for each ribosome, eliminating temporal averaging and allowing temporal correlation of tRNA dynamics and ribosomal conformational changes (Fig. 2b). tRNA binding to the non-rotated conformation of the ribosome and peptide bond formation drives the transition to the rotated state, consistent with previous findings7,20,23. By post-synchronizing all the observed ribosome molecules to the counter-rotation transition, we show that all E-site tRNA departure is correlated with the counter-rotation of the ribosome (Fig. 2b), within the time resolution of our experiment. Since E-site tRNA dissociation must occur post-translocation and intersubunit rotation is required for translocation28,29, our results suggest that translocation occurs concurrent with the intersubunit conformational counter-rotation, and that these transitions may be the driving mechanism for translocation29. Ribosome counter-rotation translocates the tRNAs from the hybrid A and P sites to the P and E sites, from which the E-site tRNA rapidly departs25.
This mechanism is in support of the spontaneous E-site tRNA departure model25,30,31, where EF-G binding and subsequent GTP hydrolysis drives the tRNA from the hybrid A and P states to the P and E sites, at which point the E-site tRNA rapidly dissociates. Furthermore, prolonged (Cy5)tRNAPhe residency on the ribosome after ribosome translocation and counter-rotation or the accumulation of Cy5 fluorescence that results in two (Cy5)tRNAPhes in the A and E sites of the ribosome that may suggest an allosteric linkage between A-site tRNA arrival with E-site tRNA departure were not observed31. Although in this case, only tRNAPhe was studied, so for other tRNAs encoding CG rich codons, it is possible that departure is much slower, allowing for the simultaneous overlap of A-site and E-site tRNA occupancy. Furthermore, high Mg2+ concentration may result in over-stabilizing of the E-site tRNA, delaying its release. However, the allosteric nature of E-site departure with A-site tRNA arrival is likely not supported, though further experiments will be required to confirm the mechanism.
Correlating EF-G with ribosome conformation
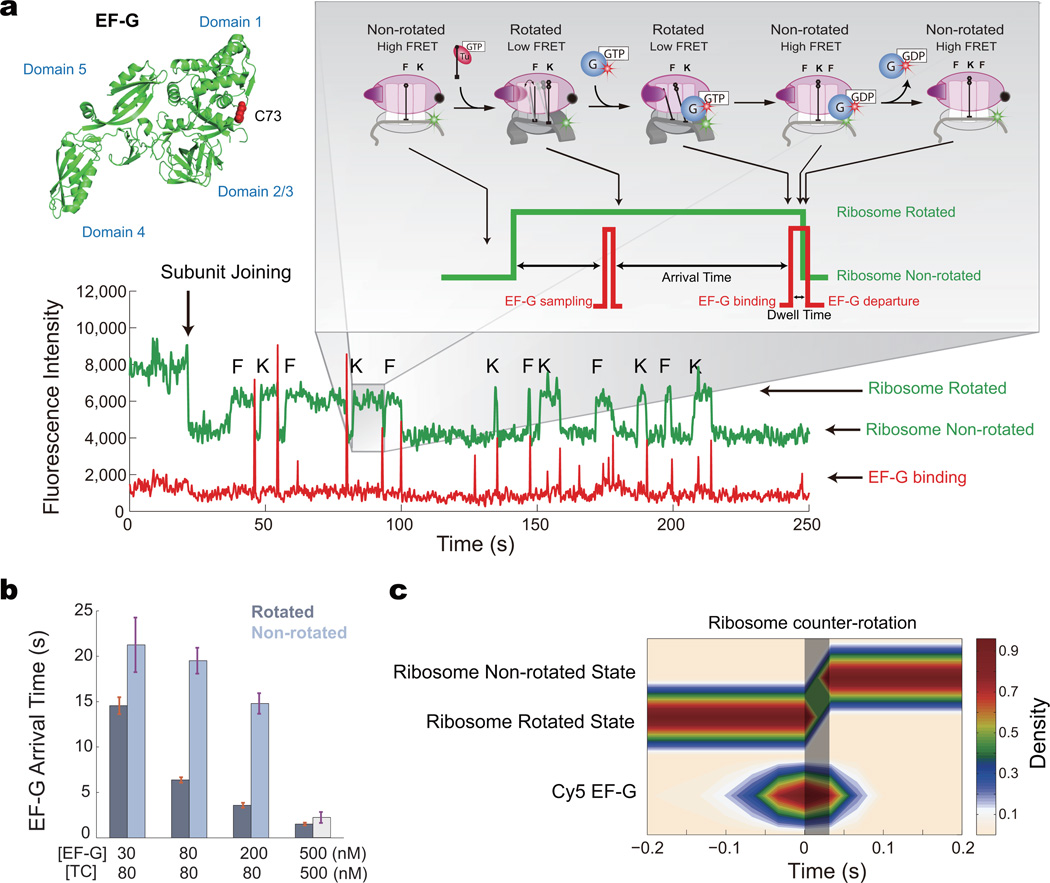
We next explored the relationships between EF-G dynamics and ribosomal conformation during elongation. To probe how EF-G binding and GTP hydrolysis affect translocation and ribosomal intersubunit conformation, we delivered a mixture of 30 – 500 nM Cy5-EF-G-GTP, 200 nM BHQ-50S, 1 µM IF2-GTP, and 80 – 500 nM Phe-tRNAPhe ternary complex and Lys-tRNALys ternary complex to pre-initiation complex with Cy3B-30S. The non-rotated state lifetime depends on the ternary complex concentration, while the rotated-state lifetime depends on the EF-G concentration (Supplementary Fig. 2). Along with each transition from the rotated to the non-rotated state (ribosome counter-rotating), a burst of red (Cy5) fluorescence is observed, corresponding to the arrival and rapid departure of EF-G (τ = 122 ms) (Fig. 3a). EF-G occupancy of the ribosome, unlike that for tRNA, is very transient. In addition, EF-G also samples nonproductively to both states of the ribosome. As we increase EF-G concentration, we correspondingly decrease the EF-G arrival time, as expected (Fig. 3b). Though, the arrival time of EF-G to the rotated state is consistently lower than the arrival time to the non-rotated state, for all EF-G concentrations, suggesting an intrinsic conformational selection mechanism of the ribosome for EF-G (see below). Here, we directly observe that transient EF-G binding drives ribosome counter-rotation and translocation.
Figure 3. EF-G regulates ribosome conformational dynamics.
a, Representative trace of Cy3B and BHQ ribosome elongating superimposed with Cy5-EF-G occupancy signal showing EF-G correlating with the ribosome counter-rotation. A single cysteine mutant of EF-G (S73C) was labeled with Cy5-maleimide, with the labeling site distant from all EF-G functional domains. Each ribosome conformational transition from rotated to non-rotated state (high to low green intensity change) is correlated with a red pulse, corresponding to the arrival and rapid departure of Cy5-EF-G. Non-productive sampling events are observed to both ribosome conformations.
b, The mean arrival times to the two conformations of the ribosome, at different concentrations of tRNA ternary complex (TC) and EF-G-GTP, showing the arrival times decrease with increasing EF-G concentration. The arrival times of EF-G-GTP to the rotated state are lower than to the non-rotated state, suggesting that EF-G-GTP binds with higher affinity to the rotated state. The arrival time to the non-rotated state at 500 nM EF-G and TC is only a lower estimate, due to the decreased non-rotated state lifetime from the increased TC concentration. From left to right, n = 139, n = 216, n = 126, n = 103. Error bars are standard error.
c, Post-synchronization of ribosome counter-rotating with EF-G, at 30 frames per second (n = 106). EF-G arrives prior to the intersubunit conformation transition (as emphasized by the shaded area) and departs rapidly after.
