Abstract
Why do populations remain genetically variable despite strong continuous natural selection? Mutation reconstitutes variation eliminated by selection and genetic drift, but theoretical and experimental studies each suggest that mutation-selection balance insufficient to explain extant genetic variation in most complex traits. The alternative hypothesis of balancing selection, wherein selection maintains genetic variation, is an aggregate of multiple mechanisms (spatial and temporal heterogeneity in selection, frequency-dependent selection, antagonistic pleiotropy, etc.). Most of these mechanisms have been demonstrated for Mendelian traits, but there is little comparable data for loci affecting quantitative characters. Here, we report a three-year field study of selection on intra-population Quantitative Trait Loci (QTL) of flower size, a highly polygenic trait of Mimulus guttatus. The QTL exhibit antagonistic pleiotropy: alleles that increase flower size reduce viability but increase fecundity. The magnitude and direction of selection fluctuates yearly and on a spatial scale of meters. This study provides direct evidence of balancing selection mechanisms on QTL of an ecologically relevant trait.
Keywords: balancing selection, Mimulus guttatus, pleiotropy, QTL
Introduction
Two broad classes of explanations are typically invoked to explain how quantitative genetic variation is maintained, mutation-selection balance and balancing selection. With mutation-selection balance, selection acts to eliminate variation that is continually reconstituted by mutation. This includes models where quantitative trait variation is the pleiotropic consequence of recurrent, unconditionally deleterious mutations (Barton 1990; Keightley & Hill 1990; Kondrashov & Turelli 1992; Mackay et al. 1992; Caballero & Keightley 1994), as well as ‘conditional models’ in which mutations are deleterious only under present environmental circumstances and genetic backgrounds (Lande 1975; Turelli 1984). In contrast, balancing selection mechanisms actively maintain genetic variation through a variety of selective processes (Clausen et al. 1940; Rose 1982; Gillespie & Turelli 1989). Simple overdominance (heterozygote superiority across environments) is the standard example, but other mechanisms are probably more important for polygenic traits. These include heterogeneous selection (fitness differs in time and space), genotype by environment interaction (genotypes differing in the environment-dependent expression), antagonistic pleiotropy (negative genetic correlations between fitness components), conflicting levels of selection (e.g. gametic versus zygotic selection) and frequency dependent selection (fitness is a function of genotype frequencies).
Phenotypic methods, such as selection gradient studies (Kingsolver et al. 2001), provide some information relevant to the question of variation. For example, changes in the direction and magnitude of phenotypic selection are clearly consistent with balancing selection models based on environmental heterogeneity. However, fluctuating selection on the phenotype does not imply fluctuating selection on alternative alleles, or at least not that the rank order of fitness of alleles changes with environment. Genotypic or quantitative genetic experiments, in which phenotypes are associated with a clone or family, are more informative. Associations of genotypic values with fitness measurements are less prone to bias (Rausher 1992; Mojica & Kelly 2010) and can directly test predictions of variation models. For example, antagonistic pleiotropy models predict a negative genetic correlation between fitness components (Radwan 2008). Unfortunately, it can be difficult to detect such correlations given that the overall covariance between fitness components is an aggregate of contributions of many different genetic and environmental factors (Fry 1993). These difficulties can be overcome by measuring fitness variation at the scale of individual QTL.
A small but growing collection of field experiments have successfully demonstrated natural selection at the scale of QTL (e.g. Bradshaw & Schemske 2003; Lexer et al. 2003; Li et al. 2003; Weinig et al. 2003; Verhoeven et al. 2004; Korves et al. 2007). The evolutionary interpretation of results from these studies is contingent on the source of genetic materials and on the field site (or sites) in which genotypes are evaluated. Questions about the maintenance of local variation are most directly addressed by considering intra-population variation. However, field QTL experiments have focused mainly on recombinant genotypes between divergent populations or species, e.g. Mimulus (Bradshaw & Schemske 2003; Hall & Willis 2006), Helianthus (Lexer et al. 2003), Avena (Gardner & Latta 2006), Hordeum (Verhoeven et al. 2004), and Boechera (Anderson et al. 2011). While these data are clearly relevant to the question of variation, it is an open question as to whether QTL distinguishing populations are of the same character as intra-population QTL. A particular concern is that intra-population QTL may have smaller effects than inter-population QTL. Indeed, field studies of intra-population variation where selection is measured at the scale of genetic loci are largely limited to Mendelian traits (e.g. Dobzhansky & Levene 1948; Ford 1971; Greaves et al. 1977; Subramaniam & Rausher 2000).
