We developed an easy-traceable TRV vector, TRV2-GFP, by tagging a GFP to the coat protein. TRV2-GFP-infected plants could be identified efficiently by GFP monitoring. TRV2-GFP is useful for functional genomics in many plants, especially for non-Solanaceae plants, like rose
Key words: Arabidopsis thaliana, gene silencing, Nicotiana benthamiana, Rosa sp., TRV–GFP, VIGS.
Abstract
Virus-induced gene silencing (VIGS) is a useful tool for functional characterization of genes in plants. Unfortunately, the efficiency of infection by Tobacco rattle virus (TRV) is relatively low for some non-Solanaceae plants, which are economically important, such as rose (Rosa sp.). Here, to generate an easy traceable TRV vector, a green fluorescent protein (GFP) gene was tagged to the 3’ terminus of the coat protein gene in the original TRV2 vector, and the silencing efficiency of the modified TRV–GFP vector was tested in several plants, including Nicotiana benthamiana, Arabidopsis thaliana, rose, strawberry (Fragaria ananassa), and chrysanthemum (Dendranthema grandiflorum). The results showed that the efficiency of infection by TRV–GFP was equal to that of the original TRV vector in each tested plant. Spread of the modified TRV virus was easy to monitor by using fluorescent microscopy and a hand-held UV lamp. When TRV–GFP was used to silence the endogenous phytoene desaturase (PDS) gene in rose cuttings and seedlings, the typical photobleached phenotype was observed in 75–80% plants which were identified as GFP positive by UV lamp. In addition, the abundance of GFP protein, which represented the concentration of TRV virus, was proved to correlate negatively with the level of the PDS gene, suggesting that GFP could be used as an indicator of the degree of silencing of a target gene. Taken together, this work provides a visualizable and efficient tool to predict positive gene silencing plants, which is valuable for research into gene function in plants, especially for non-Solanaceae plants.
Introduction
Virus-induced gene silencing (VIGS) is an attractive genetic tool for functional genomics in plants. Usually, VIGS targets a specific endogenous gene for silencing by constructing a recombinant virus carrying a fragment of the target gene (Kumagai et al., 1995). Once the virus invades the host plant, the host defence machinery, such as post-transcriptional gene silencing (PTGS), will be activated and combat the virus. PTGS will target double-stranded RNA (dsRNA), which is commonly used for replication of most plant viruses, and lead to cleavage of dsRNA and sequential generation of small interfering RNA (siRNA). The siRNAs are loaded into the RNAi silencing complex (RISC) to degrade any endogenous transcript which shows sequence homology to the siRNA (Ruiz et al., 1998; Baulcombe, 2004; Burch-Smith et al., 2004). Since VIGS is a transient method, it is an easy and rapid way to study the function of genes, even for those genes whose mutation is lethal. Moreover, despite stable plant transformation not being required for VIGS, VIGS was found to be able to be transmitted to progeny plants, namely non-integration-based transmissible PTGS (Senthil-Kumar and Mysore, 2011).
To date, VIGS vectors have been developed from several plant viruses, such as Apple latent spherical virus (ALSV) (Yamagishi and Yoshikawa, 2009; Sasaki et al., 2011), Barley stripe mosaic virus (BSMV) (Bruun-Rasmussen et al., 2007), and Tobacco rattle virus (TRV) (Liu et al., 2002b ). TRV is one of the most widely used vectors and has been successfully used in various plants, such as Nicotiana benthamiana leaf (Ratcliff et al., 2001; Liu et al., 2002b, 2004), Arabidopsis (Turnage et al., 2002), tomato fruit (Liu et al., 2002a ; Fu et al., 2005), pepper leaf (Chung et al., 2004), petunia flower (Chen et al., 2004), Aquilegia (Gould and Kramer, 2007), Thalictrum flower (Di Stilio et al., 2010), rose flower (Ma et al., 2008), strawberry fruit (Jia et al., 2011), California poppy (Wege et al., 2007), Jatropha curcas (Ye et al., 2009), and Mirabilis jalapa (Singh et al., 2012).
Since VIGS vectors were developed from plant viruses, application of VIGS is species dependent. For instance, the TRV vector is able to silence genes in all tested Solanaceae plants, such as N. benthamiana, tomato, pepper, and petunia. It is, however, less effective in some non-Solanaceae plants crops, such as rose (Ma et al., 2008), which is an important economic and cultural ornamental crop throughout the world.
In the present work, a modified TRV vector, TRV–GFP, was generated by fusing the enhanced green fluorescent protein (EGFP) open reading frame (ORF) sequence to the 3’ terminus of the coat protein (CP) gene in the TRV2 plasmid. The TRV–GFP vector was tested in several plants, including N. benthamiana, Arabidopsis, rose, strawberry, and chrysanthemum. The results showed that TRV–GFP could be detected by using a hand-held UV lamp, and the efficiency of infection by TRV–GFP was equal to that of the original TRV vector. This work provides an effective tool for monitoring the efficiency of VIGS in functional genomics of non-Solanaceae plants.
Materials and methods
Plant materials
Nicotiana benthamiana and wild-type Arabidopsis ecotype Col-0 plants were grown in pots in a growth chamber at 23 °C with a 16h/8h photoperiod and ~60% relative humidity.
For cuttings, Rosa sp. cv. ‘Samantha’ branches were rooted for 4 weeks in vermiculite under intermittent mist. After vacuum infiltration, the plants were transplanted into plastic pots containing a steam-sterilized medium consisting of peat:horticultural grade perlite:washed silica (1:1:1) and were grown in a growth chamber (23 °C, 16/8h photoperiod, ~60% relative humidity).
For rose seedlings, seeds of Rosa sp. cv. ‘Magic Meidiland’ were stored in wet sand at 1 °C for 2 months, and then were transferred to plates to geminate at room temperature. The VIGS experiment was conducted when the seedlings had two cotyledons, then they were transplanted into pots containing a mixture of 1:1 (v/v) peat:vermiculite, and transferred to a greenhouse (23°C, relative humidity 60–70%, 16h/8h light/dark).
For strawberry, Fragaria×ananassa cv. ‘Benihoppe’ plants were used. The plants were propagated by in vitro culture for 65 d. After vacuum infiltration treatment, strawberry plants were rinsed with distilled water and then transplanted into pots containing a mixture of 1:1 (v/v) peat:vermiculite. The plants were grown in a greenhouse (23 °C, relative humidity 60–70%, 14h/10h light/dark cycles).
For chrysanthemum (Dendranthema grandiflorum), a popular ground cover-type cultivar ‘Fall color’ (hereafter, chrysanthemum), was used. Plants were propagated by in vitro culture for 2 months, then transplanted into pots containing a mixture of 1:1 (v/v) peat:vermiculite, and transferred to a greenhouse (23 °C, relative humidity 60%–70%, 16h/8h light/dark).
Vector construction
For generation of TRV–GFP, the EGFP-tagged CP fragment was generated by overlap PCR and was inserted into the pTRV2 vector digested by HindIII and BamHI.
For silencing the phytoene desaturase (PDS) gene in N. benthamiana, a 368bp fragment of the N. benthamiana NtPDS gene (EU165355) was amplified and cloned into pTRV2/pTRV2-GFP to form pTRV2-NtPDS/pTRV2-GFP-NtPDS. For silencing PDS in A. thaliana, a 1000bp fragment of the Arabidopsis AtPDS gene (NM117498) was amplified and was cloned into pTRV2/pTRV2-GFP to form pTRV2-AtPDS/pTRV2-GFP-AtPDS. For silencing PDS in rose, a 486bp fragment of the RhPDS (RU05877, Rose Transcriptome Database, http://bioinfo.bti.cornell.edu/cgi-bin/rose_454/index.cgi) gene was amplified from rose cDNA and cloned into pTRV2/pTRV2-GFP to form pTRV2-RhPDS/pTRV2-GFP-RhPDS. The primers used are listed in Supplementary Table S1 available at JXB online.