EF-G-GTP binding to the rotated state does not immediately induce ribosomal transitions back to the non-rotated state. Arrival of Cy5-EF-G-GTP is correlated with each transition from rotated to non-rotated conformation by post-synchronization of these events, with EF-G arriving ~50 ms prior to the conformational transition and departing rapidly (mostly within 10 ms) afterwards (Fig. 3c), consistent with translocation times measured in bulk3, with optical tweezers32, and single-molecule fluorescence19. To decipher whether EF-G dissociation plays a role in ribosome counter-rotation, we added 50 µM of fusidic acid (an antibiotic that locks EF-G on the ribosome post GTP hydrolysis22), which extended the dwell time of EF-G on the ribosome by 80-fold (8.4 s). In the presence of fusidic acid, EF-G-GTP binds about 50 ms before the ribosome counter-rotates, similar to the behavior in the absence of drugs, but EF-G-GDP remains on the ribosome for an extended period of time following the conformational transition (Supplementary Fig. 3), indicating that EF-G departure is uncoupled from ribosome counter-rotation. Addition of 100 µM of thiostrepton, which binds near the factor-binding site on the 50S subunit, inhibited EF-G binding19 and increased the arrival times for EF-G to both ribosome conformations (Supplementary Fig. 3). Binding of EF-G in the presence of thiostrepton led to no observed correlated conformational changes within the ribosome, consistent with thiostrepton inhibition of translocation19. The addition of Mg2+ to 15 mM total Mg2+ from 5 mM inhibited rapid translation. The rotated state lifetime remains the same at 5 mM and 15 mM Mg2+, whereas the non-rotated state lifetime is increased at 15 mM Mg2+ (Supplementary Fig. 4). This is consistent with the stabilization of the classical tRNA state at high Mg2+ concentrations33. High Mg2+ concentrations also stabilize EF-G on the ribosome, increasing the EF-G dwell time driving counter-rotation. Increasing Mg2+ thus slows the overall translation rate by stabilizing the non-rotated state of the ribosome (Supplementary Fig. 5).
The role of EF-G GTP hydrolysis
We next sought to clarify the role of GTP hydrolysis by EF-G in driving ribosome conformational changes and translocation3,34. Toeprinting assays in high Mg2+ previously suggested efficient translocation in the presence of EF-G-GDPNP (a non-hydrolyzable analog of GTP) after 10 minutes35, whereas rapid mixing assays showed a ca. 50-fold acceleration of translocation by GTP compared to GDPNP3. We first explored the endpoint of translocation (Supplementary Fig. 6) with a single-molecule translocation assay analogous to toeprinting by quantifying the number of ribosomes that translocated and can thus accept the next elongator tRNA under different conditions (see Online Methods and Supplementary Fig. 6 for details). At 5 mM Mg2+ (~1.3mM free Mg2+ due to chelation by GTP) and 1 µM unlabeled EF-G, most of the ribosomes translocated in the presence of GTP after 5 minutes, while only a fraction of the ribosomes translocated in the presence of GDPNP. At 15mM Mg2+ (~11 mM free Mg2+), nearly equal number of ribosomes translocated in the presence of GTP as with GDPNP (Supplementary Fig. 6c, see figure legend for discussion). Over 5 minutes and at high EF-G and Mg2+ concentrations, translocation can be driven by EF-G-GDPNP, likely because Mg2+ stabilizes the non-rotated state and possibly lowers the energy barrier for translocation. These results frame the kinetic endpoints for translocation, and suggest the importance of both GTP hydrolysis and Mg2+ in controlling translocation rates.
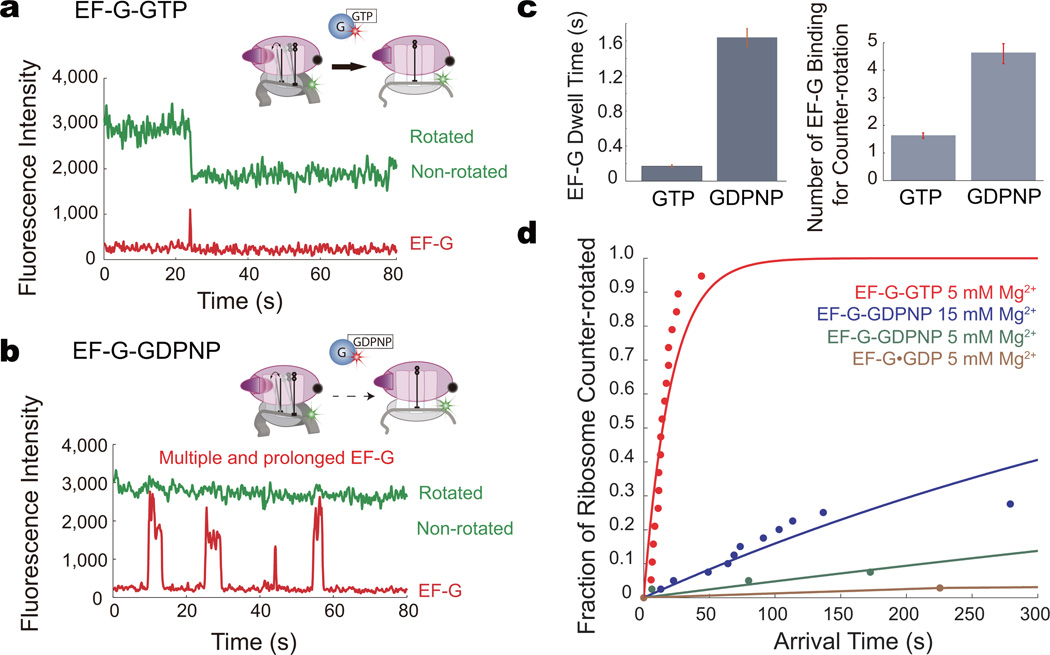
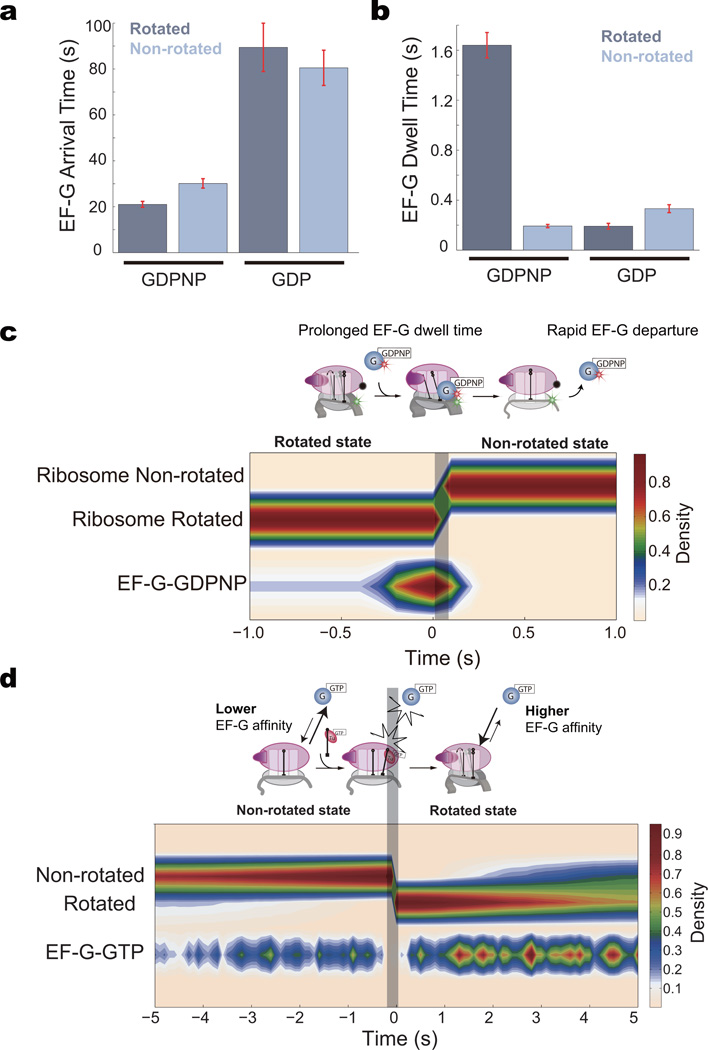
We then sought to determine whether binding alone by EF-G-GTP can drive ribosome conformational counter-rotation and translocation. We delivered 200 nM Cy5-EF-G-GDPNP, the non-hydrolyzable analog of GTP, or Cy5-EF-G-GTP to the ribosome in the rotated state (Online Methods). In contrast to EF-G-GTP, where binding is rapidly followed by translocation (Fig. 4a), EF-G-GDPNP binding to the ribosome results in a longer-lived complex (mean lifetime = 1.62 s, Fig. 4b, c) but no immediate ribosome counter-rotation. In the presence of GDPNP and 5 mM Mg2+, only ~7% of the ribosomes successfully counter-rotated within the 5-minute observation time compared to that in the presence of GTP (Fig. 4d). At 15 mM Mg2+, ~27% of the ribosomes counter-rotated, compared with EF-G-GTP. For the ribosomes that successfully counter-rotated with EF-G-GDPNP, multiple EF-G binding events were required to yield an eventual conformational change. At 15 mM Mg2+, on average 4.63 binding events are required for ribosome counter-rotation in GDPNP (Fig. 4). An extended total residence time of EF-G-GDPNP on the ribosome, achieved through multiple long-lived (>1s) binding events, is required overcome the energy barrier to counter-rotate the ribosome and translocate. In the presence of GTP, EF-G binding efficiently translocates the ribosome, requiring only on average 1.63 binding events and a total residence time of 352 ms on the ribosome (Fig. 4). These results indicate that there is a ~25 fold decrease in translocation rate without GTP hydrolysis at 11 mM free Mg2+. At 1.3 mM free Mg2+, which is the physiological concentration of Mg2+, the apparent rate of translocation is decreased by >50 fold. These data explain the prior observation that GTP hydrolysis accelerates translation by 30–50-fold, and suggests why long-timescale assays would show translocation in the presence of EF-G-GDPNP.