Flower size is a genuine quantitative trait within the Iron Mountain (IM) population of Mimulus guttatus (IM is located in Oregon, USA). Variation in this trait is caused by environmental effects combined with many QTL (Lee 2009) and flower size is under strong natural selection at IM (Willis 1996; Mojica & Kelly 2010). Despite this, abundant additive genetic variation persists and the quantity and nature of this variation cannot be explained by mutation-selection balance (Kelly & Willis 2001; Kelly 2003). Balancing selection emerges as a viable alternative explanation, but what specific form or forms of balancing selection are important? Antagonistic pleiotropy is a viable explanation. Fecundity is positively related to flower size because larger flowers produce more ovules and pollen and might be more attractive to pollinators (Mojica & Kelly 2010). However, survival to flowering is much lower in large flowered genotypes than in small flowered genotypes because they fail to flower before the onset of summer drought. Unfortunately, it is unclear if this viability/fecundity trade-off is maintaining polymorphism because the phenotypic variation in our previous field study (Mojica & Kelly 2010) was due to the aggregate effects of many, unknown QTL.
In this paper, we describe results from a three-year field study measuring phenotypic and fitness effects of two intra-population QTL of M. guttatus. We first ask whether the phenotypic effects of these QTL, first demonstrated in the greenhouse, are reiterated in the field. Do intra-population QTL have measurable fitness effects in nature, and if so, are patterns of selection consistent with the maintenance of polymorphism? Is the trade-off between survival to flowering and fecundity (antagonistic pleiotropy) evident for flower size QTL? Is QTL specific selection variable among microsites within the population or between years of the experiment, i. e. is environmental heterogeneity a potential mechanism maintaining variation?
Materials and methods
Overview
We measured survivorship and fecundity effects of QTL by transplanting and monitoring four nearly-isogenic genotypes in the field. These genotypes were created by introgressing alleles that increase flower size at two unlinked QTL into the uniform genetic background of a single inbred line, IM62. The IM62 allele is labeled L (low) while the alternative is H (high) at each locus. Here, L and H refer to effects on corolla width, although these alleles affect multiple traits. To avoid the effects of inbreeding depression, we produced hybrid isogenic genotypes by crossing the four IM62 genotypes to a different inbred line from the same population, IM767. We designate the IM767 alleles X1 and X8, where the subscript is the linkage group of that QTL. The X allele at each QTL may be functionally equivalent to one of the alternatives (L or H) but not to both, and it may differ from both. For brevity, the four genotypes H1H8/X1X8, L1L8/ X1X8, H1L8/ X1X8, and L1H8/ X1X8, are denoted HH, LL, HL, and LH, respectively (the first letter denotes allele at the QTL on linkage group 1). Experimental genotypes were transplanted as seedlings into the Oregon field site in three successive years (2008-2010); HH and LL in each year, HL in two seasons (2009-2010) and LH only in 2010. The absences of HL in 2008 and LH in 2008-2009 were due to the delays in producing these genotypes within a clean genetic background (see next section).
QTL isolation
QTL 1 and QTL 8 were mapped in two distinct but related experiments. We first created two phenotypically divergent populations through six generations of bi-directional artificial selection on corolla width (Kelly 2008; Lee 2009). The selection experiment was founded by a large collection of genotypes sampled from Iron Mountain (Oregon, USA). After six generations of selection, three large flowered plants were selected from the high population and each was crossed to a distinct small flowered plant from the low population. Each of the three F1s was self-fertilized to produce three F2 populations where individuals measured for phenotype and genotyped at markers across the genome. QTL 1 and QTL 8 were mapped in this replicated F2 study (Lee 2009) where alternative QTL genotypes were assayed in a broad range of genetic backgrounds.