Agroinfiltration and vacuum infiltration
The Agrobacterium strain GV3101 containing TRV-VIGS vectors was grown at 28 °C in Luria–Bertani medium supplemented with 10mM MES, 20mM acetosyringone, 50mg l–1 gentamicin, and 50mg l–1 kanamycin for ~24h. Agrobacterium cells were harvested and suspended in the infiltration buffer (10mM MgCl2, 200mM acetosyringone, and 10mM MES, pH 5.6). A mixture of Agrobacterium cultures containing pTRV1 and pTRV2 and its derivatives, in a ratio of 1:1 (v/v), were placed in the dark at room temperature for 4h before inoculation. For vacuum infiltration, Silwet L-77 was added to a final concentration of 0.01% (v/v).
Nicotiana benthamiana infiltration was performed as described by Liu et al. (2002a ). Agrobacterium cultures containing pTRV1 and pTRV2 or its derivatives (1:1, OD600=1.0) were injected into the lower leaf of four-leaf stage plants by using a 1ml needleless syringe. For axil injection, 20 μl of bacterial suspension (1:1, OD600=4.0) were injected per plant into the leaf axil using a 1ml syringe with a needle at the four-leaf stage period.
Since rose leaves and axils are hard to inject, the vacuum infiltration method was used for rose cuttings, and seedlings were infiltrated as described previously (Ma et al., 2008; Yan et al., 2012). Four-week-old rose cuttings were removed from the soil, the roots were rinsed with distilled water, and whole plants were submerged in infiltration mixture containing pTRV1 and pTRV2 or its derivatives (OD600=1.0) and subjected to a vacuum at –25 kPa twice, each for 60 s. This was repeated once. Treated plants were briefly washed with distilled water and then planted in pots. For seedlings, the seeds generated were submerged in infiltration mixture containing pTRV1 and pTRV2 or its derivatives (OD600=1.0) and subjected to a vacuum at –25 kPa twice, each for 60 s. The infiltrated seeds were briefly washed with distilled water and placed on wet filter paper for several days, and then the seedlings were planted in pots.
Strawberry was vacuum infiltrated in the same way as rose cuttings and seedlings. After the release of the vacuum, the plants were briefly washed with distilled and planted in pots.
Arabidopsis and chrysanthemum infiltration was performed as described by Burch-Smith et al. (2006). A mixture of Agrobacterium culture containing pTRV1 and pTRV2-GFP (OD600=1.5) was infiltrated into the two leaves of 2-week-old Arabidopsis plants and the lower leaves of six- to eight-leaf-stage chrysanthemum plants using a needleless syringe. The infected chrysanthemum plants were transferred into pots.
Total RNA extraction and RT–PCR
Total RNA was extracted from control and infected plants individually and treated with RNase-free DNase I (Promega, USA). First-strand cDNA was synthesized using 1 μg of total RNA with TRV-specific primers (pYL156 R for TRV1 and OYL198 for TRV2), and oligod(T) (for target genes) or random primer (for CP-GFP). Semi-quantitative reverse transcription–PCR (RT–PCR) was replicated for at least three biological replicates. The PCR products were separated on a 1.2% agarose gel and the images were scanned and analysed using AlphaImager 2200 (Alpha Innotech, San Leandro, CA, USA). The primers used are listed in Supplementary Table S1 at JXB online.
GFP imaging
GFP imaging in N. benthamiana cells was conducted using a laser scanning confocal microscope (Nikon C1). For visualization of the whole plant, the samples were illuminated with a 100W hand-held long-wave ultraviolet lamp (UV products, Upland, CA, USA; Black Ray model B 100AP/R) and were photographed using a Kodak wratten filter 15.
Western blot
Western blot analysis was performed using GFP-specific antibody (Abcam Inc.). Protein was extracted from leaves of individual plant of N. benthamiana, A. thaliana, rose, and strawberry with 300 μl of extraction buffer [100mM TRIS, pH 6.8, 2.5% SDS, 100mM dithiothreitol (DTT), 100mM NaCl, 10% glycerol]. For Arabidopsis, protein was extracted from leaves of individual plants with 300 μl of extraction buffer (125mM TRIS-HCl, pH 6.8, 4% SDS, 20% glycerol, 2% β-mercaptoethanol). For chrysanthemum, protein was extracted from leaves of individual plants with 300 μl of extraction buffer [200mM HEPES, pH 8.0, 20mM EDTA, 100mM DTT, 200mM phenylmethylsulphonyl fluoride (PMSF), 0.2mg ml–1 leupeptin, 50mg ml–1 Polyclar VT]. Bradford assays were used to determine protein quantities, and equal amounts of proteins (10 μg) were separated by 10% SDS–PAGE. Proteins were transferred to a polyvinylidene difluoride membrane (GE Healthcare). CP–GFP was detected after an overnight incubation at room temperature with a 1:10 000 dilution of an anti-GFP antibody conjugated to alkaline phosphatase (Bedoya et al., 2010). Alkaline phosphatase was detected using a chemiluminescent substrate (CSPD; Roche) and exposed to X-ray film (Kodak X-OMAT BT Film/XBT-1).
Results
Validation of the TRV–GFP vector in Nicotiana benthamiana
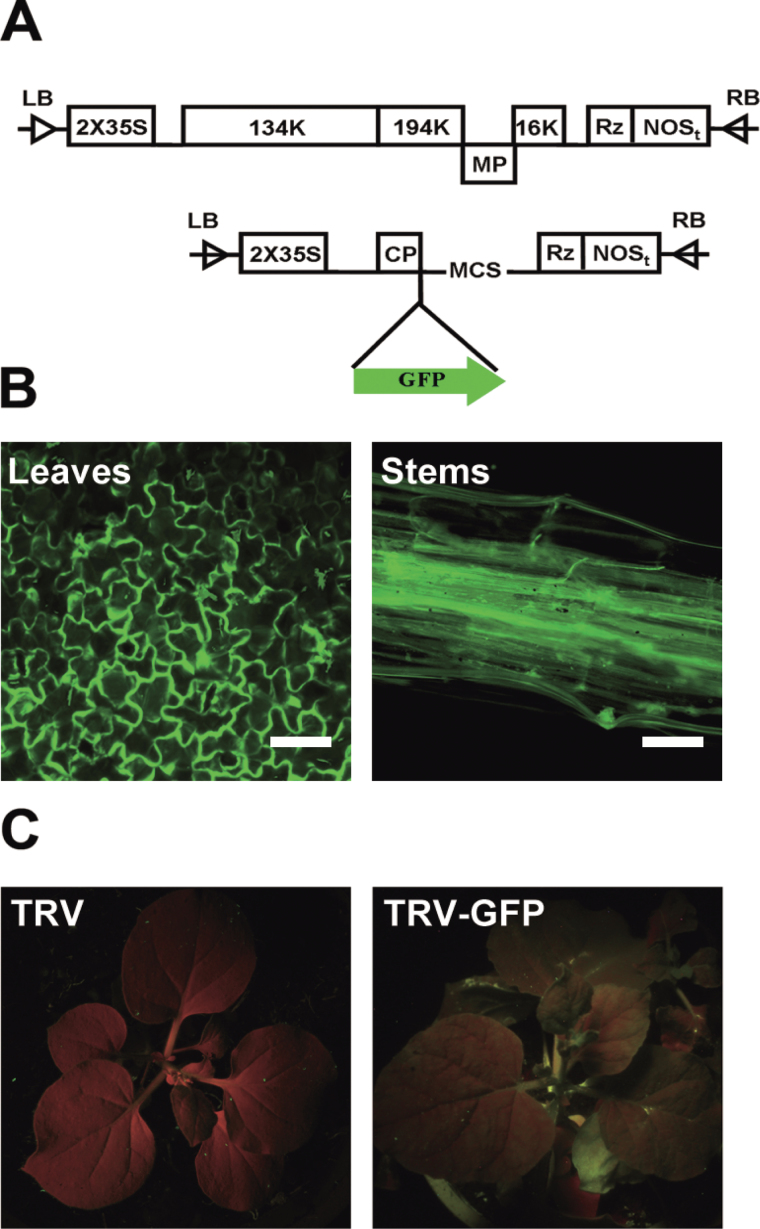
In order to generate a visualizable TRV vector, the stop codon of the ORF of CP in the TRV2 vector was deleted and the vector was tagged with the EGFP ORF sequence by overlap PCR. The modified version of the TRV2 vector was named TRV–GFP (Fig. 1A). The empty TRV–GFP was transferred into Agrobacterium strain 3101 and introduced into N. benthamiana plants by injection on leaves using a needleless syringe. As illustrated in Fig. 1B, GFP fluorescence was observed in injected leaves including stems at 5 dpi (days post-inoculation), indicating that the virus spread in the plants. At 7 dpi, GFP fluorescence could be observed in the upper young leaves using a long-wave UV lamp (Fig. 1C).
