Abstract
Epidemiological studies have established a positive correlation between human mortality and increased concentration of airborne particulate matters (PM). However, the mechanisms underlying PM related human diseases, as well as the molecules and pathways mediating the cellular response to PM, are not fully understood. This study aims to investigate the global gene expression changes in human cells exposed to PM10 and to identify genes and pathways that may contribute to PM related adverse health effects. Human bronchial epithelial cells were exposed to PM10 collected from Saudi Arabia for 1 or 4 days, and whole transcript expression was profiled using the GeneChip human gene 1.0 ST array. A total of 140 and 230 genes were identified that significantly changed more than 1.5 fold after PM10 exposure for 1 or 4 days, respectively. Ingenuity Pathway Analysis revealed that different exposure durations triggered distinct pathways. Genes involved in NRF2-mediated response to oxidative stress were up-regulated after 1 day exposure. In contrast, cells exposed for 4 days exhibited significant changes in genes related to cholesterol and lipid synthesis pathways. These observed changes in cellular oxidative stress and lipid synthesis might contribute to PM related respiratory and cardiovascular disease.
Keywords: particulate matter, gene expression, human bronchial epithelial cells, Microarray, Ingenuity pathway analysis
Introduction
Airborne particulate matter, comprised of a mixture of dust, dirt, smoke, and liquid droplets, has been linked to many adverse health effects including respiratory and cardiovascular problems. Numerous epidemiological studies have established an association between short- or long-term exposure to various airborne particulates and human mortality and morbidity (Rückerl et al. 2011; Schwarze et al. 2006; Chen and Lippmann 2009). Studies have found a significant correlation between excess mortality and short-term exposure to high concentration of ambient particulate matters (Bell and Davis 2001; Schwartz and Marcus 1990). Also, a population-based study on data collected in 6 US cities suggested an association between long-term exposure and human mortality (Dockery et al. 1993; Laden et al. 2000). In addition to mortality, airborne particulates were found to connect to increased morbidity. A number of studies reported an association between respiratory and cardiovascular symptoms and short- or long-term exposure to high level of ambient particles (Pope 1989; Brook et al. 2010; Zanobetti and Schwartz 2007). As a result, the mechanisms underlying the PM induced adverse health effects have become an area of active research.
Both in vitro and in vivo studies have established the role of reactive oxygen species (ROS) in PM-induced respiratory and cardiovascular diseases (Ghio et al. 2012; Araujo and Nel 2009; Møller et al. 2010). Following PM exposure, ROS can be generated by some PM components, such as transition metals and organics (Cho et al. 2005; Becker et al. 2005). High level of ROS production could lead to cell apoptosis and tissue damage, which may contribute to lung injury as seen in human or animal models following acute exposure to PM. In addition, ROS are able to initiate a series of redox signaling cascades to induce inflammatory responses and cytokine production found in both acute and chronic PM exposures (Akhtar et al. 2010; Diabaté et al. 2011).
Although PM exposure causes adverse health effects, the magnitude of these effects depends on many factors: particle size, source, chemical composition, and exposure duration, etc (Araujo and Nel 2009; Gordon 2007). While coarse PM (aerodynamic size between 2.5 and 10 μm) can deposit in the upper respiratory tract and lung, fine (≤2.5 μm) and ultrafine (≤0.1 μm) PM penetrate deeper into the alveolar region of the lung resulting in more profound effects on the cardiovascular system. The major sources of PM include soil, dust, crustal particles, fuel combustion, oil refinery and metal processing facilities, and are normally linked to many geographical and meteorological variables (Simkhovich et al. 2008; Araujo and Nel 2009). The chemical composition, which varies depending on the source, is a major factor that contributes to the adverse health effects of PM. Several in vitro and in vivo studies reported the effects of metals, in particular transition metals, on PM induced inflammatory response and cytotoxic activities (Chen and Lippmann 2009).
In the Middle East, dust and sand storm are important factors contributing to airborne particulates. Chronic inhalation of sand dust has been shown to lead to the development of Desert Lung Syndrome, a rare non-progressive non-occupational lung disease found in desert inhabitants (Waness et al. 2011; Nouh 1989). Silicosis and asthma are also linked to desert sand exposure. In the past decade, with the rapid industrialization and increased oil combustion, air pollution has become a serious problem. When combined with sand dust, air pollution likely contributes to significant increases in public health problems, such as cardiovascular and respiratory disease, cancer, and diabetes mellitus. In 2010, the top three causes of death in Saudi Arabia were coronary heart disease (CHD, 24%), hypertension (12%) and diabetes mellitus (6.7%) (http://www.worldlifeexpectancy.com/country-health-profile/saudi-arabia).
Recently, we have conducted a multi-week, multiple site sampling campaign to study the source apportionment and elemental composition of PM10 and PM2.5 in Jeddah, the second largest city in Saudi Arabia. The results showed that the major source factors for PM10 or PM2.5 were soil resuspension, oil combustion, mixed industrial sources, traffic sources, and marine aerosols (manuscript submitted). To investigate the impact of PM exposure on human bronchial cells, we analyzed global gene expression profiles following exposure of cells to PM10. Given that lungs are the major organs targeted by the various sized PM particles, human bronchial epithelial BEAS-2B cells were chosen to expose to the PM10 samples (including coarse, fine and ultrafine PM) for either a short-term (1 day) or longer-term (4 days) duration. Differentially expressed genes following PM10 exposure were identified and analyzed for networks and pathways that may contribute to the cellular response to PM.
Materials and Methods
Particle sample collection
Dust samples were collected from the campus of King Abdulaziz University located in south Jeddah. For metal composition analysis, PM10 (≤10 μm) was collected for 24 hours through Automated Cartridge Collector Unit (ACCU) sampler onto pre-weighed Teflon (GelmanTeflo, 37mm, 0.2 um pore). For in vitro exposure, particles were collected for 48 hours on 5300 Polypropylene filters using Staplex high volume air sampler (Staplex Air Sampler Division, USA) with PM10 inlet (serial NO. 2840) at a fixed flow rate of 900 l/min.
Element metal analysis
Metal concentration was analyzed as follows: The mass on the Teflon filter was measured using a microbalance (model MT5, Mettler-Toledo Inc.) in a temperature and humidity controlled weighing room at NYU laboratory. The concentration of metals was analyzed using a nondestructive X-ray Fluorescence (XRF) Spectrometer (EX-6600-AF, Jordan Valley) with five secondary fluorescers (Si, Ti, Fe, Ge, and Mo) and spectral software XRF2000v3.1 (U.S. EPA and ManTech Environmental Technology, Inc.) as described previously (Maciejczyk and Chen 2005).
Particle extraction
PM10 particles were extracted from polypropylene filters using a modified aqueous extraction protocol (Duvall et al. 2008). Briefly, each filter was wetted with 25 ml of 70% ethanol followed by sonication in 100 ml of distilled water for 2 hours. The particles were dried by lyophilization, then weighed and resuspended in the sterile distilled water, and stored at −80°C.
Cell culture and particle exposure
Normal human bronchial epithelial BEAS-2B cells were cultured in DMEM (Invitrogen) supplemented with 10% FBS and 100 U/ml penicillin and 100 mg/ml streptomycin (Invitrogen). For particle exposure, cells were seeded one day prior to exposure. A small aliquot of particle suspension was mixed with culture medium by sonication for 20 minutes, and then applied evenly to the cultured cells. Untreated and particle treated cells were cultured in 37°C incubator with 5% CO2 until harvesting at the indicated time intervals.
Colony survival assay
BEAS-2B cells were treated with various concentrations of particles (0, 10, 25, 50, 100, and 200 μg/cm2) for 1, 2, 7, and 14 days. Control and particle treated cells were plated at 300 cells/dish in 100-mm cell culture dishes, and cultured for two weeks. Cell colonies were stained with Giemsa solution, and the number of colonies was counted.