We used Recurrent Selection with Backcrossing to produce Nearly Isogenic Lines (NILs) containing QTL 1 and QTL 8, (Figure 1; Wright 1952; Hill 1998). F1 plants from the three crosses described above were backcrossed (BC) for four generations to IM62. Concurrent with backcrossing, we performed selection for flower size in each generation, thereby enriching the final set of NILs for QTL with consistent effects on corolla width in the IM62 genetic background. QTL 1 and QTL 8 were among the flower size loci to be fine mapped in this procedure (see Lee 2009 and the Electronic Supplementary Materials of Scoville et al 2011 for a full description of the Recurrent Selection with Backcrossing experiment). The genotypes used in the field study were descended from a single BC4F1 plant that was heterozygous at both QTLs and for three residual regions on other linkage groups (LG4, LG7, and LG13). This BC4F1 plant was selfed to produce a large collection of BC4F2 seeds. BC4F2 plants were grown and genotyped in numerous subsequent experiments over several years (see below) to identify plants that were homozygous for H alleles at the QTL with relatively clean backgrounds (where residually heterozygous regions had segregated to homozygous for the IM62 allele). Measurements of flower size in these experiments also confirmed that none of the residual regions contained flower size QTL.
Fig. 1.
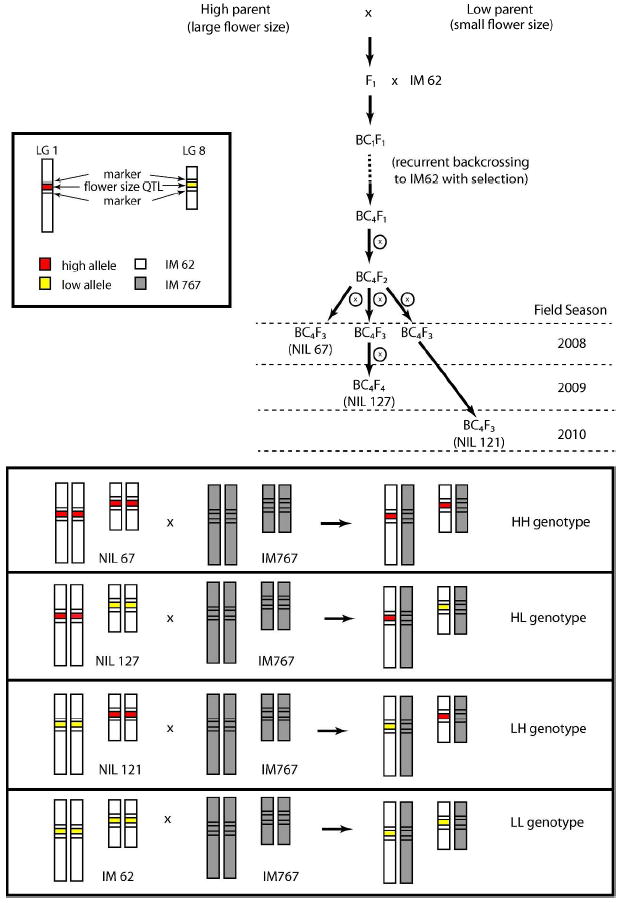
The breeding design to produce genotypes for field transplant: After four generations of recurrent selection for flower size with backcrossing to IM62, we selfed (symbol: circled ×) a single BC4F1 plant producing a family of BC4F2 progenies. Among the selfed BC4F2, we identified a NIL (BC4F3) fixed for both high alleles at QTLs 1 and 8 (NIL 67) and another NIL that is fixed for the low allele at QTL 1 and high allele at QTL 8 (NIL 121). We further selfed selected BC4F3 individuals and identified a NIL homozygous at both high and low alleles on QTL 1 and QTL 8, respectively (NIL 127). These NILs were then crossed to IM767 and genotyped to verify the HH, HL and LH genotype. The cumulative length of each arrow leading to each NIL reflects the relative time it took to identify a specific NIL with a clean background.