Fig. 1.
Construction and validation of the TRV–GFP vector. (A) Schematic diagram of the TRV–GFP vector. (B) GFP imaging of TRV–GFP-infiltrated N. benthamiana by fluorescence microscopy. (C) Imaging of TRV- (left) and TRV–GFP- (right) infiltrated N. benthamiana by a UV lamp. The TRV 5′- and 3′-untranslated regions (UTRs) are indicated by lines. Open boxes, TRV 2×35S promoter, Rz (self-cleaving ribozyme), nopaline synthase terminator (NOSt), and CP. Green arrow, GFP cistron,. Scale bar=100mm.
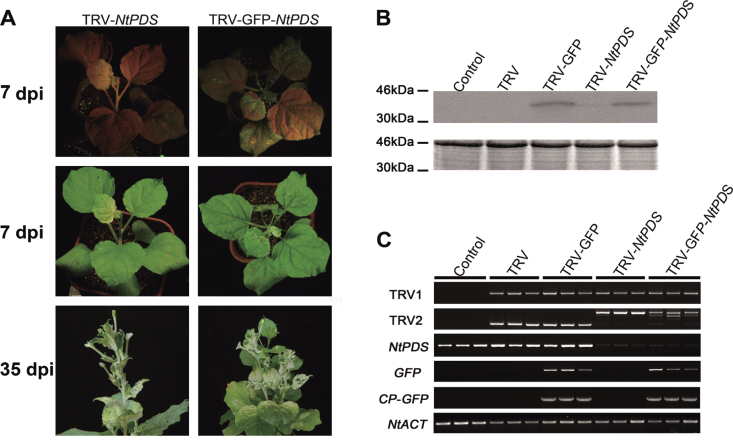
The efficiency of TRV–GFP and TRV vectors in silencing the endogenous PDS in N. benthamiana was compared. As expected, GFP fluorescence could be observed in the upper young leaves by using a long-wave UV lamp at 7 dpi (Fig. 2A). Nicotiana benthamiana plants infected by TRV-NtPDS exhibited the typical photobleached phenotype in the top young leaves at 7 dpi with a silencing efficiency of 97.9% (Table 1). The phenotype developed and lasted until flowering (35 dpi) (Fig. 2A). Plants infected by TRV–GFP-NtPDS also exhibited a very similar phenotype to that of plants infected by TRV-NtPDS. Furthermore, the percentage of positive plants for TRV–GFP-NtPDS (95.8%) was very close to that for TRV-NtPDS (97.9%) (Table 1), indicating that GFP tagging did not alter the gene silencing ability of the TRV vector.
Fig. 2.
Validation of the TRV–GFP vector in N. benthamiana. (A) TRV- and TRV–GFP-infiltrated N. benthamiana plants. The four-leaf-stage plants were inoculated on a leaf with a mixture of pTRV2-PDS/pTRV2-GFP-PDS and pTRV1 (1:1, OD600=1.0) using a needleless syringe. Plants were photographed under UV illumination and normal light. (B) CP–GFP protein levels in upper leaves of N. benthamiana 7 d after TRV vector inoculation. A 10 μg aliquot of protein was used for western blot in each lane, and anti-GFP was used as antibody to detect CP–GFP fusion protein. Coomassie blue staining was used for confirmation of equal loading in each lane. (C) Semi-quantitative RT–PCR of TRV1, TRV2, NtPDS, GFP, and CP-GFP in control and inoculated N. benthamiana plants. NtACT was used as an internal control. The TRV2 fragment was larger in plants infected by TRV-NtPDS and TRV–GFP-NtPDS due to the inserted NtPDS fragment.
Table 1.
Silencing efficiency of TRV and TRV–GFP in N. benthamiana
| TRV | TRV–GFP | |
|---|---|---|
| Total | 48 | 48 |
| GFP positive | – | 46 |
| PDS silencing | 47 | 46 |
| GSEI | 97.9% | 95.8% |
| GSEG | – | 100.0% |
GSEI, gene silencing efficiency of all infected plants; GSEG, gene silencing efficiency of GFP-positive plants.
Using western blot, GFP protein was detected in the top young leaves in TRV–GFP- and TRV–GFP-NtPDS-infected plants, but was not seen in TRV- or TRV-NtPDS-infected plants, further supporting the movement of the modified TRV–GFP virus (Fig. 2B). Semi-quantitative RT–PCR showed that the abundance of endogenous NtPDS transcript was greatly decreased in both TRV–GFP-NtPDS- and TRV-NtPDS- infected plants, indicating that the modified TRV–GFP vector was as effective as the original TRV (Fig. 2C). In addition, N. benthamiana plants were infected by TRV–GFP using the axil injection method (for details, see the Materials and methods). Similarly, the effects of the TRV–GFP vector were indistinguishable from those of the original TRV vector (Supplementary Fig. S1 at JXB online). Collectively, the modification of TRV–GFP does not jeopardize the function of TRV in gene silencing.
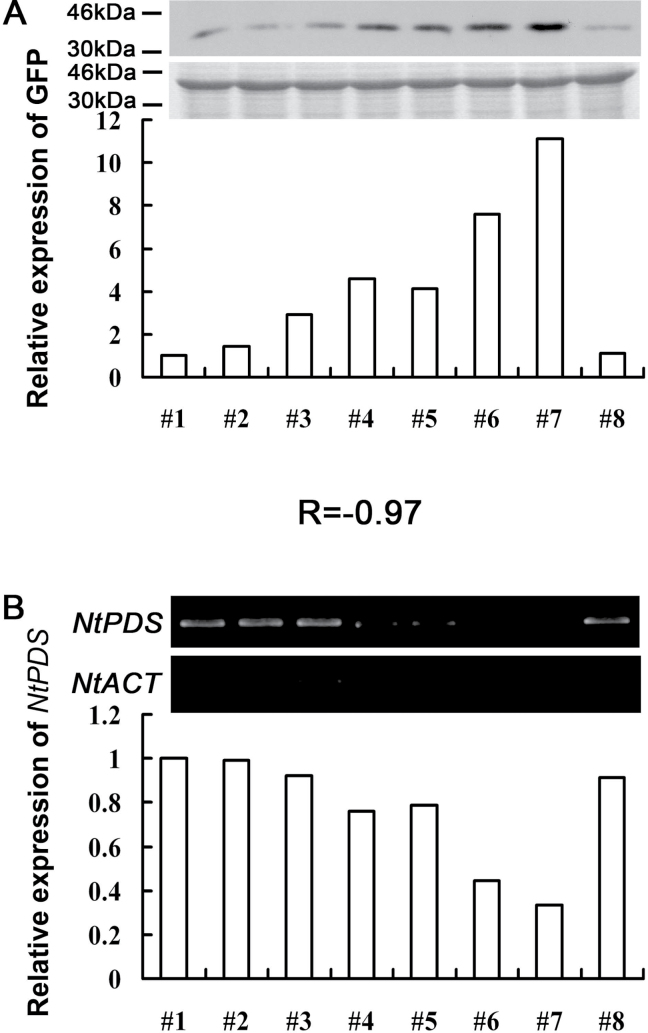
Since GFP protein was fused to the CP protein in the TRV–GFP vector, it was assumed that the accumulation level of GFP protein might reflect the concentration of the TRV virus in plants and indicate the degree of gene silencing, because a higher concentration of virus usually corresponds to the degree of gene silencing. As shown in Fig. 3, the relationship between the level of GFP protein and gene silencing efficiency was analysed, and it was found that the concentration of CP–GFP protein, which indicated the virus concentration, was negatively related to the abundance of the NtPDS gene [correlation coefficient (r)= –0.97].