Figure 4. Role of EF-G GTP hydrolysis.
a, Representative trace of ribosome counter-rotating with EF-G-GTP. EF-G-GTP efficiently drives ribosome counter-rotation.
b, Representative trace of ribosome counter-rotating with EF-G-GDPNP. EF-G-GDPNP drives ribosome counter-rotation less efficiently; only after multiple prolonged binding events does translocation occur.
c, Mean dwell time and the number of EF-G binding events required for a successful ribosome counter-rotation for EF-G-GTP and EF-G-GDPNP. Error bars are standard error.
c, The fraction of ribosomes that counter-rotated in the presence of EF-G with the GTP, GDPNP, or GDP, at 5 and 15 mM Mg2+, within the 5 minute observation window (normalized to GTP) with the arrival time of the counter-rotation. The data is fit to a single exponential for visualization. EF-G-GTP catalyzes ribosome counter-rotation efficiently, with most of the ribosomes counter-rotating within the first 50 s. The translocation efficiency of EF-G-GTP is the same at 5 and 15 mM Mg2+. EF-G-GDPNP catalyzes ribosome counter-rotation >50 fold less efficiently than EF-G-GTP. At 15 mM Mg2+, however, counter-rotation is only 25 fold less efficient with GDPNP. From top to bottom, n = 44, n = 143, n = 151, n = 76.
Conformational selection for EF-G
During translation, EF-G-GTP samples the rotated conformation of the ribosome more rapidly than the non-rotated state. From our EF-G arrival times and dwell times, we can determine the binding and dissociation rate constants kon and koff. For the rotated conformation, we get kon, rotated = ~1.9 µM−1s−1, and for the non-rotated state, we get kon, non-rotated = ~ 0.84 µM−1s−1. This is lower than the published bulk value (~150 µM−1s−1)36, but similar to other single-molecule values (1.5 µM−1s−1)19,37. This may be due to our limited frame rate, in which EF-G-GTP rapidly interacts and dissociates nonproductively from the ribosome faster than the experiment imaging rate. The koff for EF-G after translocation is 13.2 s−1, which is close to the published dissociation value (20 s−1)36 and is independent of EF-G concentrations. Upon EF-G engagement with the ribosomal A site, intrinsic EF-G ribosome dynamics limit translocation rates.
To probe how ribosome and EF-G conformational states result in these differential rates, we delivered Cy5-EF-G-GDPNP or Cy5-EF-G-GDP to pre-formed 70S ribosomes in the non-rotated conformation or the rotated conformation (Online Methods). EF-G-GDPNP binds to both ribosome conformations, with a dwell time to the rotated state that is 754% longer than to the non-rotated state (τrotated = 1.64 s vs. τnon-rotated =192 ms). EF-G-GDPNP also binds more frequently to the rotated state of the ribosome, with an ~50% faster arrival time when binding to the non-rotated state. EF-G-GDP binds infrequently to the ribosome in either the rotated state or non-rotated state with arrival times that are ~150% longer than EF-G-GDPNP and bound-state lifetimes τrotated = 191 ms and τnon-rotated = 332 ms (Fig. 5a, b). Thus, the ribosome probably distinguishes between EF-G-GTP binding to the two conformations, with EF-G-GTP binding more stably to the rotated state38; EF-G-GDP binds more slowly and weakly to both states of the ribosome and thus rapidly dissociates when formed as a result of EF-G-bound GTP hydrolysis and phosphate release16. EF-G departure is possibly not driven directly by GTP hydrolysis, but rather through the reduced affinity of EF-G-GDP for the non-rotated ribosome. Similarly, during translocation in the presence of EF-G-GDPNP, EF-G also departs rapidly after the ribosome rotates back to the non-rotated conformation (Fig. 5c).
Figure 5. Conformational selection for EF-G.
a, The arrival times for EF-G-GDPNP (200 nM) and EF-G-GDP (200 nM) to the two different ribosome conformations.
b, Dwell times for EF-G-GDPNP and EF-G-GDP binding to the two different ribosome conformations. For both panels a and b, from left to right, n = 115, n = 371, n = 162, n = 135. Error bars are standard error.
c, Post-synchronization of ribosome counter-rotation correlated with EF-G-GDPNP. For the ribosomes that successfully translocated by EF-G-GDPNP within the 5 minute observation window, EF-G binds to the ribosome for a prolonged period of time. After the ribosome counter-rotates, EF-G departs rapidly due to the decreased affinity of the ribosome’s non-rotated state with EF-G. Extended occupancy of EF-G-GDPNP on the ribosome surmounts the translocation energy barrier partially by locking the ribosome in an intermediate state of ratcheting, enabling thermal energy to eventually complete translocation at high concentrations of Mg2+.
d, Post-synchronization of the ribosome rotation to Cy5-EF-G-GTP. There is a window when the ribosome rotates that EF-G does not bind, emphasized by the shaded area, which is when tRNA-EF-Tu-GTP binds and accommodates in the ribosome. Only post EF-Tu GTP hydrolysis and EF-Tu departure is EF-G able to bind to the A-site. The higher density of EF-G binding events in the rotated state further suggests that EF-G binds with higher affinity to the rotated state of the ribosome.