The HH, LH, and HL genotypes were derived from three distinct plants in the BC4F2 progeny set (Figure 1). LL was generated by crossing IM767 to IM62. In 2008, we planted descendants of NIL 67 which had broad introgressions homozygous for H alleles at QTL 1 and 8. NIL 67 was homozygous IM62 for the LG4 and LG13 residual regions, but homozygous donor for the LG 7 region (not a flower size QTL). After the 2008 field season, we fine-mapped both QTLs in independent NIL lines to identify flanking markers. By 2009 we had confirmed effects on flower size and identified flanking markers (MgSTS781 and MgSTS271) for QTL 1 in the descendants of NIL 127. These markers span approximately 1 Mbp on the current draft genome sequence for M. guttatus. Using those flanking markers, we generated a fixed NIL line with a completely clean background for QTL 1 (LG4, LG7, and LG13 regions homozygous for IM62). By 2010, we had also identified flanking markers also for QTL 8 (yw71 and MgSTS59 span 180 kbp). Using those flanking markers, we identified NIL 121 from the original selfed BC4F2 population as homozygous for the H allele at QTL 8. The donor allele at the LG4 residual region was heterozygous in NIL 121 and hence may have been segregating in the field population, but again, this region does not affect flower size.
Field Transplant
Seeds were sown in replicated (four per genotype) and randomized 53×27×6 cm seeding flats for two weeks prior to transplanting. Seedlings were transplanted (n2008 = 400, n2009= 900, n2010= 1600) into five-meter transects at the Browder Ridge site (Oregon, USA; 44°220 7.319400 N, 122°20 57.8400 W) in June of each year. This site is within 10 km of Iron Mountain and is similar in habitat. Transplants were done at Browder Ridge to avoid contamination of the source population. At both sites, M. guttatus is annual and seeds germinate either in the autumn (and persist as seedlings under the snow) or during the spring immediately following snowmelt in late May or early June. They develop rapidly, and the peak of flowering is typically from mid- to late June. Plants set seed and die by mid-July in most years due to low soil moisture caused by summer drought.
In each year, transects were established in two spatially distinct microsites. The Wet and Dry microsites are approximately 10 m apart. A low density of native M. guttatus spans these sub-populations and pollinating insects traverse the entire population regularly. In each year, transplants were monitored daily. We recorded the day when the first flower opened (flowering time) and measured corolla width for each flower produced. At the end of the growing season (mid-July), we collected and counted seed from all transplants that survived to flower and noted failure of plants that did not reach flowering.
Statistical analysis
Given that the number of assayed genotypes as well as environmental conditions differed among years, we first conducted a data analysis for each year separately. Within years, there is a factorial organization of two causal factors, genotype and microsite. The dependent variables are survival to flowering, corolla width (square root transformed), days to flower, fecundity of flowering plants (seed set), and total fitness (the absolute seed set of all transplants with dead plants included as zeros). For the very small number of plants with multiple flowers, we averaged Sqrt(corolla width) to obtain the individual trait value. Mature plants yielding a single flower are routine in alpine populations of M. guttatus, although longer-lived individuals growing at lower elevations routinely produce many flowers. We applied a normal residuals model (standard least squares) to corolla width and days to flower. Survival to flower was treated as a binary response variable in a generalized linear model fit using the logit link function. Fecundity of flowering plants and total fitness were analyzed as raw counts using the generalized linear model with an overdispersed Poisson distribution for the dependent variable (log link function). We used REML for generalized linear model fits with the Firth adjustment to maximum likelihood values. All calculations were done using JMP v8©.
For the 2010 data, we parsed “genotype” into two factors, QTL1 and QTL8, and considered the interaction between loci. We also conducted a combined analysis for the LL and HH data because these genotypes were transplanted in all years. The explanatory factors for analysis of the combined dataset were genotype, year, and microsite. Across analyses, hypothesis tests were conducted using F-ratios for normal residual variables and likelihood ratios (X2) for generalized linear model tests. In each case, we first fit a model with all possible pairwise interactions of factors. When interactions were clearly non-significant (p > 0.1), we dropped these terms and refit the reduced model. Test results from this reduced model are reported in the Results while the full collection of tests (with parameter estimates from each model fit) is reported in Supplemental Appendix 1.