Fig. 3.
Correlation between CP–GFP protein level and abundance of the NtPDS gene in TRV–GFP-NtPDS-infected N. benthamiana plants. (A) Level of accumulation of CP–GFP protein. Protein was extracted from the third leaf above the inoculated leaves at 7 dpi. A 10 μg aliquot of protein was used for western blot in each lane and anti-GFP was used to detect CP–GFP fusion protein. Molecular sizes are indicated. Coomassie blue staining was used for confirmation of equal loading in each lane. (B) Abundance of the NtPDS gene. Total RNA was extracted from the top young leaves of the N. benthamiana plant which was used for western blot analysis at 14 dpi. NtACT was used as an internal control. The expression level of CP–GFP protein and the NtPDS gene was quantified by AlphaImager 2200. R is the correlation coefficient. Numbers indicate individual plants. The CP–GFP protein and NtPDS expression level in plant #1 was defined as 1, respectively.
Collectively, these results indicted that GFP protein fused with CP protein does not affect the silencing efficiency of the modified TRV–GFP vector, although the movement of the TRV–GFP virus is somewhat slower than that of the original TRV vector. Furthermore, the level of CP–GFP protein could be used to evaluate the degree of silencing of the target gene, which would further increase the efficiency of the VIGS approach.
Validation of the TRV–GFP vector in Arabidopsis thaliana
The TRV vector has been successfully used for functional genomics of A. thaliana (Burch-Smith et al., 2006). To investigate whether the TRV–GFP vector could induce gene silencing in A. thaliana plants, an attempt was made to suppress the expression of the endogenous PDS gene in A. thaliana. A mixture of Agrobacterium cultures containing pTRV2-AtPDS/pTRV2-GFP-AtPDS and pTRV1 was infiltrated onto the entire leaf of two- to three-leaf-stage plants.
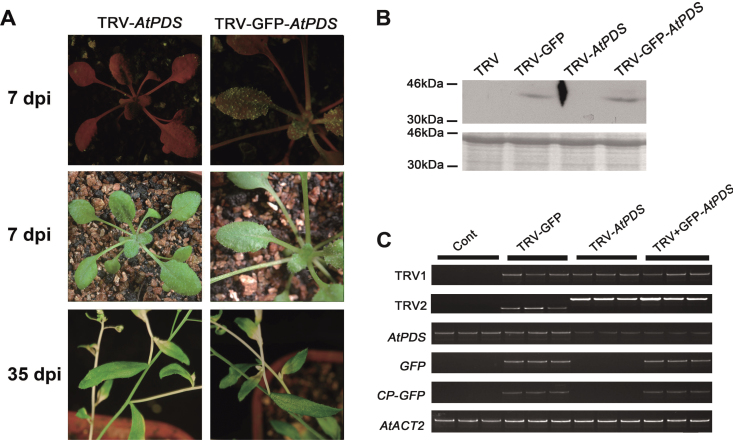
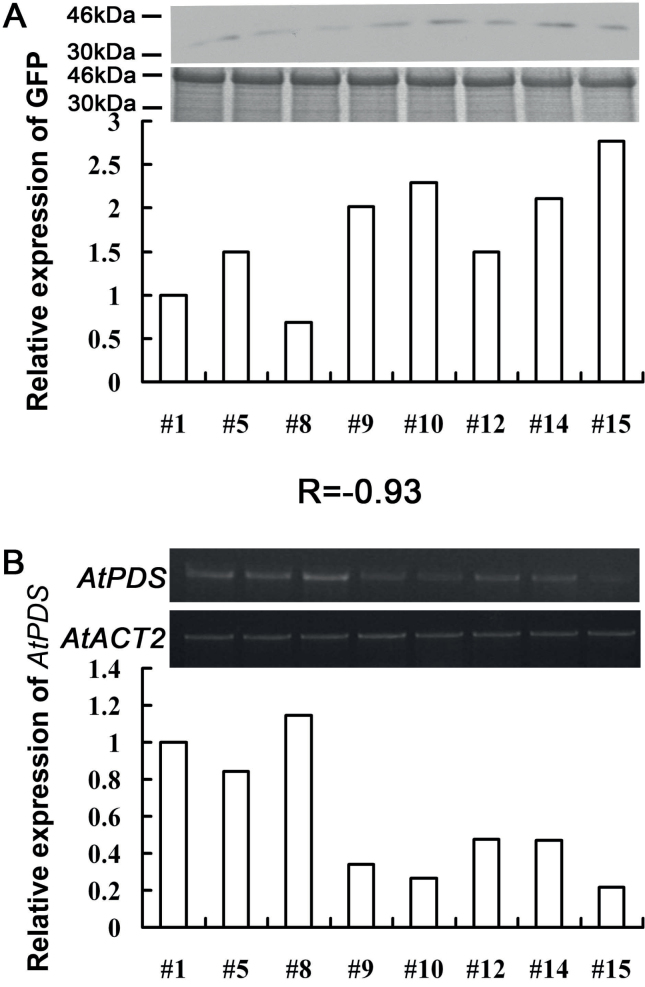
About 7 d after agroinfiltration, GFP fluorescence was observed in the upper young leaves and injected leaves (Fig. 4A). A typical photobleached phenotype was observed in the inflorescence stem of Arabidopsis plants infected with pTRV2-GFP-AtPDS and pTRV2-AtPDS at 30 d after infiltration (Fig. 4A). Western analysis confirmed that GFP proteins were only accumulated in TRV–GFP- and TRV–GFP-AtPDS-infected plants by using GFP antibody (Fig. 4B). Consistently, semi-quantitative RT–PCR showed that the AtPDS level was substantially reduced in both TRV-AtPDS-and TRV–GFP-AtPDS-infiltrated plants when compared with plants infiltrated with TRV and TRV–GFP (Fig. 4C). As in N. benthamiana, it was found that the GFP protein accumulation was also negatively correlated with the expression level of the target gene in A. thaliana [correlation coefficient (r)= –0.93] (Fig. 5). In addition, the silencing efficiency of pTRV2-GFP-AtPDS (93.8% photobleached phenotype) was very close to that of pTRV2-AtPDS (95.8%), indicating that the modified TRV vector was not different from the original vector (Table 2).
Fig. 4.
Validation of the TRV–GFP vector in A. thaliana. (A) TRV- and TRV–GFP-infiltrated plants. Plants were photographed under UV illumination and normal light. (B) CP–GFP protein levels in upper leaves 7 d after TRV vector inoculation. A 10 μg aliquot of protein was used for western blot in each lane and anti-GFP was used as antibody to detect CP–GFP fusion protein. Coomassie blue staining was used for confirmation of equal loading in each lane. (C) Semi-quantitative RT–PCR of TRV1, TRV2, AtPDS, GFP, and CP-GFP in control and inoculated A. thaliana plants. AtACT was used as an internal control. The TRV2 fragment was larger in plants infected by TRV-AtPDS and TRV–GFP-AtPDS due to the inserted AtPDS fragment.
Fig. 5.
Correlation between CP–GFP protein level and abundance of the AtPDS gene in TRV–GFP-AtPDS-infected A. thaliana plants. (A) Level of accumulation of CP–GFP protein. Protein was extracted from the third leaf above the inoculated leaf at 7 dpi. A 10 μg aliquot of protein was used for western blot in each lane and anti-GFP was used to detect CP–GFP fusion protein. Molecular sizes are indicated. Coomassie blue staining was used for confirmation of equal loading in each lane. (B) Abundance of the AtPDS gene. Total RNA was extracted from the top young leaves of the A. thaliana plant which was used for western blot analysis at 14 dpi. AtACT was used as an internal control. The expression level of CP–GFP protein and the AtPDS gene was quantified by AlphaImager 2200. R is the correlation coefficient. Numbers indicate individual plants. The CP–GFP protein and AtPDS expression level in plant #1 was defined as 1, respectively.