EF-G continuously samples the ribosome in both conformations, but only when the ribosome is in the rotated conformation does EF-G binding lead to GTP hydrolysis and translocation. Binding of EF-G to the rotated state is highly efficient in driving translocation, with a mean of ~1.3 binding events per successful ribosome counter-rotation, independent of EF-G concentration. EF-G-GTP has a lower kon to the non-rotated state, and a higher kon to the rotated state38,39. For the non-rotated ribosome awaiting the arrival of tRNA-EF-Tu, EF-G sampling is in competition with tRNA sampling: upon successful binding and accommodation of the tRNA to the A site, EF-G is blocked from binding for a period of ~100 ms, as revealed by the post-synchronization plot of the ribosome counter-rotation transition40 (Fig. 5d). Our data supports the conformational selection model for EF-G and tRNA-EF-Tu, however the biological significance in vivo of such a model will require further studies.
State-specific sampling dynamics of EF-G
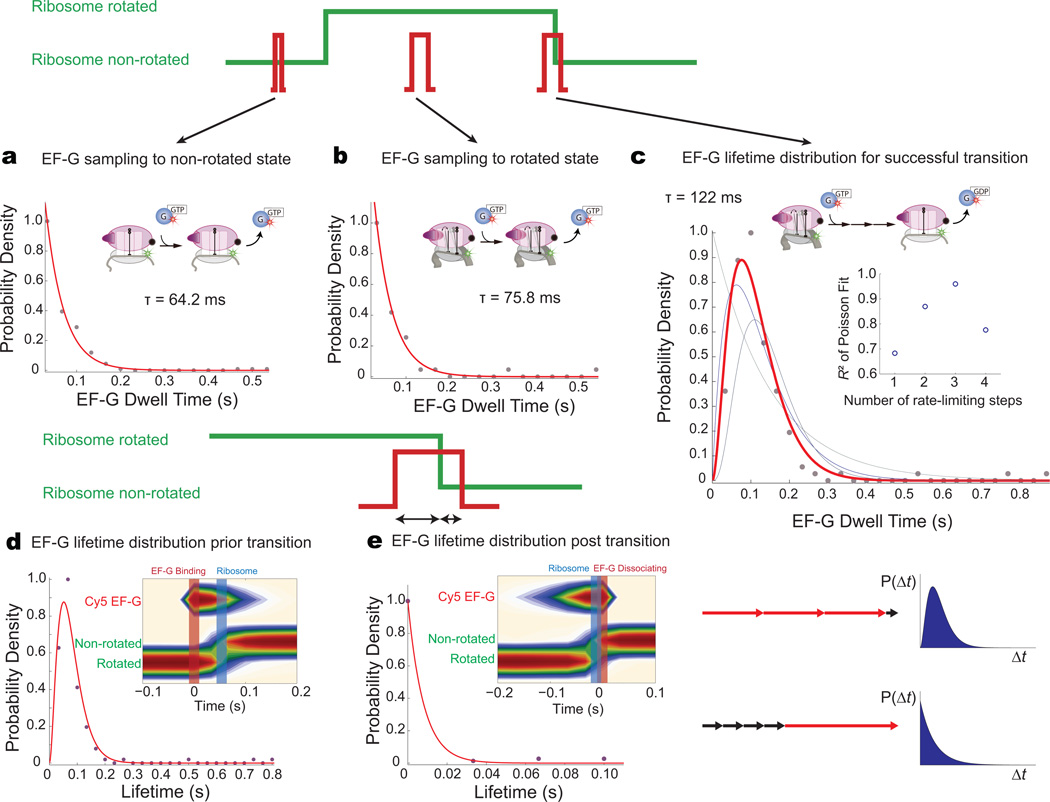
The ability to correlate ribosome conformation and composition through multiple rounds of elongation reveals the state-specific dynamics of EF-G. EF-G-GTP binds to both the rotated and non-rotated ribosomal conformation, with drastically different functional outcomes. EF-G binding to the rotated state results in efficient intersubunit counter-rotation, with an average of ~1.3 binding events for a successful transition (Supplementary Fig. 2), whereas EF-G-GTP binding events to the non-rotated state never result in intersubunit rotation. We compared the dwell time distributions of EF-G-GTP on the ribosome in three cases: (1) binding to the non-rotated state (sampling to the wrong conformation), (2) binding to the rotated state that does not lead to conformational change (unproductive sampling to the correct conformation), or (3) binding to rotated state that leads to counter-rotation. EF-G sampling of the non-rotated (τnon-rotated = 64.2 ms) or rotated (τrotated = 75.8 ms) conformations have shorter dwell times on the ribosome compared to binding events that lead to successful transition from rotated to non-rotated state (τtransition =122 ms). Dwell-time distributions for EF-G binding to both the non-rotated and rotated conformations that do not lead to any conformational transitions follow a single exponential decay (Fig. 6a, b), indicating that these events are probably non-productive binding and rapid dissociation events (single rate-limiting processes).
Figure 6. State-specific dynamics of EF-G to different ribosome conformations.
a, Dwell-time distribution of EF-G binding to the non-rotated state is a single exponential decay, suggesting rapid binding and dissociation with a single rate-limiting step process.
b, Dwell-time distribution for EF-G binding to the rotated state is a single exponential decay, also implying a single rate-limiting step process.
c, EF-G dwell-time distribution for events that lead to successful ribosome counter-rotation. The probability density is best fit by a Poisson distribution with n = 3, implying a process with multiple rate-limiting steps. The inset shows the R2 values for the Poisson fits with different n.
d, e, Relative timing of EF-G binding and dissociation with ribosome counter-rotation. Post-synchronization to either EF-G binding or EF-G dissociation shows that EF-G binds before ribosome counter-rotation (~50 ms), and departs rapidly after (~10 ms). Partitioning of the EF-G dwell time pre- and post-ribosome counter-rotation reveals that after conformational transition, EF-G departs rapidly, with only a single rate-limiting step process. All the kinetically significant steps and conformational changes of EF-G occur prior the ribosome counter-rotation.
In contrast, EF-G binding events that result in an intersubunit conformational transition are best fit by a Poisson dwell-time distribution with n = 3 (R2 = 0.96), suggesting that multiple kinetically significant steps with similar rates underlie the process (Fig. 6c). The kinetic processes identified here could involve structural rearrangement of EF-G upon accommodation to the A site and conformational changes linked to GTP hydrolysis and phosphate release. Thus, EF-G sampling events to either the non-rotated or rotated conformation likely do not result in GTP hydrolysis; only binding events that lead to successful translocation and intersubunit transition involve multiple steps with GTP hydrolysis. Rate-limiting steps with EF-G occur prior to ribosome conformational transition, as shown by partitioning EF-G occupancy lifetimes to the pre-transition and post-transition state; after the transition, EF-G rapidly dissociates (~10 ms), following single-exponential behavior (Fig. 6d, e). At 15 mM Mg2+, similar multi-step processes and EF-G sampling dynamics are observed. These results constitute a direct real-time observation of EF-G dynamics correlated with conformational dynamics through multiple elongation cycles.