Results
Transplant year 2008
HH exhibited significantly lower survivorship than LL (X2 [1] = 15. 2, p<0.0001). Among survivors, HH had larger flowers (F1,49 = 21.44, p<0.0001), require more days to reach flowering (F1,53 = 7.11, p = 0.01), and had higher fecundity (X2 [1] = 8.33, p<0.004). The difference between HH and LL for corolla width and survivorship, but not for fecundity, was consistently reiterated in subsequent years (Figure 1). Owing to the opposing directions of viability and fecundity selection in 2008, total fitness did not differ significantly between HH and LL (X2 [1] = 0.82, p>0.05). However, there was a nearly significant interaction between genotype and microsite for total fitness (X2 [1]= 3.54, p=0.06). Table 1 summarizes total fitness data for each genotype across microsites and years.
Table 1.
Temporal and spatial variation in total fitness of genotypes is reported. Total fitness is the average of absolute seed set of all transplants of a genotype. SD is the standard deviation and N is the sample size.
| Year | Microsite | Genotype | Mean | SD | N |
|---|---|---|---|---|---|
| 2008 | Wet | HH | 0.29 | 2.41 | 120 |
| HL | -- | -- | -- | ||
| LH | -- | -- | -- | ||
| LL | 1.93 | 10.29 | 120 | ||
| Dry | HH | 12.45 | 41.22 | 80 | |
| HL | -- | -- | -- | ||
| LH | -- | -- | -- | ||
| LL | 6.80 | 28.25 | 80 | ||
| 2009 | Wet | HH | 0.50 | 4.33 | 150 |
| HL | 0.46 | 5.63 | 150 | ||
| LH | -- | -- | -- | ||
| LL | 8.13 | 26.10 | 150 | ||
| Dry | HH | 0.00 | 0.00 | 150 | |
| HL | 0.00 | 0.00 | 150 | ||
| LH | -- | -- | -- | ||
| LL | 1.00 | 11.61 | 150 | ||
| 2010 | Wet | HH | 0.83 | 6.50 | 240 |
| HL | 1.82 | 13.20 | 240 | ||
| LH | 0.50 | 4.81 | 240 | ||
| LL | 0.98 | 10.54 | 240 | ||
| Dry | HH | 0.19 | 1.68 | 160 | |
| HL | 0.02 | 0.24 | 160 | ||
| LH | 0.00 | 0.00 | 160 | ||
| LL | 0.34 | 4.27 | 160 |
Transplant year 2009
Survivorship differed significantly among genotypes (X2 [2] = 9.48, p<0.01) with HL lowest and LL highest. Survivorship was much higher in Wet than Dry microsite (X2 [1] = 14.98, p = 0.0001). As a consequence of low survivorship in the Dry microsite, we could not evaluate Microsite × Genotype interactions for corolla width, days to flower, or fecundity of survivors. Genotypes did not differ significantly for these measurements in model fits without an interaction (Supplemental Appendix 1). Total fitness did differ significantly among genotypes (X2 [2] = 37.48, p<0.0001) with LL much greater than HL and HH (Table 1).
Transplant year 2010
Survival differed among genotypes (X2 [3] = 12.59, p < 0.006) in a complicated way with HL > (HH, LL) > LH. Among survivors, flower size differed in a predictable way given genotype (Figure 2; F3,84 = 36.98, p < 0.0001). Differences in fecundity of survivors were non-significant but the LL average was about half that of other genotypes. As in 2008, the generally opposing effects of viability and fecundity effects of genotypes resulted in non-significant differences in total fitness (X2 [3] = 3.38; p > 0.05). Because all four genotypes were assayed in 2010, we can treat each locus as a factor and test for an interaction (epistasis). For survival, the high allele at QTL 8 had a significantly negative direct effect (X2 [1] = 25.7, p<0.0001), but the interaction of high alleles was significantly positive (X2 [1] = 4.25, p < 0.04). In other words, survivorship of HH individuals was higher than predicted given the direct (single locus) effects of each H allele. Other interaction tests were non-significant.