RNA extraction and microarray hybridization
BEAS-2B cells were treated with particles (50 μg/cm2) that were collected either during normal or storm weather conditions for 1- or 4- days. Total RNA was extracted from control and particle treated cells using Trizol (Invitrogen) and further purified using RNeasy Plus Micro Kit (Qiagen). 100 ng of total RNA was used to synthesize double-stranded cDNA (dsDNA). cRNA was synthesized from dsDNA template, and subsequently used to produce sense single-stranded cDNA (ssDNA) with incorporated deoxyuridine triphosphate. The ssDNAs were fragmented, end-labeled, and hybridized to Affymetrix Human Gene 1.0 ST Array (Affymetrix). Hybridization and scanning of the arrays was performed using a standard procedure.
Microarray data analysis
Microarray data analysis was performed using GeneSpring v12.0 (Agilent Technologies). All microarray data is MIAME compliant and the raw data has been deposited in NCBIs Gene Expression Omnibus (GEO ID: GSE38172). The expression value of each probe set was determined after quantile normalization using RMA algorithm and baseline transformation to the median levels of control samples. Differentially expressed genes were identified using one-way ANOVA (p<0.05). Principal component analysis (PCA) was used to visualize the gene expression pattern of all samples. Hierarchical cluster analysis using Euclidean distance was performed to cluster genes and samples to generate a heat map. Functional annotation was analyzed with the Gene Ontology (GO) classification system using DAVID software (http://david.abcc.ncifcrf.gov/home.jsp). Gene network and pathway analysis was performed using Ingenuity Pathway Analysis (http://www.ingenuity.com).
Real time PCR
Total RNA extracted from control and treated cells was converted to single stranded cDNA using Superscrip® III (Invitrogen). Quantitative real-time PCR analysis was performed using SYBR green PCR system (Applied Biosystems) on ABI prism 7900HT system (Applied Biosystems). Relative gene expression levels were normalized to ACTB expression. All PCR reactions were performed in triplicate. Results were presented as fold change to the level expressed in control BEAS-2B cells.
Results
Metal concentration of PM samples
Jeddah, the second largest city in the Kingdom of Saudi Arabia, is bordered by the Red Sea in the west and the Al-Sarawat Mountains in the northeast, east and southeast. As in many other cities in the Arabian Peninsula, sand storms are common in Jeddah, and normally peak during the summer season. A total of 30 PM10 samples on Teflon filters were collected from the campus of King Abdulaziz University between June and September 2011, a time period that most faculty and students were on summer vacation. The PM10 that was captured in 3 air filters was collected during the sand storms (designated as the storm group), and PM10 captured in 27 other air filters represented the air particles for the typical summer vacation days (designated as the normal group). By using nondestructive XRF, we analyzed the concentration of 27 elements in PM10 (Table 1). Silicon concentration was the highest among 27 elements in both normal and storm samples, followed by calcium, sulfur, aluminum and iron (Table 1), which represented a typical source category of resuspended soil. Except for sulfur, the concentrations of silicon, calcium, aluminum and iron were increased in the storm samples, suggesting that the soil resuspension was the major source factor that accounted for the increased mass during the sand storm. In addition, the storm group exhibited a slight decrease in the concentration of most other metal elements, including those representing marine aerosol (sodium, chlorine), fuel combustion (nickel, vanadium, sulfur), mixed industrial (zinc, copper) and traffic sources (lead, bromine, selenium). Interestingly, several metal elements including cadmium, strontium, titanium and cobalt were increased in the storm group, probably originating from a local industrial source.
Table 1.
The concentrations of elements (ng/mg mass) in particles
| Elements (ng/mg) | Normal | Storm | ||
|---|---|---|---|---|
| Mean | S.D. | Mean | S.D. | |
| Sodium | 21335 | 13621 | 11425 | 9351 |
| Magnesium | 16465 | 4167 | 16050 | 511 |
| Aluminium | 35286 | 11955 | 41589 | 16386 |
| Silicon | 116194 | 32378 | 135499 | 37820 |
| Phosphorus | 1768 | 714 | 1884 | 1018 |
| Sulfur | 47620 | 15311 | 31932 | 8777 |
| Chlorine | 18722 | 16545 | 16487 | 18881 |
| Potassium | 8218 | 2417 | 7702 | 946 |
| Calcium | 55405 | 17661 | 67780 | 35787 |
| Titanium | 2819 | 1013 | 3781 | 1851 |
| Vanadium | 375 | 132 | 246 | 53 |
| Chromium | 95 | 27 | 109 | 53 |
| Manganese | 1079 | 471 | 1235 | 63 |
| Iron | 32152 | 9934 | 38044 | 11484 |
| Cobalt | 319 | 119 | 400 | 133 |
| Nickel | 132 | 38 | 100 | 12 |
| Copper | 243 | 111 | 141 | 96 |
| Zinc | 850 | 823 | 560 | 124 |
| Gallium | 54 | 81 | 29 | 16 |
| Germanium | 47 | 86 | 27 | 45 |
| Arsenic | 148 | 228 | 74 | 101 |
| Selenium | 47 | 38 | 38 | 36 |
| Bromine | 199 | 78 | 121 | 63 |
| Rubidium | 39 | 23 | 42 | 9 |
| Strontium | 304 | 75 | 476 | 257 |
| Cadmium | 1179 | 1122 | 1947 | 2101 |
| Lead | 3247 | 6778 | 943 | 1408 |
Gene expression profiles in BEAS-2B cells exposed to PM10
To investigate the effects of PM in human lung cells, PMs collected in polypropylene filters were extracted and used to treat immortalized human bronchial epithelial BEAS-2B cells in vitro. One filter from each group was selected based on the metal concentration closest to the mean concentration. BEAS-2B cells were exposed to PM10 at various doses (0, 10, 25, 50, 100, and 200 μg/cm2) for 1, 2, 7 and 14 days, and cytotoxicity was measured by cell viability using colony formation assay. Surprisingly, cells exposed to PM10, even at the highest dose, did not exhibit any significant cytotoxicity (data not shown). It is possible that collected PM10 represents the normal daily exposure in a non-occupational setting, which may not be sufficient to reduce cell survival at the tested doses and time durations. Since microarray analysis is capable of detecting subtle changes in gene expression that occur at lower doses than those that trigger a significant phenotypic alteration, we performed a whole transcripts analysis using Affymetrix Human Gene 1.0 ST Array. The median dose (50 μg/cm2) of PM10 from the normal or storm group was chosen to treat BEAS-2B cells for either short-term (1 day) or longer-term (4 days) exposure. Two independent sets of untreated control cells and PM10 treated cells were harvested, and subjected to gene expression analysis.
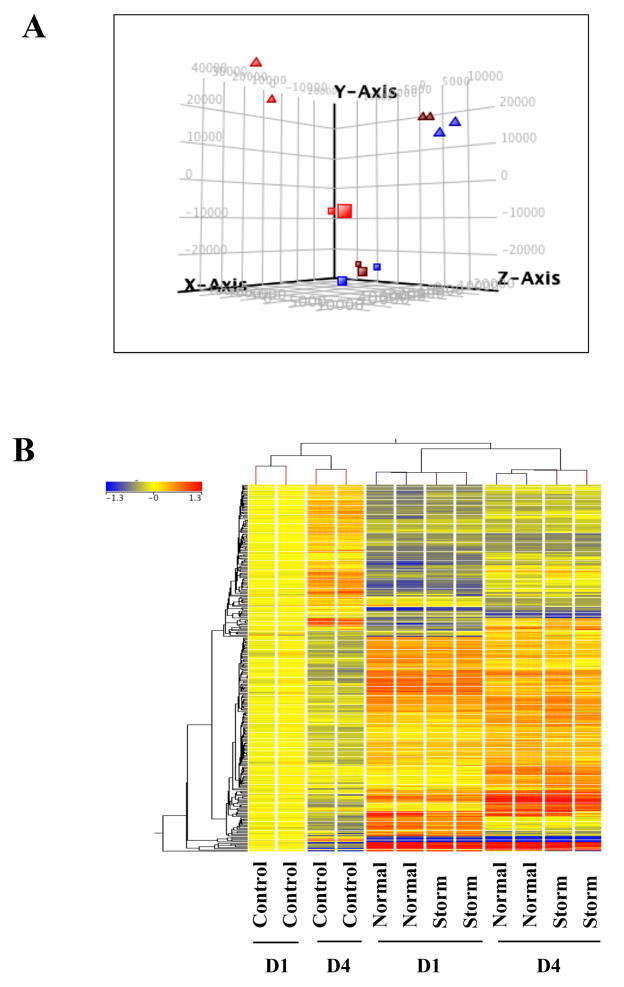
To explore the global impact of PM10 exposure on gene expression, we first performed a principal component analysis (PCA) to visualize the profile of all genes without any filtering. As shown in Figure 1A, the samples treated with PM10 were clearly separated from untreated control samples, indicating a detectable difference in the gene expression pattern between control and PM10 treated samples. In contrast, samples from the normal (normal air pollution in the absence of sand storm) group clustered closely to those from the storm group, suggesting a similar effect on gene expression profiles. Interestingly, samples from 1-day exposure (D1) were clustered separately from the 4-day exposure (D4) group, suggesting the impact of exposure duration on gene expression profiles. Similar effects can be seen in hierarchical clustering analysis of genes changed more than 1.5 fold (Figure 1B). Fold change analysis (one-way ANOVA, alpha level only) identified 140 and 251 entities that changed more than 1.5 fold in D1 and D4, respectively, from either the normal group or the storm group versus the untreated control group. It is important to note that the changed entities in normal versus control were almost identical to those in storm versus control, both in D1 and D4 (Supplemental Material, Figure 1A & 1B), further supporting the conclusion that PM10 from the normal or the storm group induced similar gene changes in cells (Figure 1). Thus, a common gene list shared between both groups was used to analyze the networks and pathways changed by PM10 exposure.