Perturbation of EF-G sampling dynamics by antibiotics
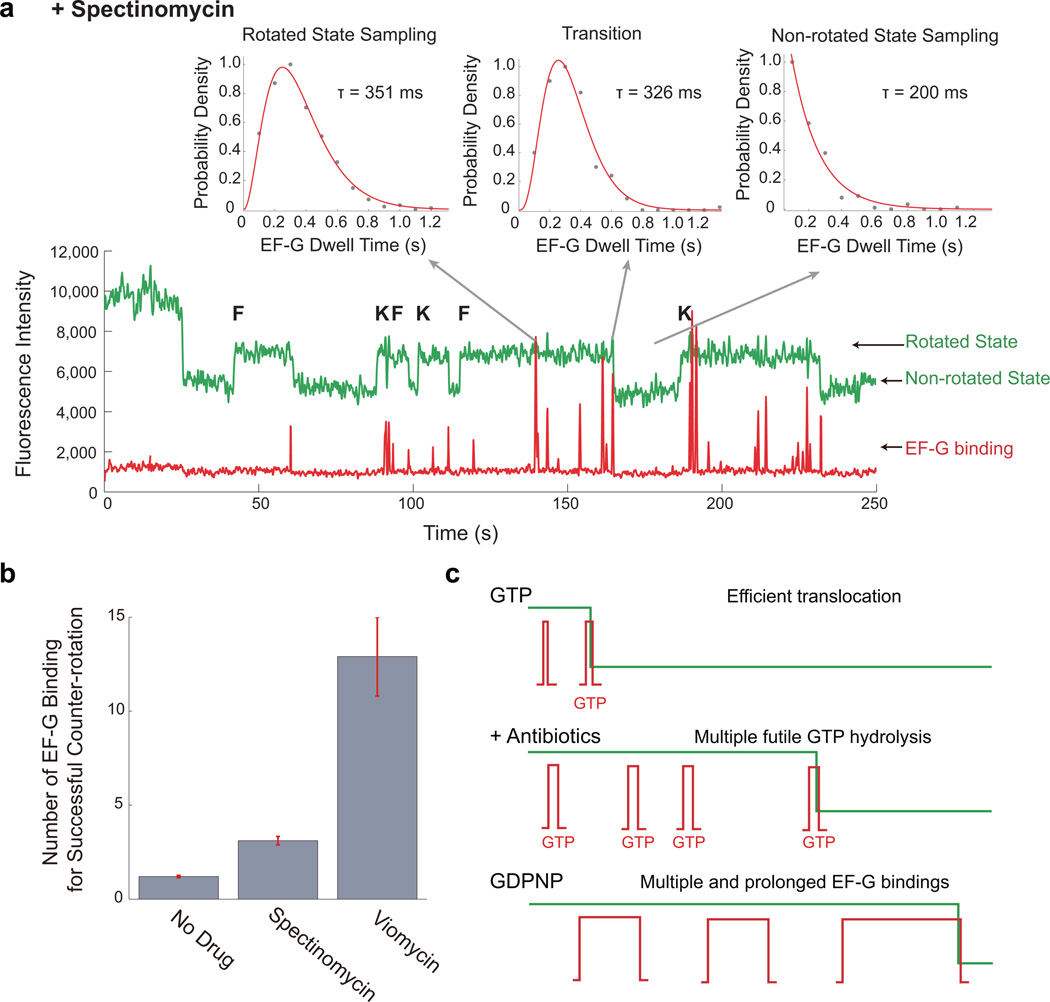
EF-G-ribosome dynamics are dramatically perturbed by antibiotics that inhibit translocation. Spectinomycin inhibits EF-G-catalyzed translocation without affecting GTP hydrolysis7,19, whereas viomycin stabilizes the hybrid conformation of the tRNA and disrupts communication at the subunit interface necessary to mediate ribosome rotation41,42. The presence of either antibiotic increases the number of EF-G sampling events to the rotated state prior to a successful counter-rotation (from a mean number of 1.30 to 3.11 for 100 µM spectinomycin and 12.9 for 500 µM viomycin) (Fig. 7), with a corresponding increase in the rotated state lifetime, as the ribosome awaits EF-G to translocate. EF-G sampling (to the rotated conformer) in the presence of antibiotics follows a Poisson dwell-time distribution, suggesting multiple kinetically rate-limiting steps and possibly GTP hydrolysis, consistent with prior observations that spectinomycin and viomycin do not inhibit GTP hydrolysis by EF-G19 (Supplementary Fig. 7, Fig. 7). The mean dwell-time for EF-G with spectinomycin increased to 351 ms for sampling to the rotated state and to 326 ms for successful translocation events. Sampling to the non-rotated state remained single-exponential in the presence of both drugs, reinforcing that antibiotics dramatically perturb EF-G dynamics when sampling the pre-translocation state of the ribosome. Thus, spectinomycin and viomycin likely inhibit late steps after EF-G GTP hydrolysis, stabilizing ribosomal conformations and increasing the barrier height for translocation7. This leads to futile translocation cycles that probably involve GTP hydrolysis, slowing translation and enhancing its energetic cost (Fig. 7). Our results suggest that translational use of GTP for translocation is tuned to approximately 1 GTP per codon, which is dramatically altered in the presence of drugs.
Figure 7. Perturbations of EF-G dynamics by antibiotics.
a, Representative trace of Cy3B and BHQ ribosome elongating with 80 nM Cy5-EF-G-GTP, 80 nM Phe-tRNAPhe ternary complex, and 80 nM Lys-tRNALys ternary, in the presence of 100 µM spectinomycin. The EF-G dwell time distribution for a binding that led to a successful counter-rotation is a multi-step process same as observed without antibiotics. The distribution for sampling to rotated state shifted to be a multi-step distribution, indicative that these sampling events likely involve futile GTP hydrolysis. The distribution for sampling to the non-rotated state remains exponential.
b, Mean number of EF-G binding events for a successful ribosome counter-rotation without drugs and in the presence of translocation inhibiting antibiotics, spectinomycin and viomycin. These futile sampling apparently involve GTP hydrolysis, enhancing the energetic cost of translation in the presence of these drugs. From left to right, n = 216, n = 161, n = 125. Error bars are standard error.
c, Schematic of ribosome counter-rotating with EF-G-GTP, EF-G-GTP in the presence of translocation inhibiting antibiotics, and EF-G-GDPNP. Translocation with EF-G-GTP is efficient, with sampling events not hydrolyzing GTP. In the presence of antibiotics, multiple EF-G binding events are required, with the sampling events representing futile GTP hydrolysis cycles. With EF-G-GDPNP, multiple prolonged EF-G events are required to eventually translocate the ribosome.
DISCUSSION
We have illustrated here the intricate interplay of ligand composition and ribosome conformation during translation. Using single-molecule approaches, we have directly correlated EF-G and tRNA arrival and departure dynamics with global ribosomal conformation during translation through multiple codons, further enhancing models of translocation built from numerous kinetic16,18,36, single-molecule1,6,19,43, and structural reports4,14,22. Multiple intermediates states of ratcheting have been previously observed structurally, starting first with the rotation of the 30S body, followed by rotation of the head domain and other conformational rearrangements44. This is consistent with our FRET probe positions reporting on the rotational movement of the 30S body and the ribosome rotating upon peptide bond formation, releasing energy to surmount the barrier to intersubunit rotation20. This complements other single-molecule studies showing spontaneous fluctuations of tRNA between the hybrid and classical state, of the L1 stalk, and of various 30S head rotational movements upon peptide bond formation11–13,45. In the rotated state, EF-G-GTP continuously samples the ribosome, with EF-G-GTP binding more rapidly and stably to the rotated state with a deacylated P-site tRNA than the non-rotated conformation4,38 (Fig. 8). The two conformers of the ribosome do not interconvert rapidly on the timescale of translation, consistent with recent cryo-electron microscopy structures46 and molecular dynamics simulations that indicate energy barriers between the two rotational states of the ribosome greater than kT. Prior single-molecule analysis of ribosomal fluctuations using protein labeling had suggested spontaneous inter-subunit ratcheting, but the distinct positions of probes in different studies are likely reporting on different rotational movements during the ratcheting trajectory47.