Fig. 2.
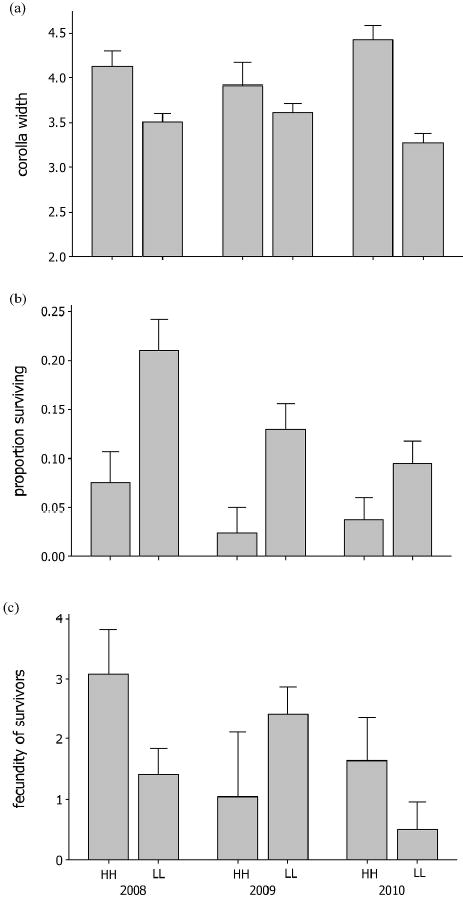
Means are reported for (a) corolla width, (b) proportion surviving, and (c) fecundity of survivors for each Mimulus guttatus genotype (HH and LL), in each year of transplant. (a) Corolla width is square root transformed (√mm). (b) Proportion surviving is the number of individuals that survived to flower. (c) Fecundity of survivors is ln (seed set +1). Error bars are 95 % confidence intervals of each mean.
Genotype by environment interaction and heterogeneity in selection
Temporal fluctuations in the strength and direction of selection are evident from the year-to-year analysis reported above (also see Table 1). For the LL/HH contrast, we can simultaneously test for effects of genotype, microsite, year, and their interactions. Corolla width exhibited a highly significant year by genotype interaction (F2,140 = 10.1, p < 0.0001). Genotype (X2 [1] = 38.1, p<0.0001) and year (X2 [2] = 26. 2, p<0.0001) each had significant direct effects on survivorship, but there was also a significant interaction between genotype and microsite (X2 [1] = 5.38, p = 0.02) and between microsite and year (X2 [2] = 60.1, p<0.0001). Fecundity of survivors (X2 [2] = 8.94, p = 0.01) and total fitness (X2 [2] = 16.3, p = 0.0003) each exhibited significant year by genotype interactions. Significant interactions do not necessarily imply effect reversal in which a genotype superior in one circumstance (year and/or microsite) is inferior in another. For example, fecundity selection on LL vs. HH apparently reversed in 2009 relative to 2008 and 2010 (Figure 1), but this reversal is tentative because the difference within 2009 is not quite significant.
Discussion
Recent surveys of genomic variation in humans have identified molecular signatures of natural selection (Sabeti et al. 2007; Tishkoff et al. 2007). Counter to initial expectations, hard sweeps wherein a novel mutation is immediately favored and spreads to fixation, e.g. (Coop et al. 2009; Pritchard et al. 2010; Simonson et al. 2010), seem to be relatively infrequent. In contrast, polygenic adaptation (divergence in allele frequencies at many loci) is likely common (Hancock et al. 2010; Pritchard & Di Rienzo 2010). The infrequency of hard sweeps has prompted renewed focus on adaptation via selection on standing variation. Unfortunately, the signature of this kind of selection can be very subtle at the gene sequence level (Kelly 2006; Pennings & Hermisson 2006; Teshima et al. 2006) and direct experimental approaches are required.