Table 2.
Infection efficiency of TRV and TRV–GFP in A. thaliana
| TRV | TRV–GFP | |
|---|---|---|
| Total | 48 | 48 |
| GFP positive | – | 45 |
| PDS silencing | 46 | 45 |
| GSEI | 95.83% | 93.75% |
| GSEG | – | 100.00% |
GSEI, gene silencing efficiency of all infected plants; GSEG, gene silencing efficiency of GFP-positive plants.
Application of the TRV–GFP vector in rose (Rosa sp.)
Rose (Rosa sp.) is an important horticultural crop both economically and culturally. Unfortunately, the infection efficiency of TRV is relatively low in rose, which means that it is more time-consuming to identify positive TRV-infected plants. Since TRV–GFP infection was visualizable, the use of TRV–GFP in rose was tested.
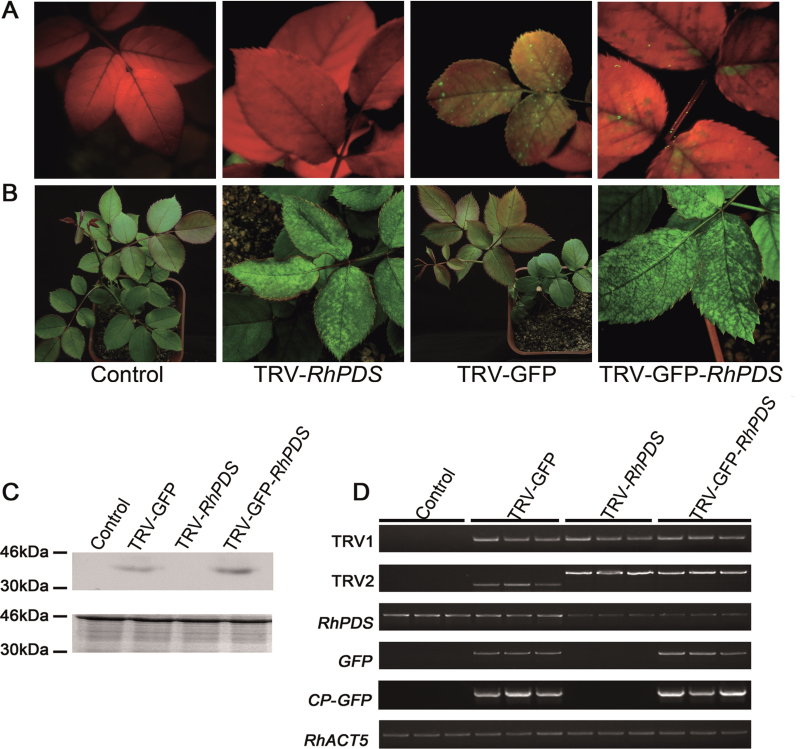
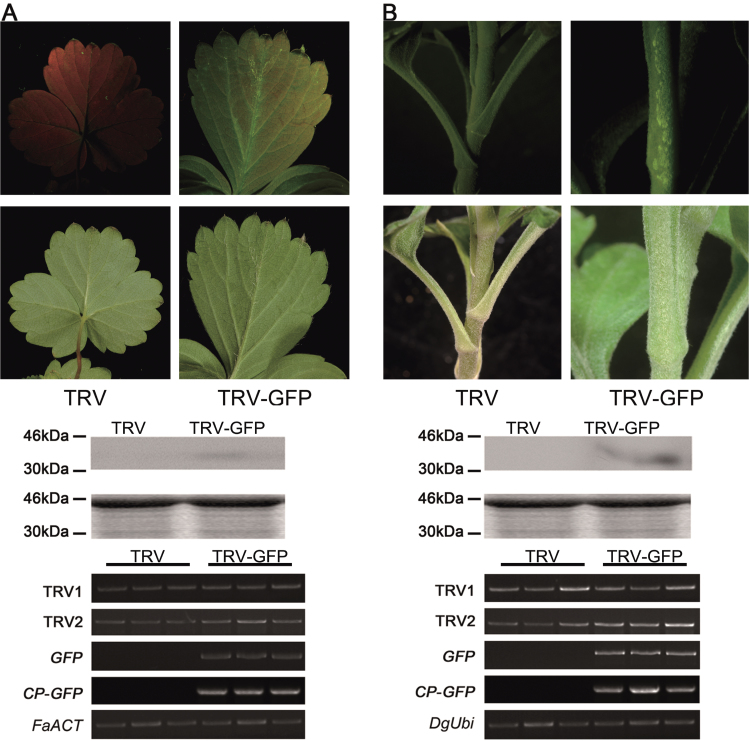
At ~30 dpi, green fluorescence was observed in the top young leaves of pTRV2-GFP-RhPDS-infected rose plants (Fig. 6A). The typical photobleached phenotype appeared in the top young leaves at ~45 dpi, which was slower than that in N. benthamiana (Fig. 6B). Semi-quantitative RT–PCR and western blot showed the existence of the CP-GFP transcript and CP–GFP fusion protein in the top young leaves of plants infected by TRV–GFP and TRV–GFP-RhPDS, indicating that the modified TRV virus successfully moved to the newly emerged leaves (Fig. 6C, D). Also, the abundance of the RhPDS transcript was substantially reduced in the top young leaves of plants infected by TRV–GFP and TRV–GFP-RhPDS (Fig. 6D). Moreover, detached rose petals were infected using the vacuum infiltration method, and the results demonstrated that GFP fluorescence was very strong in petals at 7 dpi (Supplementary Fig. S2 at JXB online). Since rose petals are important for investigating floral scent and colouring, TRV–GFP would be valuable for characterization of genes which are involved in these biological processes.
Fig. 6.
Silencing of RhPDS in rose cuttings. (A) TRV- and TRV–GFP-infiltrated rose plants. Rose cuttings were inoculated with a mixture of pTRV2-RhPDS/pTRV2-GFP-RhPDS and pTRV1 (1:1, OD600=1.0) using vacuum infiltration. The plants were photographed under UV illumination and normal light at 30 and 45 dpi, respectively. (B) CP–GFP protein levels in the upper leaves 30 d after inoculation. A 10 μg aliquot of protein was used for western blot in each lane and anti-GFP was used to detect CP–GFP fusion protein. Coomassie blue staining was used for confirmation of equal loading in each lane. (C) Semi-quantitative RT–PCR of TRV1, TRV2, RhPDS, GFP, and CP-GFP in control and inoculated plants. RhACT5 was used as an internal control. Total RNA and protein were extracted from the top young leaves at 45 dpi, which did not exist when the cuttings were infiltrated and grew out at ~15 dpi.
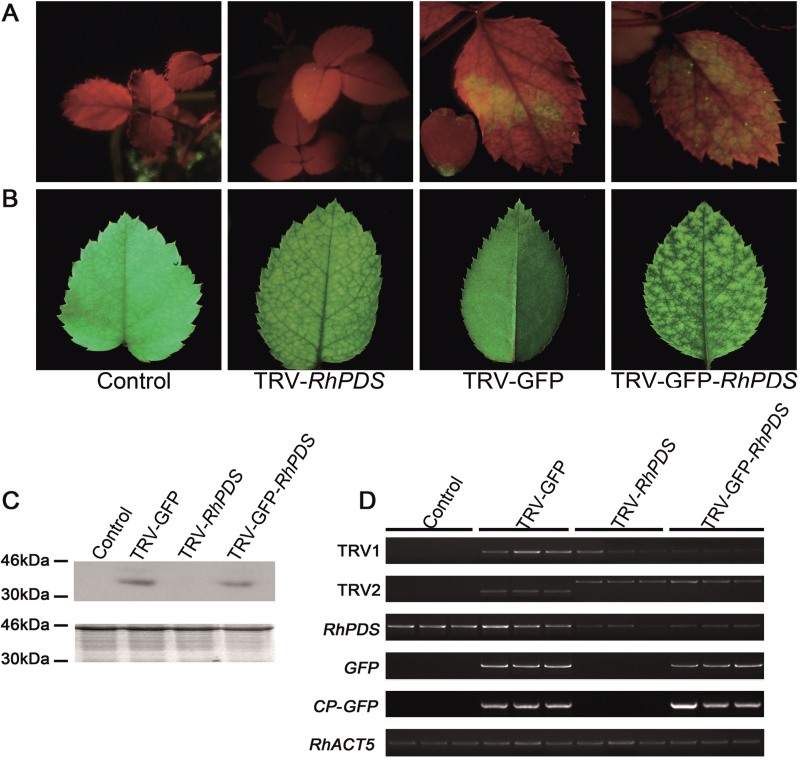
A previous report showed that the VIGS approach could be used to silence target genes in seeds to investigate the gene function in the earlier development period (Yamagishi and Yoshikawa, 2009; Senthil-Kumar and Mysore, 2011). Thus experiments were carried out to test whether the modified TRV–GFP was also compatible with rose seeds. After 2 months storage at 1 °C in wet sand, seeds of Rosa sp. cv. ‘Magic Meidiland’ were placed on wet paper in a plate to geminate at room temperature. Germinated seeds were used to perform the vacuum agroinfiltration. As shown in Fig. 7A, GFP fluorescence was observed in the top young leaves at ~25 dpi. Interestingly, when the plants were in bloom, GFP fluorescence could even be observed in petals (Fig. 7A, inset).