Figure 8. Schematic of elongation.
a, The two conformations of the ribosome (rotated and non-rotated) are regulated by peptide bond formation and EF-G GTP hydrolysis. The two conformations of the ribosome, in turn, allow the ribosome to selectively discriminate between binding of tRNA-EF-Tu-GTP or EF-G-GTP.
b, The ribosome is separated into two global conformations, the “unlocked” state and the “locked” state. Upon peptide bond formation, the ribosome “unlocks” (with the rotation of the 30S body), releasing fluctuations of the L1 stalk between open and closed states, fluctuations of tRNAs between the classical and hybrid states, and spontaneous rotations in the 30S head domain. EF-G-GTP binding then stabilizes the L1 stalk in the closed state and tRNA in the hybrid state, as well as causing the head of the 30S subunit to rotate11. GTP hydrolysis by EF-G unlocks mRNA movement36, followed by translocation of tRNAs to the P and E sites (driven by back-rotation of the 30S body and head domains) and relocking of the ribosome and mRNA movement to preserve the reading frame. The E-site tRNA and EF-G-GDP departs rapidly, returning the ribosome to the original state.
Our results show that EF-G-GTP binding alone does not induce efficient translocation. At low concentrations of Mg2+, translocation with EF-G-GDPNP is > 50 fold slower than with GTP, consistent with prior bulk observations3,16,18. We show that at long timescales (5 min) and high Mg2+, EF-G-GDPNP binding can induce significant translocation, consistent with prior bulk toe-printing results35. Increasing the Mg2+ concentration stabilizes both the non-rotated state of the ribosome and the classical state of tRNAs1 and likely modulates the energy barrier for translocation. Binding of EF-G-GDPNP surmounts the barrier partially by locking the ribosome in an intermediate state of ratcheting29,44, enabling thermal energy to eventually complete translocation at high concentrations of magnesium. This process is inefficient, requiring multiple long-lived EF-G binding events, yielding a long integrated residence time of EF-G on the ribosome. Thus, the binding free energy of EF-G-GTP-ribosome interaction is not sufficient to induce rapid translocation at physiological Mg2+ concentrations.
Efficient and rapid ribosome counter-rotation and translocation require GTP hydrolysis by EF-G. During this process, EF-G undergoes several rate-limiting processes, possibly conformational changes induced by binding and GTP hydrolysis18. Structural changes between free and ribosome-bound EF-G suggest that a 20–40-Å movement of EF-G’s IV–V domain toward the A site is important to translocation48. GTP hydrolysis releases 8 kcal/mol of free energy and leads to conformational rearrangements in Domain IV of EF-G4,49. This may promote further conformational changes of the ribosome (possibly an “unlocking” step for mRNA and tRNA translocation36) and drives ribosome intersubunit counter-rotation to the non-rotated state, in addition to various reverse-rotations along the back-ratcheting motion46. The decreased affinity of the non-rotated ribosome for EF-G-GDP as well as conformational events in the post-translocation complex may be required for EF-G dissociation50. The free energy for rotation and counter-rotation are provided mainly by irreversible processes (peptide bond formation and GTP hydrolysis) that provides directionality to translation. The molecular nature of the coupling of GTP hydrolysis to the various ribosomal conformational changes will require further investigation. Nonetheless, our data support a “power stroke” mechanism for EF-G in translocation.
Ribosome and factor conformation control ligand occupancy of the ribosome during translation. EF-G-GTP has higher arrival rates and slower off rates to the rotated versus non-rotated conformation of the ribosome39. EF-G-GDP binds slowly and briefly to both conformers. Aminoacyl-tRNA-EF-Tu-GTP binds rapidly and stably only to the classical state. This conformational selection model provides an explanation of why EF-Tu and EF-G do not competitively inhibit translation even though their cellular concentrations are high38,39.
Our data show how antibiotics distort the free energy landscape of ribosomal conformational rearrangements to perturb the efficiency of energy usage in translocation. Translocation inhibitors viomycin and spectinomycin lengthen the lifetime of the rotated state, and increase the number of EF-G sampling events that are required for successful translocation. The dwell-time distributions of these sampling events strongly suggest that GTP hydrolysis is occurring in futile cycles of EF-G binding and attempted translocation. Prior single-molecule experiments have shown how aminoglycoside antibiotics slow translocation through mechanisms that stabilize tRNA or ribosomal conformational states39,51. Antibiotic action is enhanced through multiple rounds of the translation cycle.
The results presented here showcase the power of single-molecule methods combining the use of FRET and ZMWs to correlate conformational and compositional dynamics within distinct sub-steps in complex biological processes such as translation. We have shown that translocation requires the correlated interactions of factors and tRNA and their control of ribosomal conformation. These experiments represent steps towards real-time observation of the molecular choreography that underlies translation, and are widely applicable to a range of biological systems.
ONLINE METHODS
1. Sample Preparation
Escherichia coli ribosomal subunits and translation factors were prepared and purified as described before20. Hairpin loop extensions were introduced into phylogenetically-variable, surface-accessible loops of the E. coli 16S rRNA in helix 44 and 23S rRNA in helix 101 using previously described site-directed mutagenesis21. The 70S ribosomes were then purified from SQ380 cells expressing these mutant ribosomes, and the 30S and 50S subunits were prepared from dissociated 70S particles using previously described protocols20. IF2, EF-Tu, EF-G, EF-Ts, and ribosomal protein S1 from E. coli were purified from overexpressing strains as previously described20.
3’-dye labeled DNA oligonucleotides (labeled with Cy3B or BHQ-2) complementary to the mutant ribosome hairpins20,21 were ordered from Trilink. Right before each experiment, purified 30S and 50S ribosomal subunits (final concentration = 1 µM) were mixed in 1:1 ratio with the 3’ dye-labeled oligonucleotides specific for the hairpin extensions in each subunit for 37°C for 10 min and then at 30°C for 20 min in a Tris-based polymix buffer system (50 mM Tris-acetate pH 7.5, 100 mM potassium chloride, 5 mM ammonium acetate, 0.5 mM calcium acetate, 5 mM magnesium acetate, 0.5 mM EDTA, 5 mM putrescine-HCl, and 1 mM spermidine).
The importance of using the nonfluorescent quencher BHQ-2 for the FRET acceptor has been well discussed in our previous work23. Cy3B and Cy3 with Cy5 (originally used as FRET pairs) have stable photophysics with high quantum yield and long lifetimes. The other spectral dyes (Cy2, Cy3.5, and Cy5.5) generally have short lifetimes with low quantum yield, making correlation studies difficult. The use of BHQ-2 frees up the Cy5 dye to label other protein factor or tRNA.
fMet-tRNAfMet, Lys-tRNAlys, and Phe-tRNAPhe are charged and purified according to published protocols1,2. Phe-(Cy5)tRNAPhe were labeled with Cy5-NHS (GE Lifesciences) at the elbow position (U47), purified, and aminoacylated as previously described25.
The 6(FK) mRNA used consists of a 5’-biotin followed by a 5’-UTR and Shine-Dalgarno sequence derived from gene 32 of the T4 phage, an AUG start codon, 6 alternating repeats of Phe and Lys codons, an UAA stop codon, and four spacer Phe codons. The mRNA was chemically synthesized by Dharmacon.