Field transplant experiments directly measure the fitness consequences of genetic variation and have been successfully applied at the QTL level in a number of plant species. These experiments are mainly of two types, common garden studies and reciprocal transplant experiments. Common garden studies assay phenotype and fitness of recombinant genotypes within a single natural location that may or may not be related to the sites of origin of founding genotypes. For example, Lexer et al. (2003) monitored hybrids of two sunflower species, Helianthus annuus and H. petiolaris, within a novel salt marsh habitat. Salt marsh is the natural environment of Helianthus paradoxus, a homoploid hybrid species derived from H. annuus and H. petiolaris. Three QTL had large effects on survivorship suggesting that the genetic composition of the hybrid population facilitated rapid evolution of ecological specialization. Common garden studies can also involve experimental manipulation. For example, Li et al. (2003) showed that QTL of rice (Oryza sativa) exhibit reversal of effect across different water availability treatments. In Arabidopsis thaliana, Weinig et al. (2003) demonstrated significant QTL by season interactions for herbivory resistance while Korves et al. (2007) showed that seasonally varying selection on FRIGIDA (a flowering time QTL) is itself contingent on genetic background.
Reciprocal transplant experiments assay recombinant genotypes in field sites that are at or near the locations from which founding genotypes were sampled (Anderson et al. 2011). A major objective with this design is to decompose local adaptation into its genomic components. Verhoeven et al. (2004) mapped survivorship and fecundity QTL in F3 hybrids between inland and coastal genotypes of wild barley. Selection on QTL differed in magnitude but not direction between locations. Gardner and Latta (2006) obtained comparable results with hybrid genotypes of a cross between ecotypes of Avena barbata. There was minimal evidence for selection favoring alternative alleles of the same QTL in xeric versus mesic transplant sites. Lowry and colleagues (2009) identified salt-spray tolerance QTL of Mimulus guttatus that affect fitness in a coastal habitat but not in the alternative alpine environment (see also Hall et al. 2010). In each of these cases, QTL by location interactions manifest as directional selection within one habitat but apparent neutrality in the other.
The current collection of field QTL studies is based on genetic materials derived from crosses between divergent populations, ecotypes, or species. In contrast, the present study evaluates intra-population QTL. The paucity of intra-population data likely reflects an enduring skepticism about successfully measuring natural selection on intra-population QTL. If such loci contribute in a minor way to variation, fitness effects may simply be undetectable. A surprising and important result from this experiment is that phenotypic differences between QTL genotypes were actually magnified in the field (Figures 1-2). For corolla width measured in the greenhouse, the estimated additive effects are 1.8 and 1.0 mm for QTL1 and QTL8, respectively (Scoville et al. 2011). In the 2010 field season, the additive effect for each locus was approximately 4 mm (Figure 2). The proximate cause for increased allelic effects may be that larger flower sizes are associated with delayed flowering. Differences in developmental timing that are fairly subtle under benevolent, nutrient rich conditions become more pronounced under harsh, nutrient-poor field conditions (Figure 3).
Fig. 3.
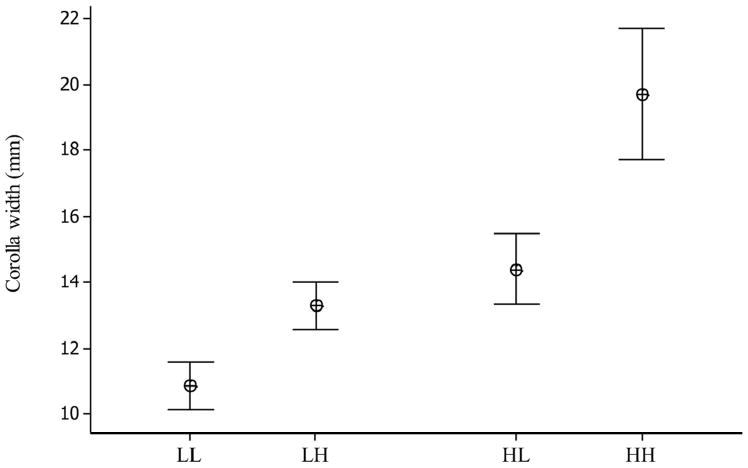
The mean and 95% confidence interval for flower size of each genotype in 2010.