Fig. 7.
Silencing of RhPDS in rose seedlings. (A) TRV- and TRV–GFP-infiltrated rose plants. Rose seedlings were inoculated with a mixture of pTRV2-RhPDS/pTRV2-GFP-RhPDS and pTRV1 (1:1, OD600=1.0) using vacuum infiltration. The plants were photographed under UV illumination and normal light at 25 and 30 dpi, respectively. The inset image in the fluorescent photo of TRV–GFP-infected plant shows a petal with GFP green fluorescence. (B) CP–GFP protein levels in upper leaves 30 d after TRV vector inoculation. A 10 μg aliquot of protein was used for western blot in each lane and anti-GFP was used as antibody to detect CP–GFP fusion protein. Coomassie blue staining was used for confirmation of equal loading in each lane. (C) Semi-quantitative RT–PCR of TRV1, TRV2, RhPDS, GFP, and CP-GFP in control and inoculated plants. RhACT5 was used as an internal control. Total RNA and protein were extracted from the top young leaves.
The existence of CP–GFP in the top leaves of rose seedlings was confirmed by semi-quantitative RT–PCR and western blot (Fig. 7 C, D). As expected, the photobleached phenotype was observed in the top young leaves at 30 dpi, and the abundance of RhPDS was substantially reduced in plants infected by TRV-RhPDS and TRV–GFP-RhPDS (Fig. 7D).
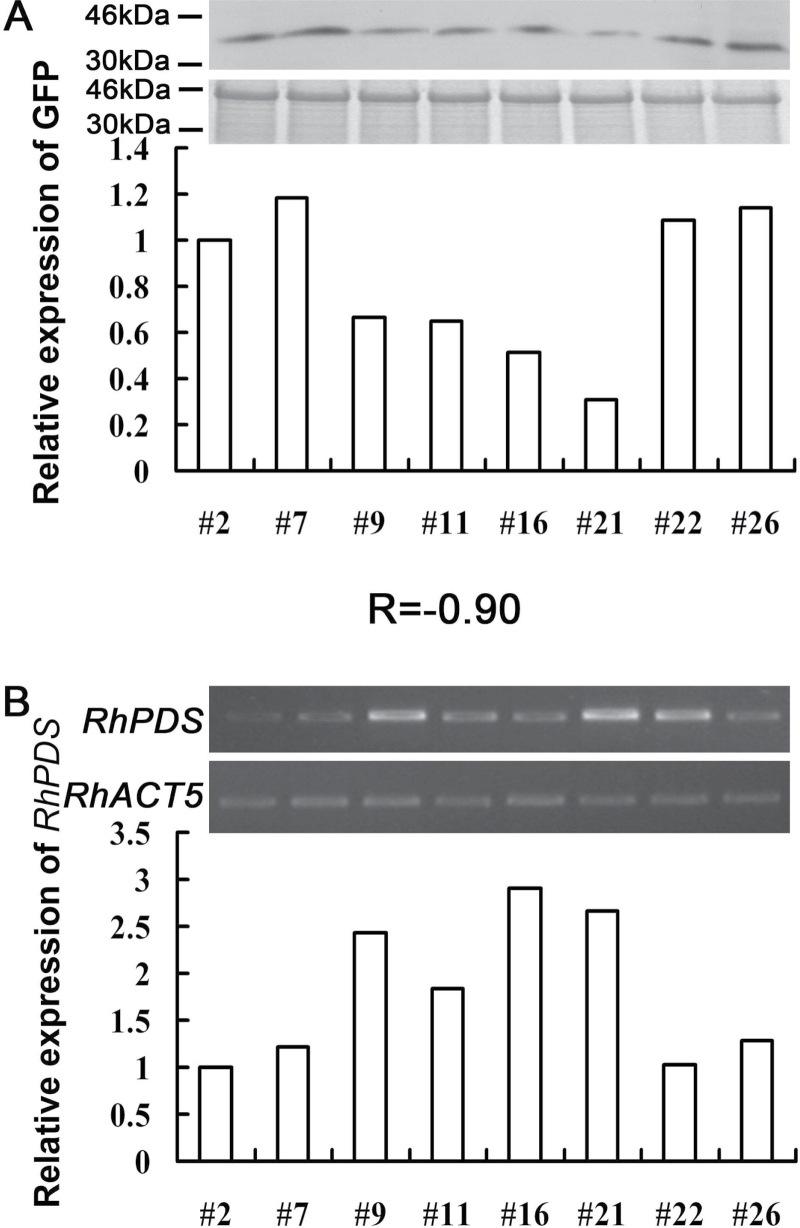
The number of plants positively infected by TRV and TRV–GFP was compared. A PDS-positive plant was defined as one that exhibited the typical photobleached phenotype and a substantially decreased abundance of RhPDS transcript, while GFP-positive plants were defined as plants exhibiting GFP fluorescence under UV light. The percentage of PDS-positive plants was similar between TRV-RhPDS (33.3% for cuttings and 34.3% for seedlings) and TRV–GFP-RhPDS (34% for cuttings and 29.4% for seedlings) plants, indicating that fusion with GFP did not obviously influence the silencing efficiency of the TRV vector in rose (Table 3). For GFP-positive plants, 80% (for cuttings) and 75% (for seedlings) of plants appeared to be PDS positive (Table 3). These results indicated that application of TRV–GFP could greatly increase the screening efficiency when compared with the original TRV vector, especially for plants which are less compatible with the TRV virus, such as rose. Interestingly, it was found that the photobleached phenotype appeared earlier in seedlings (30 dpi) than in cuttings (45 dpi). Moreover, as proved in N. benthamiana and A. thaliana, it was found that the level of CP–GFP protein was negatively correlated with the abundance of RhPDS transcript in the top leaves of rose cuttings infected by TRV–GFP-RhPDS [correlation coefficient (r)= –0.90] (Fig. 8).
Table 3.
Silencing efficiency of TRV and TRV–GFP in Rosa sp
| TRV | TRV–GFP | ||
|---|---|---|---|
| Cuttings | Total | 30 | 35 |
| GFP positive | – | 15 | |
| PDS silencing | 9 | 12 | |
| GSEI | 33.3% | 34.0% | |
| GSEG | – | 80.0% | |
| Seedlings | Total | 32 | 51 |
| GFP positive | – | 20 | |
| PDS silencing | 11 | 15 | |
| GSEI | 34.3% | 29.4% | |
| GSEG | – | 75.0% |
GSEI, gene silencing efficiency of all infected plants; GSEG, gene silencing efficiency of GFP-positive plants.
Fig. 8.