Figure 1.
Gene expression profiles of PM exposed cells. (A) Principal Components Analysis revealed distinct separation between control cells and PM exposed cells. Square: 1 day exposure; triangle: 4 day exposure; Red: control group; blue: normal group; brown: storm group. (B) Hierarchical cluster analysis of genes with more than 1.5-fold change in expression in one out of two groups (normal, storm) compared to untreated control cells. The bar relates the color code to the expression value determined after quantile normalization and baseline transformation to the median levels of control samples.
Gene expression profiles in BEAS-2B cells exposed to PM10 for 1 day
After removing unannotated and repeated entities, there was a total of 125 genes that significantly changed more than 1.5 fold in cells exposed to PM10 for 1 day compared to untreated control cells (p≤0.05). Among these genes, 58 were up-regulated, and 67 were down-regulated. Table 2 shows the top 15 up-regulated genes and top 15 down-regulated genes. Several genes known to be involved in the cell response to oxidative stress were significantly up-regulated, including HMOX1 (3.0), SLC7A11 (2.53), STC2 (2.01), SRXN1 (2.0), GCLM (1.89), and SQSTM1 (1.8). Interestingly, the genes related to TGF-β signaling, including ligand BMP4 (−2.06), transcription factor SMAD6 (−1.98), as well as targets ID1 (−1.81) and ID2 (−1.89), were all decreased. The full gene list containing 125 of changed genes is presented as Supplemental Material, Table 1.
Table 2.
Top 15 up- and down-regulated genes (p<0.05) in day 1
| Affymetrix Id | Genesymbol | Gene Name | Fold Changes | |
|---|---|---|---|---|
| Normal | Storm | |||
| 7951271 | MMP1 | matrix metallopeptidase 1 | 6.36 | 4.56 |
| 8021635 | SERPINB2 | serpin peptidase inhibitor, clade B, member 2 | 3.92 | 3.21 |
| 7909271 | IL24 | interleukin 24 | 3.50 | 3.43 |
| 8072678 | HMOX1 | heme oxygenase 1 | 3.03 | 2.23 |
| 8131844 | GPNMB | glycoprotein nmb | 2.81 | 2.83 |
| 8128123 | RRAGD | Ras-related GTP binding D | 2.68 | 2.69 |
| 8102800 | SLC7A11 | solute carrier family 7, member 11 | 2.53 | 1.65 |
| 8005475 | TRIM16L | tripartite motif-containing 16-like | 2.37 | 1.76 |
| 7942123 | CCND1 | cyclin D1 | 2.12 | 1.82 |
| 8084630 | LOC344887 | NmrA-like family domain containing 1 pseudogene | 2.08 | 1.35 |
| 8115851 | STC2 | stanniocalcin 2 | 2.04 | 1.87 |
| 8092578 | ETV5 | ets variant 5 | 2.03 | 1.65 |
| 8064375 | SRXN1 | sulfiredoxin 1 | 2.00 | 1.55 |
| 8093104 | TM4SF19 | transmembrane 4 L six family member 19 | 1.97 | 1.90 |
| 7964460 | DDIT3 | DNA-damage-inducible transcript 3 | 1.94 | 1.88 |
| 8059279 | EPHA4 | EPH receptor A4 | −1.81 | −1.57 |
| 8152512 | TNFRSF11B | tumor necrosis factor receptor superfamily, member 11b | −1.82 | −2.09 |
| 8131666 | ITGB8 | integrin, beta 8 | −1.83 | −1.61 |
| 8115099 | PDGFRB | platelet-derived growth factor receptor, beta polypeptide | −1.88 | −1.71 |
| 7945232 | ADAMTS15 | ADAM metallopeptidase with thrombospondin type 1 motif, 15 | −1.89 | −1.66 |
| 8040103 | ID2 | inhibitor of DNA binding 2 | −1.89 | −1.65 |
| 7984353 | SMAD6 | SMAD family member 6 | −1.98 | −1.87 |
| 8092169 | TNFSF10 | tumor necrosis factor superfamily, member 10 | −2.03 | −1.65 |
| 7979241 | BMP4 | bone morphogenetic protein 4 | −2.06 | −1.71 |
| 7976567 | BDKRB1 | bradykinin receptor B1 | −2.18 | −2.17 |
| 8101429 | PLAC8 | placenta-specific 8 | −2.18 | −1.98 |
| 8043244 | ATOH8 | atonal homolog 8 (Drosophila) | −2.26 | −1.90 |
| 8139488 | IGFBP3 | insulin-like growth factor binding protein 3 | −2.27 | −2.36 |
| 8146863 | SULF1 | sulfatase 1 | −2.28 | −2.19 |
| 7980485 | DIO2 | deiodinase, iodothyronine, type II | −4.19 | −3.61 |
To investigate the biological function of genes differentially modulated by PM10 exposure, the list of genes changed more than 1.5-fold was uploaded into the Ingenuity Pathway Analysis (IPA) tool. The top two biological function categories (ranked by p-value) are “cellular growth and proliferation” and “cell death”, suggesting that one of the early events in PM exposed cells is a change in cell growth property. Genes associated with top biological functions include ID1-3, CDKN2B, CCND1, etc.
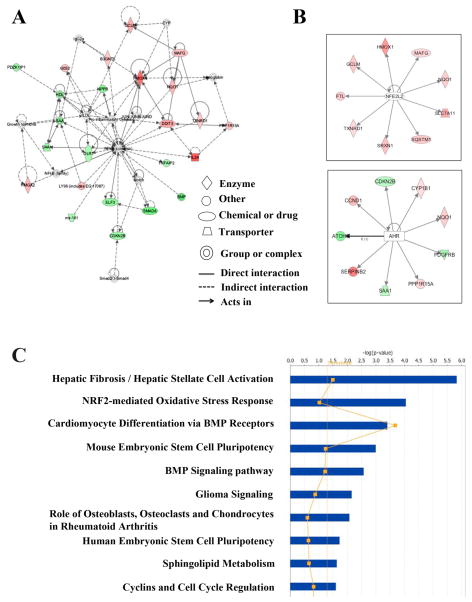
Gene network analysis of these differentially expressed genes revealed 16 significant networks. Among these networks, “cellular compromise, drug metabolism, cell death” were the top related network with 21 focus molecules and significance score of 43 (the negative log of p value) (Figure 2A). Other interesting networks are “gene expression, cellular function and maintenance, skeletal and muscular system development and function” (score: 34); “cell death, cellular development, hematological system development and function” (score: 23); “nutritional disease, cancer, organismal development” (score: 21); and “connective tissue disorders, inflammatory disease, cancer” (score: 21). The top five gene networks, along with their significance scores, number of genes involved, and names of differentially expressed genes, were listed as Supplemental material, Table 2.