A cysteine-free mutant of EF-G (C114D, C266A, and C398S) was used to create a single-cysteine EF-G (S73C) by QuickChange site-directed mutagenesis (Strategene). The EF-G S73C mutant protein was overexpressed in E. coli BL21(DE3) cells. Cells were lysed using a French press, and the lysate clarified by centrifugation was loaded onto a 5-mL HiTrap Ni2+ column (GE Healthcare). The fractions containing protein was pooled together and purified on size-exclusion column (Superdex 200). Purified protein was labeled with monomaleimide-Cy5 (GE Lifesciences) by incubating the protein with 10-fold excess of the Cy5-maleimide dye for 2 hours at room temperature. After the reaction, the mixture was passed through two 10 DG desalting gravity columns (Bio Rad) to remove free dye. The labeled EF-G was stored in storage buffer with 50% glycerol at −20°C.
2. Immobilization of PIC on ZMW
To assemble 30S pre-initiation complexes (PICs), we mixed 0.25 µM Cy3B-30S, pre-incubated with stoichiometric amount of S1, 1 µM initiation factor-2 (IF2), 1 µM fMet-tRNAfMet (or fMet-(Cy3)tRNAfMet), 1 µM biotinylated mRNA 6(FK), and 4 mM GTP, in a 5 mM Mg2+ Tris-based polymix buffer without reducing agents and incubated reaction mixture at 37°C for 5 min.
ZMW chips were used and prepared as described previously25. The ZMW surface was derivatized with 16.6 µM neutravidin for 3 minutes at room temperature, and rinsed with polymix buffer to remove unbound neutravidin. Before surface immobilization, we diluted assembled PICs to 25 nM in 5 mM Mg2+ polymix buffer containing 1 µM IF2 and 4 mM GTP. PICs were immobilized by delivering the diluted PIC mixture to the ZMW surface and incubating at room temperature for 3 minutes. Afterwards, surface was rinsed with 5 mM Mg2+ polymix buffer containing 1 µM IF2, 4 mM GTP, and an oxygen scavenging system containing 1 mM Trolox, 2.5 mM PCA, and 50 nM PCD. Immobilized PICs were identified by the Cy3B-30S fluorescence and were distributed in ZMW holes according to Poisson statistics. Control experiments without mRNA demonstrated the absence of non-specific surface interactions at concentrations up to 1 µM of labeled tRNAs and ribosomes, and up to 500 nM of labeled EF-G.
All experiments were performed at a room temperature of 22°C. All experiments were dual illuminated with 532 nm laser (0.5 µWµm−2) and 647 nm laser (0.5 µWµm−2 or 0.25 µWµm−2 where appropriate), unless stated otherwise.
3. Observing elongation on ZMW
30S PICs were preformed in bulk and immobilized on surface of the ZMW slide. Ternary complex were also preformed in bulk with Phe-tRNAPhe or Lys-tRNAlys, EF-Tu, EF-Ts, GTP, and energy regeneration system (phosphoenolpyruvate and pyruvate kinase). Slides were imaged for 5 min, and to initiate translation, 200 nM BHQ-50S, 1 µM IF2, 0 – 500 nM Cy5-EF-G or EF-G, 80 – 500 nM ternary complex (Lys-tRNAlys-EF-Tu-GTP and Phe-tRNAPhe-EF-Tu-GTP or Phe-(Cy5)tRNAPhe-EF-Tu-GTP where appropriate), 4 mM GTP, as well as oxygen scavenging system were delivered at 7 sec time-point. Experiments were either conducted at either 30 fps or 10 fps. All experiments are performed with 5 mM Mg2+ polymix buffer unless stated otherwise. For experiments performed at 15 mM Mg2+, Mg(OAc)2 is added to bring Mg2+ to the desired concentration.
The elongation experiment with Phe-(Cy5)tRNAPhe was performed at high concentrations of Phe-(Cy5)tRNAPhe and Lys-tRNAlys (200 nM) and half 647 nm laser power (0.25 µWµm−2 instead of 0.5 µWµm−2) to reduce the effects of Cy5 photobleaching.
4. Observing EF-G-GDPNP and EF-G-GDP binding
For the experiments involving EF-G-GDPNP or EF-G-GDP (and EF-G-GTP as control), we preformed ribosome 70S complexes in either the rotated or non-rotated conformation. Non-rotated ribosomes were preformed by adding 200 nM BHQ-50S, 1 µM IF2, and 4 mM GTP to the PICs in 5 mM Mg2+ Tris-based polymix buffer. Ribosomes in the rotated state were pre-formed with the addition of Phe-tRNAPhe and Lys-tRNAlys ternary complex. The resulting complexes were immobilized on the ZMW surface for 3 minutes at room temperature. Afterwards, surfaces were rinsed with polymix buffer containing 1 µM IF2, 4 mM GDP, GDPNP or GTP, 1 mM Trolox, 2.5 mM PCA, and 50 nM PCD to wash GTP and unbound factors (Mg(OAc)2 was added to 15 mM Mg2+ where appropriate). Samples were imaged for 5 min and 200 nM Cy5-EF-G-GDPNP, Cy5-EF-G-GTP or Cy5-EF-G-GDP in polymix buffer with oxygen scavenging factors (Mg(OAc)2 was added to 15 mM Mg2+ where appropriate) were delivered during acquisition at 7 sec time-point.
5. Single-molecule translocation assay
For the single-molecule end-point translocation assay under different magnesium conditions, we preformed 70S in the rotated state with (Cy3)tRNAfMet in the P site and (Cy5)tRNAPhe in the A site by adding 200 nM BHQ-50S, 1 µM IF2, 200 nM Phe-(Cy5)tRNAPhe, and 4 mM GTP to the PICs (formed with fMet-(Cy3)tRNAfMet) in 5 mM Mg2+ Tris-based polymix buffer. The resulting complexes were immobilized on the ZMW surface for 3 minutes at room temperature. Afterwards, surfaces were rinsed with polymix buffer containing 1 µM IF2, 4 mM GDPNP or GTP, with oxygen scavenging factors, to wash GTP and unbound factors (Mg(OAc)2 was added to 15 mM Mg2+ where appropriate). We then added 1 µM EF-G, 4 mM GTP or GDPNP, and 1 µM IF2, in polymix buffer (Mg(OAc)2 was added to 15 mM Mg2+ where appropriate), to the complex and incubated for 5 minutes. The surface was then rinsed again with polymix buffer containing 1 µM IF2, 4 mM GTP, and oxygen scavenging factors (Mg(OAc)2 was added to 15 mM Mg2+ where appropriate) to remove EF-G.
To test if the complex translocated, we delivered Lys-(Cy2)tRNAlys ternary complex in polymix buffer with 4 mM GTP, oxygen scavenging factors, and Mg(OAc)2 where appropriate, and imaged for 5 minutes for stable Lys-(Cy2)tRNAlys accommodation signal. For ribosomes that translocated, there will be a stable Cy5 signal with an arrival of stable Cy2 signal. For ribosomes that did not translocate, there will be a lack of stable Cy2 signal due to the A-site being blocked.
6. ZMW data collection and analysis
The ZMW fluorescence was collected on a highly parallel confocal fluorescence detection instrument, using prism-based dispersion optics and an electron-multiplying charge-coupled device camera. Fluorescence traces were collected at a rate of 30 frames per second or 10 frames per second, for 5 minutes. Using custom software writing in Matlab (MathWorks), fluorescence traces corresponding to Cy3, Cy5, Cy2, and Cy3.5 (not used) are extracted and analyzed to measure lifetimes and arrival times, as described previously25. All data analysis and plots are done with Matlab.