Correlation between CP–GFP protein level and abundance of the RhPDS gene in TRV–GFP-RhPDS-infected rose cuttings. (A) Accumulation level of CP–GFP protein. Protein was extracted from the top young leaves at 45 dpi. A 10 μg aliquot of protein was used for western blot in each lane and anti-GFP was used as antibody to detect CP–GFP fusion protein. Coomassie blue staining was used for confirmation of equal loading in each lane. (B) Abundance of the RhPDS gene. Total RNA was extracted from the top young leaves of the rose plant which was used for western blot analysis at 45 dpi. RhACT5 was used as an internal control. The expression level of CP–GFP and RhPDS was quantified by AlphaImager 2200. R is the correlation coefficient. Numbers indicate individual plants. The expression level of CP–GFP protein and the RhPDS gene in plant #2 was defined as 1, respectively.
These results further supported that it is possible to estimate th edegree of gene silencing by detecting the level of CP–GFP protein in the earlier period of VIGS. Considering that the photobleached phenotype appeared at ~30 dpi in rose cuttings, the modified TRV–GFP vector could be a valuable tool to increase the efficiency of reverse genetic research greatly in rose.
Application of the TRV–GFP vector in other plants
To confirm whether the modified TRV–GFP is suitable for other plants, the TRV–GFP vector was tested in two important horticultural crops, strawberry and chrysanthemum. For strawberry, plants generated from tissue culture were infiltrated by the vacuum infiltration method (1:1, OD600=1.0). For chrysanthemum, the plants were inoculated using a needleless syringe (1:1, OD600=1.5). GFP fluorescence signals were strong in the top new leaves above the agroinoculated leaves at ~21 dpi in strawberry (Fig. 9A). For chrysanthemum, GFP fluorescence was stronger in stems at ~14 dpi (Fig. 9B). As expected, RT–PCR and western blot confirmed that TRV–GFP could successfully work in strawberry and chrysanthemum (Fig. 9; Table 4). Collectively, these results indicated that the TRV–GFP vector could be used widely in multiple plants.
Fig. 9.
Application of TRV–GFP in strawberry (A) and chrysanthemum (B). The plants were inoculated by needleless vacuum infiltration (for strawberry) or syringe injection (for chrysanthemum). The plants were photographed at 21 dpi (for strawberry) and 14 dpi (for chrysanthemum). A 10 μg aliquot of protein was used for western blot in each lane and anti-GFP was used as antibody to detect CP–GFP fusion protein. Coomassie blue staining was used for confirmation of equal loading in each lane. The FaACT and DgUbi genes were used as internal controls for RT–PCR in strawberry and chrysanthemum, respectively.
Table 4.
Infection efficiency of TRV and TRV–GFP in strawberry and chrysanthemum
| Plant | TRV | TRV–GFP | |
|---|---|---|---|
| Strawberry | Total | – | 65 |
| GFP positive | – | 24 | |
| Infection efficiency | – | 36.9% | |
| Chrysanthemum | Total | – | 24 |
| GFP positive | – | 17 | |
| Infection efficiency | – | 70.8% |
Discussion
In the post-genomic era, more and more efforts have been made in functional genomics. As a simple and rapid tool in functional genomics, VIGS has been widely used to analyse gene function in many plant species, as well as in multiple development processes and in response to extrinsic stimuli. Therefore, in the past decade, tremendous improvements VIGS have been reported, such as developing new vectors and improving the vector delivery methods (Jiang et al., 2011; Senthil-Kumar and Mysore, 2011).
Since VIGS vectors are all constructed based on natural plant viruses, application of certain VIGS vectors would be limited by species specificity. The TRV vector is the most widely used VIGS vector so far. It is applicable for many plants because the virus has a broad host range (Bachan and Dinesh-Kumar, 2012). However, the efficiency of TRV in some non-Solanaceae plants, such as rose and strawberry, was still relatively low when compared with N. benthamiana, petunia, and tomato (Ma et al., 2008; personal communication).
Here, to increase the selection efficiency, the CP gene in TRV2 was tagged with GFP to generate a TRV–GFP vector. This modified vector exhibited similar efficiency in infection and gene silencing in N. benthamiana and A. thaliana, although the obvious phenotype of gene silencing emerged a little bit later in TRV–GFP-infected N. benthamiana plants than in plants infected with the original TRV vector. In rose, TRV–GFP was tested in cuttings and seedlings by vacuum infiltration methods. The results indicated that TRV–GFP could spread and silence endogenous genes as efficiently as the original TRV vector. Given that TRV–GFP could be readily tracked by a UV lamp, it is easy to identify GFP-positive plants from the TRV–GFP-infected population. Moreover, ~80% of GFP-positive plants were shown to be positive for gene silencing. Therefore, the modified TRV–GFP vector could promote screening efficiency, although it could not increase the authentic efficiency of gene silencing.
Previously, a dark purple transgenic tomato line was generated by overexpressing an anthocyanin regulatory gene from maize, Lc (Leaf colour) (Goldsbrough et al., 1996). It was reported that this Lc-based tomato line was successfully used as a functional analysis tool to indicate the effects of VIGS by silencing the Lc gene (Jiang et al., 2008). Similarily, a MYB-related transcription factor gene from Antirrhinum majus, Rosea1 (Ros1), which is able to activate anthocyanin biosynthesis, was also introduced into tomato under the control of the fruit-specific E8 promoter. The generated transgenic tomato line exhibited a specific phenotype of purple fruit. Then, a modified TRV2 vector carrying a fragment of Ros1 was used to perform VIGS analysis in the transgenic tomato, and the disappearance of purple colour could be used as a marker of gene silencing (Orzaez et al., 2009). Ros1 was also tagged to several viruses, including Tobacco etch potyvirus (TEV), Tobacco mosaic virus (TMV), Potato virus X (PVX), and Turnip mosaic virus (TuMV), and served as a marker to track virus infection and movement (Bedoya et al., 2012). Moreover, the concentration of anthocyanin was used to determine the viral load qualitatively and quantitatively. Interestingly, although Ros1 was tagged to TRV and TRV-Ros1 induced symptoms of virus infection, the symptomatic tissue failed to show any phenotype of anthocyanin accumulation (Bedoya et al., 2012).
Although anthocyanin accumulation is readily detectable by the naked eye, it is an important trait for many plants, especially for ornamental plants, such as petunia, rose, and chrysanthemum. Therefore, application of an anthocyanin-based system might disturb the endogenous anthocyanin biosynthesis. As a fluorescent protein from jellyfish (Aequorea victoria), GFP does not participate in plant biological processes. Recently, like Ros1, the GFP gene was also overexpressed in tomato driven by the 35S promoter, and its subsequent silencing was used as a traceable marker to indicate the occurrence of gene silencing (Quadrana et al., 2011). This GFP-based system was also shown to have only a negligible impact on the metabolism in leaves and fruits (Quadrana et al., 2011). In the present experiments, no obvious phenotypic changes were found in any tested plant (data not shown). Furthermore, similarly to anthocyanin, the concentration of GFP protein was also able to reflect the viral load and degree of silencing. Therefore, it is possible to perform high-throughout pre-screening by GFP-based enzymed-linked immunosorbent assay (ELISA) analysis.
In the past decade, a large numbers of genes were identified from economically important plants, such as tomato (Tomato Genome Consortium, 2012), cucumber (Huang et al., 2009), apple (Velasco et al., 2010), strawberry (Shulaev et al., 2011), and rose (Dubois et al., 2012; Kim et al., 2012; Pei et al., 2013). These plants all possess some special traits which cannot be investigated in model plants, such as fruit ripening, floral colour, and fragrance. Therefore, it is crucial to perform functional characterization of genes in these plants themselves instead of in model plants, such as Arabidopsis. This study suggested that the modified TRV–GFP vector is a simple but effective tool for functional genomics, especially in non-Solanaceae plants.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Validation of the TRV–GFP vector in N. benthamiana by using the axil injection method (syringe with needle).
Figure S2. Validation of the TRV–GFP vector in detached rose petals.
Table S1. Primers used for vector construction and RT–PCR.
Acknowledgements
We thank Dr Yule Liu (Tsinghua University) for providing the original TRV vector and for his excellent advice, and Dr Daqi Fu (China Agricultural University) for his kind and valuable help with VIGS. This work was supported by National Science Foundation grants to NM (grant numbers 30900991 and 31372095).