Figure 2.
Top gene networks and canonical pathways related with genes changed more than 1.5-fold after 1-day exposure. (A) Top represented gene network identified by Ingenuity Pathway Analysis (IPA). Color of each node indicates the regulation of gene expression. Red: up-regulation; green: down-regulation. (B) Most active transcription factors in genes changed more than 1.5-fold after 1-day exposure. The color of each downstream target of NRF2 or AHR indicated the regulation of gene expression. (C) Top 10 canonical pathways identified by IPA from genes changed more than 1.5-fold (p≤0.05). Bars represent −log (p-value) for significance; orange line represent the ratio of changed genes in the total number of genes in the specific pathway.
In addition to the analysis of gene networks, we also analyzed the top canonical pathways associated with differentially expressed genes. Hepatic fibrosis pathway (p= 1.46 × 10−6) and NRF2-mediated oxidative stress response (p= 8.8 × 10−5) were the top two most represented canonical pathways (Figure 2C). Further analysis of genes associated with human diseases revealed that genes changed in 1-day exposure were linked to inflammation, connective tissue disorders and respiratory disease.
Other than biological function analysis, IPA also provided a useful tool, IPA-Tox, to identify the key functions and pathways that are changed upon treatment with toxic compounds and linked the changed gene expression profiles to clinical pathology endpoints. Interestingly, the top pathway identified in up-regulated genes was NRF2-mediated oxidative stress response (p= 9.85 × 10−6), which is consistent with the canonical pathway analysis. The top pathways identified in down-regulated genes were TGF-β signaling (p= 0.0046).
IPA’s newest transcription factor analysis represents a novel approach to predict the activation or inhibition of transcription factors based on the expression pattern of genes downstream of those factors. Although there is no significant change in mRNA level of NFE2L2 gene (encoding NRF2 protein), NRF2 was identified as the most activated transcription factor (Z-score: 3.124) in PM exposed cells after 1-day exposure. Based on the expression levels of downstream targets, the second most active transcription factor predicted was AHR (Z-score: 2.54), which is known to be involved in drug metabolism and cell detoxification (Figure 2B).
Gene ontology analysis with genes changed more than 1.5-fold after PM exposure identified “response to oxidative stress” as the highly represented biological process in up-regulated genes, which further support the results obtained from IPA analysis (Supplemental Material, Table 3). Moreover, KEGG pathway analysis revealed “TGF-β signaling pathway” (p-value: 9.2 × 10−8) was the most represented in down-regulated genes. A number of genes involved were down-regulated, including BMP4, NOG, SMAD9, CDKN2B, SMAD6, ID1-3.
Gene expression in BEAS-2B cells exposed to PM10 for 4 days
A total of 230 genes were significantly changed more than 1.5-fold in 4-day exposed cells compared to untreated control cells, including 147 up-regulated genes and 73 down-regulated genes. Table 3 showed the top 15 up- and down-regulated genes, and the complete list of genes changed more than 1.5-fold can be found in Supplemental Material, Table 4.
Table 3.
Top 15 up- and down-regulated genes (p<0.05) in day 4
| Affymetrix Id | Genesymbol | Gene Name | Fold Changes | |
|---|---|---|---|---|
| Normal | Storm | |||
| 7909271 | IL24 | interleukin 24 | 10.15 | 9.48 |
| 8021635 | SERPINB2 | serpin peptidase inhibitor, clade B, member 2 | 10.12 | 7.16 |
| 8131844 | GPNMB | glycoprotein nmb | 4.05 | 4.40 |
| 7925929 | AKR1C3 | aldo-keto reductase family 1, member C3 | 3.62 | 2.99 |
| 7929816 | SCD | stearoyl-CoA desaturase (delta-9- desaturase) | 3.20 | 3.71 |
| 8154381 | C9orf150 | chromosome 9 open reading frame 150 | 3.17 | 2.45 |
| 8051583 | CYP1B1 | cytochrome P450, family 1, subfamily B, polypeptide 1 | 3.12 | 3.37 |
| 8128123 | RRAGD | Ras-related GTP binding D | 3.10 | 3.19 |
| 8005475 | TRIM16L | tripartite motif-containing 16-like | 3.05 | 2.83 |
| 7951271 | MMP1 | matrix metallopeptidase 1 | 3.04 | 2.04 |
| 8137526 | INSIG1 | insulin induced gene 1 | 2.95 | 3.54 |
| 8152522 | ENPP2 | ectonucleotide pyrophosphatase/phosphodiesterase 2 | 2.83 | 2.62 |
| 8077899 | PPARG | peroxisome proliferator-activated receptor gamma | 2.79 | 2.45 |
| 8021301 | RAB27B | RAB27B, member RAS oncogene family | 2.55 | 2.88 |
| 8171435 | PIR | pirin (iron-binding nuclear protein) | 2.54 | 2.24 |
| 8083034 | CLSTN2 | calsyntenin 2 | −1.86 | −1.79 |
| 8102938 | RNF150 | ring finger protein 150 | −1.89 | −1.81 |
| 7916584 | TACSTD2 | tumor-associated calcium signal transducer 2 | −1.89 | −1.80 |
| 7909503 | SERTAD4 | SERTA domain containing 4 | −1.94 | −2.02 |
| 8095110 | KIT | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | −1.96 | −1.80 |
| 8045664 | LYPD6B | LY6/PLAUR domain containing 6B | −2.07 | −2.13 |
| 8139488 | IGFBP3 | insulin-like growth factor binding protein 3 | −2.11 | −2.52 |
| 8069689 | ADAMTS5 | ADAM metallopeptidase with thrombospondin type 1 motif, 5 | −2.14 | −2.10 |
| 8058765 | FN1 | fibronectin 1 | −2.15 | −2.18 |
| 7980485 | DIO2 | deiodinase, iodothyronine, type II | −2.24 | −2.97 |
| 7968417 | FRY | furry homolog (Drosophila) | −2.26 | −2.08 |
| 7912537 | DHRS3 | dehydrogenase/reductase member 3 | −2.62 | −2.60 |
| 7912520 | NPPB | natriuretic peptide B | −2.91 | −3.54 |
| 8146863 | SULF1 | sulfatase 1 | −4.19 | −3.89 |
| 8152512 | TNFRSF11B | tumor necrosis factor receptor superfamily, member 11b | −5.59 | −4.99 |
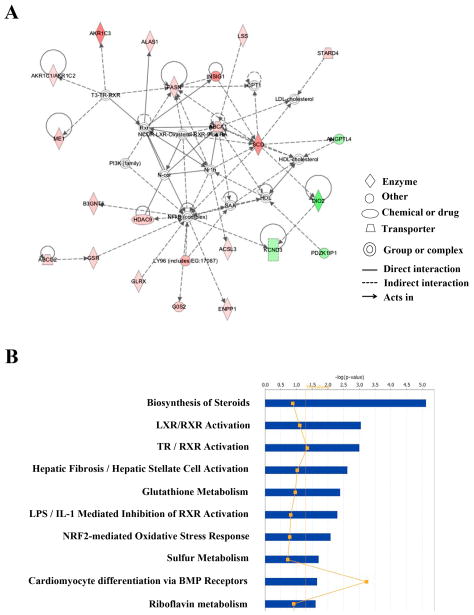
IPA analysis of differentially expressed genes revealed that the top two biological functions were “cellular movement” and “lipid metabolism”, suggesting a switch from an early response of cell growth or death to a later adaptive response including the changes involving mobility and metabolism. A total of 18 significant gene networks were associated with genes changed after a 4-day exposure. Among these networks, “lipid metabolism, small molecule biochemistry, vitamin and mineral metabolism” had the highest significant score (44) and 24 focus molecules (Figure 3A). Other interesting networks included “developmental disorder, hematological disease, immunological disease” (score: 42); “behavior, cellular development, hematological system development and function” (score: 41); “cellular growth and proliferation, tumor morphology, cardiovascular system development and function” (score: 32); and “cell morphology, nervous system development and function, cell death” (score: 30). Supplemental Material, Table 5 lists the top five gene networks, along with their significance scores, number of genes involved, and the names of differentially expressed genes.