7. Poisson fit to determine number of rate-limiting steps
At the single-molecule level, processes are stochastic, so a reaction takes a variable time τ to complete a cycle. The distribution of dwell times P(τ) contains information about the mechanism of the process. For successive rate-determining steps with equal rate constants k, the probability of observing a dwell time τ is a Poisson distribution32.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Jonas Korlach (Pacific Biosciences) for providing technical support on the instrument. This work was supported by US National Institutes of Health (NIH) grant GM51266 to J.C., A.T., and J.D.P., by NIH grant GM099687 to A.P., S.E.O’L, and J.D.P., and by a Stanford Interdisciplinary Graduate Fellowship to J.C.
Footnotes
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
AUTHOR CONTRIBUTIONS
J.C. performed all the experiments and the data analysis. J.C. and J.D.P. designed the project and wrote the manuscript. S.E.O’L. assisted with protein purification and labeling. All authors discussed the results and commented on the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 2.Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci U S A. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 4.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 5.Spirin AS. A model of the functioning ribosome: locking and unlocking of the ribosome subparticles. Cold Spring Harb Symp Quant Biol. 1969;34:197–207. doi: 10.1101/sqb.1969.034.01.026. [DOI] [PubMed] [Google Scholar]
- 6.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 7.Aitken CE, Puglisi JD. Following the intersubunit conformation of the ribosome during translation in real time. Nat Struct Mol Biol. 2010;17:793–800. doi: 10.1038/nsmb.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agirrezabala X, et al. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munro JB, Altman RB, O'Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 11.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 13.Fei J, et al. Allosteric collaboration between elongation factor G and the ribosomal L1 stalk directs tRNA movements during translation. Proc Natl Acad Sci U S A. 2009;106:15702–15707. doi: 10.1073/pnas.0908077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stark H, Rodnina MV, Wieden HJ, van Heel M, Wintermeyer W. Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell. 2000;100:301–309. doi: 10.1016/s0092-8674(00)80666-2. [DOI] [PubMed] [Google Scholar]
- 15.Agrawal RK, Heagle AB, Penczek P, Grassucci RA, Frank J. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat Struct Biol. 1999;6:643–647. doi: 10.1038/10695. [DOI] [PubMed] [Google Scholar]
- 16.Wilden B, Savelsbergh A, Rodnina MV, Wintermeyer W. Role and timing of GTP binding and hydrolysis during EF-G-dependent tRNA translocation on the ribosome. Proc Natl Acad Sci U S A. 2006;103:13670–13675. doi: 10.1073/pnas.0606099103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J Mol Biol. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 18.Hauryliuk V, et al. The pretranslocation ribosome is targeted by GTP-bound EF-G in partially activated form. Proc Natl Acad Sci U S A. 2008;105:15678–15683. doi: 10.1073/pnas.0807912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munro JB, Wasserman MR, Altman RB, Wang L, Blanchard SC. Correlated conformational events in EF-G and the ribosome regulate translocation. Nat Struct Mol Biol. 2010;17:1470–1477. doi: 10.1038/nsmb.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marshall RA, Dorywalska M, Puglisi JD. Irreversible chemical steps control intersubunit dynamics during translation. Proc Natl Acad Sci U S A. 2008;105:15364–15369. doi: 10.1073/pnas.0805299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dorywalska M, et al. Site-specific labeling of the ribosome for single-molecule spectroscopy. Nucleic Acids Res. 2005;33:182–189. doi: 10.1093/nar/gki151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao YG, et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Tsai A, Petrov A, Puglisi JD. Nonfluorescent Quenchers To Correlate Single-Molecule Conformational and Compositional Dynamics. J Am Chem Soc. 2012 doi: 10.1021/ja2119964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai A, et al. Heterogeneous pathways and timing of factor departure during translation initiation. Nature. 2012 doi: 10.1038/nature11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uemura S, et al. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010;464:1012–1017. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 27.Marshall RA, Aitken CE, Puglisi JD. GTP hydrolysis by IF2 guides progression of the ribosome into elongation. Mol Cell. 2009;35:37–47. doi: 10.1016/j.molcel.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horan LH, Noller HF. Intersubunit movement is required for ribosomal translocation. Proc Natl Acad Sci U S A. 2007;104:4881–4885. doi: 10.1073/pnas.0700762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ratje AH, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Semenkov YP, Rodnina MV, Wintermeyer W. The"allosteric three-site model" of elongation cannot be confirmed in a well-defined ribosome system from Escherichia coli. Proc Natl Acad Sci U S A. 1996;93:12183–12188. doi: 10.1073/pnas.93.22.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen C, et al. Allosteric vs. spontaneous exit-site (E-site) tRNA dissociation early in protein synthesis. Proc Natl Acad Sci U S A. 2011;108:16980–16985. doi: 10.1073/pnas.1106999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wen JD, et al. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee TH, Blanchard SC, Kim HD, Puglisi JD, Chu S. The role of fluctuations in tRNA selection by the ribosome. Proc Natl Acad Sci U S A. 2007;104:13661–13665. doi: 10.1073/pnas.0705988104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ermolenko DN, Noller HF. mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat Struct Mol Biol. 2011;18:457–462. doi: 10.1038/nsmb.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spiegel PC, Ermolenko DN, Noller HF. Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. RNA. 2007;13:1473–1482. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Savelsbergh A, et al. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 37.Munro JB, Altman RB, Tung CS, Sanbonmatsu KY, Blanchard SC. A fast dynamic mode of the EF-G-bound ribosome. EMBO J. 2010;29:770–781. doi: 10.1038/emboj.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zavialov AV, Ehrenberg M. Peptidyl-tRNA regulates the GTPase activity of translation factors. Cell. 2003;114:113–122. doi: 10.1016/s0092-8674(03)00478-1. [DOI] [PubMed] [Google Scholar]
- 39.Wang L, et al. Allosteric control of the ribosome by small-molecule antibiotics. Nat Struct Mol Biol. 2012;19:957–963. doi: 10.1038/nsmb.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richman N, Bodley JW. Ribosomes cannot interact simultaneously with elongation factors EF Tu and EF G. Proc Natl Acad Sci U S A. 1972;69:686–689. doi: 10.1073/pnas.69.3.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ermolenko DN, et al. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat Struct Mol Biol. 2007;14:493–497. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- 42.Stanley RE, Blaha G, Grodzicki RL, Strickler MD, Steitz TA. The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome. Nat Struct Mol Biol. 2010;17:289–293. doi: 10.1038/nsmb.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen CL, et al. Single-Molecule Fluorescence Measurements of Ribosomal Translocation Dynamics. Mol Cell. 2011;42:367–377. doi: 10.1016/j.molcel.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang W, Dunkle JA, Cate JH. Structures of the ribosome in intermediate states of ratcheting. Science. 2009;325:1014–1017. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo Z, Noller HF. Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc Natl Acad Sci U S A. 2012;109:20391–20394. doi: 10.1073/pnas.1218999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 47.Chen J, Tsai A, O'Leary SE, Petrov A, Puglisi JD. Unraveling the dynamics of ribosome translocation. Curr Opin Struct Biol. 2012;22:804–814. doi: 10.1016/j.sbi.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li W, Trabuco LG, Schulten K, Frank J. Molecular dynamics of EF-G during translocation. Proteins. 2011;79:1478–1486. doi: 10.1002/prot.22976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. The process of mRNA-tRNA translocation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19671–19678. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peske F, Matassova NB, Savelsbergh A, Rodnina MV, Wintermeyer W. Conformationally restricted elongation factor G retains GTPase activity but is inactive in translocation on the ribosome. Mol Cell. 2000;6:501–505. doi: 10.1016/s1097-2765(00)00049-6. [DOI] [PubMed] [Google Scholar]
- 51.Tsai A, et al. The Impact of Aminoglycosides on the Dynamics of Translation Elongation. Cell Rep. 2013 doi: 10.1016/j.celrep.2013.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.