References
- Baulcombe DC. 1999. Fast forward genetics based on virus induced gene silencing. Current Opinion in Plant Biology 2, 109–113 [DOI] [PubMed] [Google Scholar]
- Bachan S, Dinesh-Kumar SP. 2012. Tobacco rattle virus (TRV)-based virus-induced gene silencing. Methods in Molecular Biology 894, 83–92 [DOI] [PubMed] [Google Scholar]
- Bedoya LC, Martínez F, Orzáez D, Daròs JA. 2012. Visual tracking of plant virus infection and movement using a reporter MYB transcription factor that activates anthocyanin biosynthesis. Plant Physiology 158, 1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruun-Rasmussen M, Madsen CT, Jessing S, Albrechtse M. 2007. Stability of Barley stripe mosaic virus-induced gene silencing in barley. Molecular Plant-Microbe Interactions 20, 1323–1333 [DOI] [PubMed] [Google Scholar]
- Burch-Smith TM, Schiff M, Liu Y, Dinesh-Kumar SP. 2006. Efficient virus-induced gene silencing in Arabidopsis . Plant Physiology 142, 21–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Jiang CZ, Gookin TE, Hunter DA, Clark DG, Reid MS. 2004. Chalcone synthase as a reporter in virus-induced gene silencing studies of flower senescence. Plant Molecular Biology 55, 521–530 [DOI] [PubMed] [Google Scholar]
- Chung E, Seong E, Kim YC, Chung EJ, Oh SK, Lee S, Park JM, Joung YH, Choi D. 2004. A method of high frequency virus-induced gene silencing in chili pepper (Capsicum annuum L. cv. Bukang). Molecules and Cells 17, 377–380 [PubMed] [Google Scholar]
- Di Stilio VS, Kumar RA, Oddone AM, Tolkin TR, Salles P, McCarty K. 2010. Virus-induced gene silencing as a tool for comparative functional studies in Thalictrum. PLoS One 5, e12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois A, Carrere S, Raymond O, et al. 2012. Transcriptome database resource and gene expression atlas for the rose. BMC Genomics 13, 638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu DQ, Zhu BZ, Zhu HL, Jiang WB, Luo YB. 2005. Virus-induced gene silencing in tomato fruit. The Plant Journal 43, 299–308 [DOI] [PubMed] [Google Scholar]
- Gould B, d Kramer E. 2007. Virus-induced gene silencing as a tool for functional analyses in the emerging model plant Aquilegia (columbine, Ranunculaceae). Plant Methods 3, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsbrough A, Tong Y, Yoder J. 1996. Lc as a non-destructive visual reporter and transposition excision marker gene for tomato. The Plant Cell 9, 927–933 [Google Scholar]
- Huang S, Li R, Zhang Z, et al. 2009. The genome of the cucumber, Cucumis sativus L. Nature Genetics 41, 1275–1281 [DOI] [PubMed] [Google Scholar]
- Jia HF, Chai YM, Li CL, Lu D, Luo JJ, Qin L, Shen YY. 2011. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiology 157, 188–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang CZ, Chen JC, Reid M. 2011. Virus-induced gene silencing in ornamental plants. Methods in Molecular Biology 744, 81–96 [DOI] [PubMed] [Google Scholar]
- Jiang CZ, Lu F, Imsabai W, Meir S, Reid MS. 2008. Silencing polygalacturonase expression inhibits tomato petiole abscission. Journal of Experimental Botany 59, 973–979 [DOI] [PubMed] [Google Scholar]
- Kim J, Park JH, Lim CJ, et al. 2012. Small RNA and transcriptome deep sequencing proffers insight into floral gene regulation in Rosa cultivars. BMC Genomics 13, 657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai MH, Donson J, della-Cioppa G, Harvey D, Hanley K, Grill LK. 1995. Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proceedings of the National Academy of Sciences, USA 92, 1679–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Nakayama N, Schiff M, Litt A, Irish VF, Dinesh-Kumar SP. 2004. Virus induced gene silencing of a DEFICIENS ortholog in Nicotiana benthamiana. Plant Molecular Biology 54, 701–711 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP. 2002. a Virus-induced gene silencing in tomato. The Plant Journal 31, 777–786 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP. 2002. b Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. The Plant Journal 30, 415–429 [DOI] [PubMed] [Google Scholar]
- Ma N, Xue JQ, Li YH, Liu XJ, Dai FW, Jia WS, Luo YB, Gao JP. 2008. Rh-PIP2;1, a rose aquaporin gene, is involved in ethylene-regulated petal expansion. Plant Physiology 148, 894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzaez D, Medina A, Torre S, Fernandez-Moreno JP, Rambla JL, Fernandez-del-Carmen A, Butelli E, Martin C, Granell A. 2009. A visual reporter system for VIGS in tomato fruit based on anthocyanin accumulation. Plant Physiology 150, 1122–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei HX, Ma N, Tian J, Luo J, Chen JW, Li J, Zheng Y, Chen X, Fei ZJ, Gao JP. 2013. A NAC transcription factor controls ethylene-regulated cell expansion in flower petals. Plant Physiology 163, 775–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrana L, Rodriguez MC, López M, et al. 2011. Coupling virus-induced gene silencing to exogenous green fluorescence protein expression provides a highly efficient system for functional genomics in Arabidopsis and across all stages of tomato fruit development. Plant Physiology 156, 1278–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff F, Martin-Hernandez AM, Baulcombe DC. 2001. Tobacco rattle virus as a vector for analysis of gene function by silencing. The Plant Journal 25, 237–245 [DOI] [PubMed] [Google Scholar]
- Ruiz MT, Voinnet O, Baulcombe DC. 1998. Initiation and maintenance of virus-induced gene silencing. The Plant Cell 10, 937–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S, Yamagishi N, Yoshikawa N. 2011. Efficient virus-induced gene silencing in apple, pear and Japanese pear using Apple latent spherical virus vectors. Plant Methods 7, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil-Kumar M, Mysore KS. 2011. New dimensions for VIGS in plant functional genomics. Trends in Plant Science 16, 656–665 [DOI] [PubMed] [Google Scholar]
- Shulaev V, Sargent DJ, Crowhurst RN, et al. 2011. The genome of woodland strawberry (Fragaria vesca). Nature Genetics 43, 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Liang YC, Kumar P, Reid M, Jiang CZ. 2012. Co-silencing the mirabilis antiviral protein permits virus-induced gene silencing in Mirabilis jalapa . Journal of Horticultural Sci ence and Biotechnology 87, 334–340 [Google Scholar]
- Tomato Genome Consortium 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnage MA, Muangsan N, Peele CG, Robertson D. 2002. Geminivirus-based vectors for gene silencing in Arabidopsis . The Plant Journal 30, 107–114 [DOI] [PubMed] [Google Scholar]
- Wege S, Scholz A, Gleissberg S, Becker A. 2007. Highly efficient virus-induced gene silencing (VIGS) in California poppy (Eschscholzia californica): an evaluation of VIGS as a strategy to obtain functional data from non-model plants. Annals of Botany 100, 641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Affourtit J, et al. 2010. The genome of the domesticated apple (Malus×domestica Borkh.). Nature Genetics 42, 833–839 [DOI] [PubMed] [Google Scholar]
- Yamagishi N, Yoshikawa N. 2009. Virus-induced gene silencing in soybean seeds and the emergence stage of soybean plants with Apple latent spherical virus vectors. Plant Molecular Biology 71, 15–24 [DOI] [PubMed] [Google Scholar]
- Yan HX, Fu DQ, Zhu BZ, Liu HP, Shen XY, Luo YB. 2012. Sprout vacuum-infiltration: a simple and efficient agroinoculation method for virus-induced gene silencing in diverse solanaceous species. Plant Cell Reports 31, 1713–1722 [DOI] [PubMed] [Google Scholar]
- Ye J, Qu J, Bui HT, Chua NH. 2009. Rapid analysis of Jatropha curcas gene functions by virus-induced gene silencing. Plant Biotechnology Journal 7, 964–976 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.