Figure 3.
Top gene networks and canonical pathways related with genes changed more than 1.5-fold after 4-day exposure. (A) Top represented gene network identified by Ingenuity Pathway Analysis (IPA). Color of each node indicates the regulation of gene expression. Red: up-regulation; Green: down-regulation. (B) Top 10 canonical pathways identified by IPA from genes changed more than 1.5-fold (p≤0.05). Bars represent −log (p-value) for significance; orange line represent the ratio of changed genes in the total number of genes in the specific pathway.
The top represented canonical pathway revealed by IPA analysis was “biosynthesis of steroids” (p= 7.58 × 10−6), followed by LXR/RXR activation (p= 8.91 × 10−4) and TR/RXR activation (p= 9.94 × 10−4) (Figure 3B). The top human diseases associated with these changed genes included cancer, gastrointestinal disease and metabolic disease. While cholesterol biosynthesis (p= 1.28 × 10−7) was identified as the top pathway in IPA-Tox in up-regulated genes, hepatic fibrosis (p= 7.03 × 10−5) was the top Tox pathway in down-regulated genes. The top Tox functions of the changed gene expression were hepatocellular carcinoma (p= 4.08 × 10−4), renal tubule injury (p= 2.09 × 10−4), and liver stenosis (p= 0.00496). Consistent with pathway analysis, SREBF1 and SREBF2, the genes encoding the master regulators of lipid and cholesterol synthesis, were found to be the most active transcription factors in the analysis. As seen in Figure 4, a number of genes downstream of either SREBF1 or SREBF2, were up-regulated after 4 day PM10 exposure. Other active or inhibited transcription factors included NR1I2, NR1H3, SMAD4, MTPN, and CTNNB1.
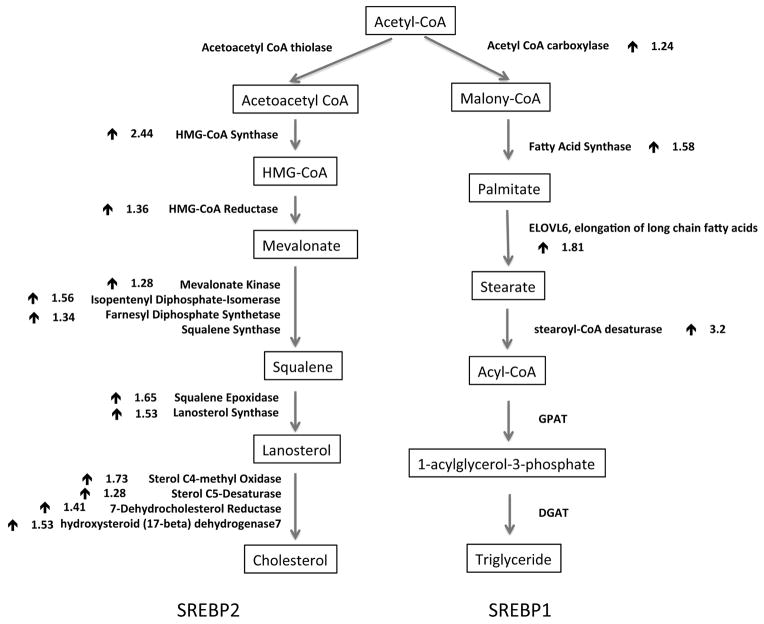
Figure 4.
Activation of genes involved in SREBPs mediated cholesterol and fatty acid synthesis in cells exposed to PM for 4 days. Downstream target genes of either SREBP2 (left) or SREBP1 (right) were listed in bold. The arrow (
 ) indicated the up-regulation of mRNA levels, and number next to the arrow represented the average fold changes from duplicate microarray results.
) indicated the up-regulation of mRNA levels, and number next to the arrow represented the average fold changes from duplicate microarray results.
Gene ontology analysis with differentially expressed genes confirmed the results of IPA analysis (Supplemental Material, Table 6). The top five represented biological processes in up-regulated genes were associated with sterol, cholesterol and lipid biosynthesis and metabolism. The KEGG pathway analysis revealed the steroid biosynthesis and PPAR signaling pathway as the top changed pathways in up-regulated genes after a 4-day exposure.
Effects of exposure duration on gene expression
Both IPA and Gene Ontological Analysis revealed distinct gene networks and pathways in D1 and D4 samples, suggesting an impact of exposure duration on gene expression changes and cellular responses. We then compared gene changes in D1 and D4 groups. As shown in Supplemental Material, Figure 1C, about 50% of the genes changed in D1, both up- and down-regulated, were also changed in D4. However, these commonly changed genes represented only 1/5 of up-regulated and 1/3 of down-regulated genes that were altered in D4. When the altered genes in both D1 and D4 were carefully examined, we found that more than 50% of gene expression correlated to the exposure duration. For example, SERPINB2, also known as Plasminogen activator inhibitor-2, increased about 3.9-fold at D1 but 10-fold in D4. TNFRSF11B is down-regulated about 1.8 fold after one day exposure but decreased about 5.6 fold after 4 day exposure. Thus, some genes exhibited exposure duration dependent expression changes.
Gene expression validation
To validate the gene expression changes observed in the microarray analysis, BEAS-2B cells were exposed to 50 μg/cm2 of PM10 from the normal or the storm group for 1 or 4 days. Total RNA were extracted from two independent sample sets, and subjected for quantitative real time PCR of selected genes. Gene fold changes were compared to those obtained from microarrays. HMOX1, IGFBP3, DDIT3 and SLC7A11 were chosen as genes significantly changed after 1-day exposure. SREBF1, SREBF2, HMGCS1, INAIG1, IDI1, CYP1B1 and IGFBP3 were analyzed for the 4-day exposure. Table 4 summarizes the average fold change of these selected genes in microarray analysis, as well as the corresponding real-time PCR results.
Table 4.
Real-time PCR validation of microarray results
| Day 1 (Fold change) | Microarray | qRT-PCRa | ||
|---|---|---|---|---|
| Normal | Storm | Normal | Storm | |
| HMOX1 | 3.03 | 2.23 | 7.22 | 4.8 |
| DDIT3 | 1.94 | 1.88 | 2.9 | 2.5 |
| SLC7A11 | 2.53 | 1.65 | 2.45 | 1.69 |
| IGFBP3 | 0.44 | 0.42 | 0.25 | 0.23 |
| Day 4 (Fold change) | Microarray | qRT-PCRa | ||
| Normal | Storm | Normal | Storm | |
| SREBF1 | 1.29 | 1.4 | 1.2 | 1.6 |
| SREBF2 | 1.21 | 1.24 | 1.16 | 1.33 |
| HMGCS1 | 2.44 | 2.59 | 3.1 | 3.9 |
| INSIG1 | 2.92 | 3.54 | 3.9 | 5.6 |
| IDl1 | 1.56 | 1.67 | 2.2 | 2.6 |
| CYP1B1 | 3.12 | 3.37 | 5.7 | 5.6 |
| IGFBP3 | 0.47 | 0.39 | 0.25 | 0.26 |
The RT-PCR data represent the means of triplicates from two independent experiments, and are presented as fold change to the level expressed in control BEAS-2B cells.
Discussion
Numerous epidemiological studies have established an association between high concentration of PM and pulmonary cardiovascular disease. Studies using either cultured cells or animal models uncovered that a series of early events, including oxidative stress, inflammation and cytokine production, were likely the mediators of PM induced adverse health problems (Akhtar et al. 2011; Brook et al. 2010; Chen and Lippmann 2009; Rückerl et al. 2011). However, the cellular and molecular mechanisms underlying PM induced adverse health effects are largely unknown. Our results indicated that PM10 exposure of cultured cells resulted in significant changes in the global gene expression profile. In addition, exposure duration had an important impact on the type of gene expression changes. While a short-term exposure (1 day) modulated genes involved in cell response to oxidative stress and drug metabolism, a more extended exposure (4 days) resulted in expression changes in genes involved in inflammation, cytokine production, and cell metabolism. Strikingly, a number of SREBPs downstream target genes that were important for cholesterol and triglyceride biosynthesis were significantly upregulated after 4-day exposure, suggesting a possible link between PM10 exposure and the dysregulation of cholesterol and fatty acid metabolism.
One of the earliest events following PM10 exposure was a cell protective response to oxidative stress that was mainly caused by ROS production (Ghio et al. 2012; Araujo and Nel 2009; Møller et al. 2010). Studies have shown that ROS can be generated by transition metals in the PM via a Fenton reaction or by cellular detoxification enzymes (Cho et al. 2005; Becker et al. 2005). When the levels of ROS are high, cells initiate a protective response by stabilizing and activating the transcription factor NRF2 (Araujo and Nel 2009; Diabaté et al. 2011). Once it has been activated, NRF2 translocates into the nucleus and induces the transcription of more than 200 downstream targets genes including antioxidants and detoxification enzymes (Malhotra et al. 2010). IPA analysis of transcription factors revealed that NRF2 was the most active transcription factor in cells exposed to PM10 for 1-day (Figure 2). Many NRF2 target genes were up-regulated (Table 2), including antioxidant enzymes HMOX1 (3.03), NQO1 (1.72), TXRND1 (1.5), SRXN1 (2.0), enzyme involved in glutathione synthesis GCLM (1.89), and cysteine-glutamate transporter SLC7A11 (2.53), indicating PM10 exposure induced oxidative stress and initiated antioxidant defense in BEAS-2B cells. Our results from 1-day exposure are consistent with several recent studies on PM induced gene expression changes (Akhtar et al. 2010; Dergham et al. 2012; Diabaté et al. 2011; Gualtieri et al. 2012; Huang et al. 2009; Huang et al. 2011; Murphy et al. 2008; Riechelmann et al. 2007; Ross et al. 2007; Watterson et al. 2007).
AHR was predicted by IPA as the second most active upstream regulator in BEAS-2B cells exposed to PM for 1-day (Figure 2), which is supported by increased levels of its downstream targets including both phase 1 (CYP1B1, 1.45) and phase II (NQO1, 1.73) detoxification enzymes. With an extended 4-day exposure, the levels of CYP1B1 and NQO1 continued to increase reaching 3.12 and 2.35, respectively, suggesting a time dependent response of AHR activation. As a major receptor for polycyclic aromatic hydrocarbons (PAHs), AHR and its downstream targets play an important role in mediating cell adaptation and detoxification in response to environmental PAHs. PM induced AHR activation and subsequent induction of detoxification enzymes have been reported in A549, BEAS-2B and other cell types (Courter et al. 2008; Dieme et al. 2012; Gualtieri et al. 2011; Gualtieri et al. 2012; Mahadevan et al. 2005).
Another interesting finding was the up-regulation of genes related to cholesterol and lipid biosynthesis in cells exposed to PM10 for 4 days. Both IPA and Gene Ontology analysis indicated that lipid metabolism, particularly cholesterol and sterol biosynthesis, were the most enriched gene network in cells exposed to PM10 for 4 days compared with untreated control cells. Consistent with functional analysis, pathways analysis revealed the biosynthesis of steroids as the most represented canonical pathway. Moreover, SREBF1 and SREBF2 genes were identified as the most active transcription factors in PM10 exposed cells based on the changes in their downstream target genes.
SREBF1 and SREBF2 genes encode three basic helix-loop-helix (bHLH) lucine zipper proteins: SREBP1a, SREBP1c and SREBP2, which are master regulators of cholesterol and fatty acid biosynthesis (Eberlé et al. 2004; Shimano 2009; Ye and DeBose-Boyd 2011). SREBPs are synthesized in their precursor forms and stored in the endoplasmic reticulum (ER), which normally forms a complex with the SREBP-cleavage activating protein (SCAP) that can function as a sterol sensor (Bengoechea-Alonso and Ericsson 2007). In sterol-depleted cells, SCAP transports SREBPs from the ER to the Golgi, where the mature form of SREBPs are released by proteolytic cleavage and subsequently translocated to the nucleus to stimulate downstream target genes. While SREBP1a and 1c preferentially regulate genes involved in triglyceride and fatty acid synthesis, SREBP2 activates genes that control cholesterol and sterol biosynthesis (Bengoechea-Alonso and Ericsson 2007; Horton et al. 2003).
Multiple lines of evidence suggested an activation of both SREBP1 and SREBP2 in cells exposed to PM10 for 4 days. First, a large number of genes that participated in almost every step of the cholesterol synthesis pathway were up-regulated in PM10 exposed cells compared to untreated control cells (Figure 5). Although the fold enrichment ranged between 1.28 and 2.44, the finding was quite striking with the activation of almost the entire pathway controlled by SREBP2. Second, the mRNA levels of several important enzymes involved in synthesis of monosaturated fatty acids from acetyl-CoA were increased in PM10 exposed cells, such as acetyl-CoA carboxylase, fatty acid synthase, long-chain fatty acyl elongase, and steroyl-CoA desaturase, which are known targets of SREBP1c (Horton et al. 2003). Moreover, the mRNA levels of three enzymes (Malic enzyme 1, G6PD, and PGDH) that are activated by both SREBP1 and SREBP2 and required for generating NADPH from various sources were increased in PM treated cells. Last and most important, the mRNA levels of SREBF1 and SREBF2 were also induced by PM exposure (Table 6, Supplemental Material, Table 3). Taken together, BEAS-2B cells following 4 days of exposure to PM10 showed increased gene expression and enhanced transcriptional activity of SREBPs.
It is not clear how PM10 activated SREBPs in BEAS-2B cells. Similar to many other transcription factors, SREBPs can be regulated at two levels: transcriptional and posttranslational. One important transcriptional regulator of SREBPs is liver X-activated receptor (LXR) (Eberlé et al. 2004; Raghow et al. 2008). LXRs form heterodimers with retinoid X receptor and bind to SREBPs promoter to activate transcription, and thus play an important role in cholesterol and fatty acid metabolism. Recent studies reported a novel role of LXR in protecting the lung from injury by activating antioxidant enzymes and inhibiting inflammatory gene expression (Birrell et al. 2007; Gong et al. 2009). Interestingly, the IPA pathway analysis identified LXR activation as the second most represented canonical pathway in PM exposed BEAS-2B cells (Figure 4b), suggesting a potential role of LXR in transcriptional regulation of SREBPs in these cells. Posttranslational regulation of SREBPs involving two rounds of proteolytic cleavage occurred in the ER-Golgi. ER stress and activation of the unfolded protein response (UFP) have been associated with dysregulation of SREBPs activation and cholesterol homeostasis (Colgan et al. 2011). Recently, several studies reported that airborne PM induced ER stress and UFP in human lung cells (Watterson et al. 2009; Laing et al. 2010). Our studies also showed increased levels of mRNA for Hsp70, Hsp90, PPP1R15A and DDIT3, suggesting a possible response to ER stress in PM10 exposed cells, which might account for the activation of SREBPs. Lastly, as the members of bHLH transcription factor family, the transcriptional activity of SREBPs can be modulated by a group of negative regulator, Ids (Moldes et al. 1999; Rahmouni and Sigmund 2008). Three members of Id proteins, Id1-3, were significantly downregulated in PM exposed BEAS-2B cells, which may have resulted in enhanced SREBPs transcriptional activity in these cells.
We do not know whether PM10 induced SREBPs activation could result in enhanced cholesterogenesis and lipogenesis, since we studied bronchial epithelial cells, while the liver is the site for much of the body’s cholesterogenesis and lipogenesis. It is worth noting that SREBPs are normally expressed at a low level in lung cells that require lipogenesis to maintain surfactant. Deregulation of lipogenesis has been reported to cause lung damage (Plantier et al. 2012; Besnard et al. 2009). In addition, BEAS-2B cells may provide a better representation of the in vivo airway epithelial when they are cultured in vitro at the air-liquid interface. It is not clear whether the results from our study represented the changes of gene and pathway that occurred in vivo. Further profiling of gene expression changes and analyzing the cholesterol and triglyceride levels in vivo following long-term PM exposure will address these questions. In summary, our study is the first to report that PM10 exposure can modulate genes related to cholesterol and lipid metabolism, providing a new insight into the mechanisms underlying PM induced cardiovascular disease.
Supplementary Material
Acknowledgments
We would like to thank NYU Cancer Institute Genomics Facility for microarray hybridization and scanning. This work was funded by King Abdulaziz University (KAU), Jeddah, under grant number 4/00/00/252.
References
- Akhtar US, Scott JA, Chu A, Evans GJ. In vivo and In vitro Assessment of Particulate Matter Toxicology. Urban Airborne Particulate Matter. 2011:427–449. [Google Scholar]
- Akhtar US, McWhinney RD, Rastogi N, Abbatt JPD, Evans GJ, Scott JA. Cytotoxic and proinflammatory effects of ambient and source-related particulate matter (PM) in relation to the production of reactive oxygen species (ROS) and cytokine adsorption by particles. Inhal Toxicol. 2010;22(Suppl 2):37–47. doi: 10.3109/08958378.2010.518377. [DOI] [PubMed] [Google Scholar]
- Araujo JA, Nel AE. Particulate matter and atherosclerosis: role of particle size, composition and oxidative stress. Part Fibre Toxicol. 2009;6:24. doi: 10.1186/1743-8977-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker S, Dailey LA, Soukup JM, Grambow SC, Devlin RB, Huang YCT. Seasonal Variations in Air Pollution Particle-Induced Inflammatory Mediator Release and Oxidative Stress. Environ Health Perspect. 2005;113:1032–1038. doi: 10.1289/ehp.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell ML, Davis DL. Reassessment of the lethal London fog of 1952: novel indicators of acute and chronic consequences of acute exposure to air pollution. Environ Health Perspect. 2001;109(Suppl 3):389–394. doi: 10.1289/ehp.01109s3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengoechea-Alonso MT, Ericsson J. SREBP in signal transduction: cholesterol metabolism and beyond. Curr Opin Cell Biol. 2007;19:215–222. doi: 10.1016/j.ceb.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Besnard V, Wert SE, Stahlman MT, Postle AD, Xu Y, Ikegami M, Whitsett JA. Deletion of Scap in alveolar type II cells influences lung lipid homeostasis and identifies a compensatory role for pulmonary lipofibroblasts. J Biol Chem. 2009;284:4018–4030. doi: 10.1074/jbc.M805388200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birrell MA, Catley MC, Hardaker E, Wong S, Willson TM, McCluskie K, Leonard T, Farrow SN, Collins JL, Haj-Yahia S, Belvisi MG. Novel role for the liver X nuclear receptor in the suppression of lung inflammatory responses. J Biol Chem. 2007;282:31882–31890. doi: 10.1074/jbc.M703278200. [DOI] [PubMed] [Google Scholar]
- Brook RD, Rajagopalan S, Pope CA, Brook JR, Bhatnagar A, Diez-Roux AV, Holguin F, Hong Y, Luepker RV, Mittleman MA, Peters A, Siscovick D, Smith SC, Whitsel L, Kaufman JD American Heart Association Council on Epidemiology and Prevention, Council on the Kidney in Cardiovascular Disease, and Council on Nutrition, Physical Activity and Metabolism. . Particulate matter air pollution and cardiovascular disease: An update to the scientific statement from the American Heart Association. Circulation. 2010;121:2331–2378. doi: 10.1161/CIR.0b013e3181dbece1. [DOI] [PubMed] [Google Scholar]
- Chen LC, Lippmann M. Effects of metals within ambient air particulate matter (PM) on human health. Inhal Toxicol. 2009;21:1–31. doi: 10.1080/08958370802105405. [DOI] [PubMed] [Google Scholar]
- Cho AK, Sioutas C, Miguel AH, Kumagai Y, Schmitz DA, Singh M, Eiguren-Fernandez A, Froines JR. Redox activity of airborne particulate matter at different sites in the Los Angeles Basin. Environ Res. 2005;99:40–47. doi: 10.1016/j.envres.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Colgan SM, Hashimi AA, Austin RC. Endoplasmic reticulum stress and lipid dysregulation. Expert Rev Mol Med. 2011;13:e4. doi: 10.1017/S1462399410001742. [DOI] [PubMed] [Google Scholar]
- Courter LA, Luch A, Musafia-Jeknic T, Arlt VM, Fischer K, Bildfell R, Pereira C, Phillips DH, Poirier MC, Baird WM. The influence of diesel exhaust on polycyclic aromatic hydrocarbon-induced DNA damage, gene expression, and tumor initiation in Sencar mice in vivo. Cancer Lett. 2008;265:135–147. doi: 10.1016/j.canlet.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dergham M, Lepers C, Verdin A, Billet S, Cazier F, Courcot D, Shirali P, Garçon G. Prooxidant and Proinflammatory Potency of Air Pollution Particulate Matter (PM(2.5-0.3)) Produced in Rural, Urban, or Industrial Surroundings in Human Bronchial Epithelial Cells (BEAS-2B) Chem Res Toxicol. 2012;25:904–919. doi: 10.1021/tx200529v. [DOI] [PubMed] [Google Scholar]
- Diabaté S, Bergfeldt B, Plaumann D, Ubel C, Weiss C. Anti-oxidative and inflammatory responses induced by fly ash particles and carbon black in lung epithelial cells. Anal Bioanal Chem. 2011;401:3197–3212. doi: 10.1007/s00216-011-5102-4. [DOI] [PubMed] [Google Scholar]
- Dieme D, Cabral-Ndior M, Garçon G, Verdin A, Billet S, Cazier F, Courcot D, Diouf A, Shirali P. Relationship between physicochemical characterization and toxicity of fine particulate matter (PM2.5) collected in Dakar city (Senegal) Environ Res. 2012;113:1–13. doi: 10.1016/j.envres.2011.11.009. [DOI] [PubMed] [Google Scholar]
- Dockery DW, Pope CA, Xu X, Spengler JD, Ware JH, Fay ME, Ferris BG, Speizer FE. An association between air pollution and mortality in six U.S. cities. N Engl J Med. 1993;329:1753–1759. doi: 10.1056/NEJM199312093292401. [DOI] [PubMed] [Google Scholar]
- Duvall RM, Norris GA, Dailey LA, Burke JM, McGee JK, Gilmour MI, Gordon T, Devlin RB. Source apportionment of particulate matter in the U.S. and associations with lung inflammatory markers. Inhal Toxicol. 2008;20:671–683. doi: 10.1080/08958370801935117. [DOI] [PubMed] [Google Scholar]
- Eberlé D, Hegarty B, Bossard P, Ferré P, Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004;86:839–848. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Ghio AJ, Carraway MS, Madden MC. Composition of air pollution particles and oxidative stress in cells, tissues, and living systems. J Toxicol Environ Health B Crit Rev. 2012;15:1–21. doi: 10.1080/10937404.2012.632359. [DOI] [PubMed] [Google Scholar]
- Gong H, He J, Lee JH, Mallick E, Gao X, Li S, Homanics GE, Xie W. Activation of the liver X receptor prevents lipopolysaccharide-induced lung injury. J Biol Chem. 2009;284:30113–30121. doi: 10.1074/jbc.M109.047753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon T. Linking health effects to PM components, size, and sources. Inhal Toxicol. 2007;19(Suppl 1):3–6. doi: 10.1080/08958370701490312. [DOI] [PubMed] [Google Scholar]
- Gualtieri M, Ovrevik J, Mollerup S, Asare N, Longhin E, Dahlman HJ, Camatini M, Holme JA. Airborne urban particles (Milan winter-PM2.5) cause mitotic arrest and cell death: Effects on DNA, mitochondria, AhR binding and spindle organization. Mutat Res. 2011;713:18–31. doi: 10.1016/j.mrfmmm.2011.05.011. [DOI] [PubMed] [Google Scholar]
- Gualtieri M, Longhin E, Mattioli M, Mantecca P, Tinaglia V, Mangano E, Proverbio MC, Bestetti G, Camatini M, Battaglia C. Gene expression profiling of A549 cells exposed to Milan PM2.5. Toxicol Lett. 2012;209:136–145. doi: 10.1016/j.toxlet.2011.11.015. [DOI] [PubMed] [Google Scholar]
- Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proceedings of the National Academy of Sciences. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YC, Karoly ED, Dailey LA, Schmitt MT, Silbajoris R, Graff DW, Devlin RB. Comparison of gene expression profiles induced by coarse, fine, and ultrafine particulate matter. J Toxicol Environ Health A. 2011;74:296–312. doi: 10.1080/15287394.2010.516238. [DOI] [PubMed] [Google Scholar]
- Huang YC, Li Z, Carter JD, Soukup JM, Schwartz DA, Yang IV. Fine ambient particles induce oxidative stress and metal binding genes in human alveolar macrophages. Am J Respir Cell Mol Biol. 2009;41:544–552. doi: 10.1165/rcmb.2008-0064OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laden F, Neas LM, Dockery DW, Schwartz J. Association of fine particulate matter from different sources with daily mortality in six U.S. cities. Environ Health Perspect. 2000;108:941–947. doi: 10.1289/ehp.00108941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing S, Wang G, Briazova T, Zhang C, Wang A, Zheng Z, Gow A, Chen AF, Rajagopalan S, Chen LC, Sun Q, Zhang K. Airborne particulate matter selectively activates endoplasmic reticulum stress response in the lung and liver tissues. Am J Physiol Cell Physiol. 2010;299:C736–C749. doi: 10.1152/ajpcell.00529.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejczyk P, Chen LC. Effects of subchronic exposures to concentrated ambient particles (CAPs) in mice. VIII. Source-related daily variations in in vitro responses to CAPs. Inhal Toxicol. 2005;17:243–253. doi: 10.1080/08958370590912914. [DOI] [PubMed] [Google Scholar]
- Mahadevan B, Keshava C, Musafia-Jeknic T, Pecaj A, Weston A, Baird WM. Altered gene expression patterns in MCF-7 cells induced by the urban dust particulate complex mixture standard reference material 1649a. Cancer Res. 2005;65:1251–1258. doi: 10.1158/0008-5472.CAN-04-2357. [DOI] [PubMed] [Google Scholar]
- Malhotra D, Portales-Casamar E, Singh A, Srivastava S, Arenillas D, Happel C, Shyr C, Wakabayashi N, Kensler TW, Wasserman WW, Biswal S. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38:5718–5734. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldes M, Boizard M, Liepvre XL, Fève B, Dugail I, Pairault J. Functional antagonism between inhibitor of DNA binding (Id) and adipocyte determination and differentiation factor 1/sterol regulatory element-binding protein-1c (ADD1/SREBP-1c) trans-factors for the regulation of fatty acid synthase promoter in adipocytes. Biochem J. 1999;344(Pt 3):873–880. [PMC free article] [PubMed] [Google Scholar]
- Murphy G, Rouse RL, Polk WW, Henk WG, Barker SA, Boudreaux MJ, Floyd ZE, Penn AL. Combustion-derived hydrocarbons localize to lipid droplets in respiratory cells. Am J Respir Cell Mol Biol. 2008;38:532–540. doi: 10.1165/rcmb.2007-0204OC. [DOI] [PubMed] [Google Scholar]
- Møller P, Jacobsen NR, Folkmann JK, Danielsen PH, Mikkelsen L, Hemmingsen JG, Vesterdal LK, Forchhammer L, Wallin H, Loft S. Role of oxidative damage in toxicity of particulates. Free Radic Res. 2010;44:1–46. doi: 10.3109/10715760903300691. [DOI] [PubMed] [Google Scholar]
- Nouh MS. Is the desert lung syndrome (nonoccupational dust pneumoconiosis) a variant of pulmonary alveolar microlithiasis? Report of 4 cases with review of the literature. Respiration. 1989;55:122–126. doi: 10.1159/000195715. [DOI] [PubMed] [Google Scholar]
- Plantier L, Besnard V, Xu Y, Ikegami M, Wert SE, Hunt AN, Postle AD, Whitsett JA. Activation of sterol-response element-binding proteins (SREBP) in alveolar type II cells enhances lipogenesis causing pulmonary lipotoxicity. J Biol Chem. 2012;287:10099–10114. doi: 10.1074/jbc.M111.303669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope CA. Respiratory disease associated with community air pollution and a steel mill, Utah Valley. Am J Public Health. 1989;79:623–628. doi: 10.2105/ajph.79.5.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghow R, Yellaturu C, Deng X, Park EA, Elam MB. SREBPs: the crossroads of physiological and pathological lipid homeostasis. Trends Endocrinol Metab. 2008;19:65–73. doi: 10.1016/j.tem.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Rahmouni K, Sigmund CD. Id3, E47, and SREBP-1c: fat factors controlling adiponectin expression. Circ Res. 2008;103:565–567. doi: 10.1161/CIRCRESAHA.108.184366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechelmann H, Deutschle T, Grabow A, Heinzow B, Butte W, Reiter R. Differential response of Mono Mac 6, BEAS-2B, and Jurkat cells to indoor dust. Environ Health Perspect. 2007;115:1325–1332. doi: 10.1289/ehp.9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AJ, Dailey LA, Brighton LE, Devlin RB. Transcriptional profiling of mucociliary differentiation in human airway epithelial cells. Am J Respir Cell Mol Biol. 2007;37:169–185. doi: 10.1165/rcmb.2006-0466OC. [DOI] [PubMed] [Google Scholar]
- Rükerl R, Schneider A, Breitner S, Cyrys J, Peters A. Health effects of particulate air pollution: A review of epidemiological evidence. Inhal Toxicol. 2011;23:555–592. doi: 10.3109/08958378.2011.593587. [DOI] [PubMed] [Google Scholar]
- Schwartz J, Marcus A. Mortality and air pollution in London: a time series analysis. Am J Epidemiol. 1990;131:185–194. doi: 10.1093/oxfordjournals.aje.a115473. [DOI] [PubMed] [Google Scholar]
- Schwarze PE, Ovrevik J, Låg M, Refsnes M, Nafstad P, Hetland RB, Dybing E. Particulate matter properties and health effects: consistency of epidemiological and toxicological studies. Hum Exp Toxicol. 2006;25:559–579. doi: 10.1177/096032706072520. [DOI] [PubMed] [Google Scholar]
- Shimano H. SREBPs: physiology and pathophysiology of the SREBP family. FEBS J. 2009;276:616–621. doi: 10.1111/j.1742-4658.2008.06806.x. [DOI] [PubMed] [Google Scholar]
- Simkhovich BZ, Kleinman MT, Kloner RA. Air pollution and cardiovascular injury epidemiology, toxicology, and mechanisms. J Am Coll Cardiol. 2008;52:719–726. doi: 10.1016/j.jacc.2008.05.029. [DOI] [PubMed] [Google Scholar]
- Waness A, El-Sameed YA, Mahboub B, Noshi M, Al-Jahdali H, Vats M, Mehta AC. Respiratory disorders in the Middle East: a review. Respirology. 2011;16:755–766. doi: 10.1111/j.1440-1843.2011.01988.x. [DOI] [PubMed] [Google Scholar]
- Watterson TL, Hamilton B, Martin R, Coulombe RA. Urban particulate matter causes ER stress and the unfolded protein response in human lung cells. Toxicol Sci. 2009;112:111–122. doi: 10.1093/toxsci/kfp186. [DOI] [PubMed] [Google Scholar]
- Watterson TL, Sorensen J, Martin R, Coulombe RA. Effects of PM2.5 collected from Cache Valley Utah on genes associated with the inflammatory response in human lung cells. J Toxicol Environ Health A. 2007;70:1731–1744. doi: 10.1080/15287390701457746. [DOI] [PubMed] [Google Scholar]
- Ye J, DeBose-Boyd RA. Regulation of cholesterol and fatty acid synthesis. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanobetti A, Schwartz J. Particulate air pollution, progression, and survival after myocardial infarction. Environ Health Perspect. 2007;115:769–775. doi: 10.1289/ehp.9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.