Antagonistic pleiotropy
Antagonistic pleiotropy is a genetic trade-off wherein an allele that increases one fitness component (e.g. survival) reduces another (e.g. mating success). A variety of these trade-offs have been proposed and they naturally emerge in traits that incur physiological costs like plant resistance to herbivores (see Strauss et al. 2002 for review). For example, trichome density and glucosinolate concentration can limit herbivore damage but reduces fitness in the absence of natural enemies (Mauricio 1998). In our study, the antagonistic pleiotropy is a trade-off between survival to flowering and fecundity of plants that manage to flower. In each year, there were significant differences among genotypes in survivorship, with higher survival usually associated with alleles that reduce flower size. However, total fitness did not differ significantly among genotypes in 2008 or 2010 owing to conflicting selection on survival and fecundity.
Antagonistic pleiotropy for the floral QTL is a direct consequence of differences among genotypes in the transition from vegetative growth to flowering. Plants with delayed transition routinely die of desiccation before they flower. In most years/microsites, H genotypes delayed flowering relative to L genotypes (Fig. 3). When H genotypes did survive to flower, they were larger plants with larger flowers. Increased fecundity of these plants is likely due to higher ovule and pollen numbers per flower, perhaps combined with increased pollinator attraction. A caveat here is that the introgressed segments (H alleles for each QTL) contained (Mojica & Kelly 2010) any genes apart from the locus or loci influencing flower size. It is possible that other polymorphisms in these segments also influence development rate or fecundity. However, data from previous field studies support a causal role for flower size loci. The pattern of selection in the current study is consistent with measurements of survival and reproduction on flower size genotypes that differ at many QTL. Transplant of outbred ‘small’, ‘medium’, and ‘large’ flowered M. guttatus plants into the same field site yielded comparable correlations with survivorship and fecundity (see Figure 1 of Mojica & Kelly 2010).
The trade-off of development rate with fecundity has also been demonstrated at a larger spatial scale in Mimulus guttatus. A reciprocal transplant between alpine and coastal populations demonstrated strong divergent selection on time to flower, consistent in direction with the phenological divergence of the parental genotypes (Hall et al 2006). While inter-population QTL are routinely interpreted as products of past adaptation, these loci may or may not reflect fixed differences between the parental populations. A locus segregating within an inter-population cross could be polymorphic within one or both ancestral populations (e.g. Colosimo et al. 2005) and thus relevant to questions about local variation. With geographically varying selection and gene flow between populations, selection may be balancing at scale of entire species but locally purifying. Migration-selection balance differs from mutation-selection balance in that deleterious migrant alleles are advantageous under some environmental conditions and because migration rates will often be orders of magnitude greater than per-locus mutation rates. As a consequence, local allele frequencies may be more intermediate with migration than mutation selection balance.
Environmental heterogeneity
This trade-off of fitness components (Fig. 1) is a necessary but not sufficient condition for antagonistic pleiotropy to maintain genetic variation. Particular dominance relationships between alleles are necessary for a stable polymorphism, at least in simple models (Hedrick 1999; Radwan 2008). We cannot evaluate dominance with the present data because our design includes only two alternative genotypes per QTL. However, stable polymorphism is more likely if per locus viability and fecundity effects are large, as they are in this study (Hedrick 1999). Perhaps more importantly, conditions for balanced polymorphism may be more permissive if the relative magnitudes and direction of viability and fertility selection vary with environment. Spatial variation in selection is a particularly powerful mechanism if there is limited gene flow between environments. Because of limited seed and/or pollen dispersal, local adaptation can occur at the scale of meters within contiguous plant populations (Stewart & Schoen 1987; for reviews see Kawecki & Ebert 2004; and Hereford 2009).
Selection on these flower size QTL did exhibit complex spatial and temporal dependencies. For survivorship, there was also a significant genotype by microsite interaction across years. Fecundity of survivors and total fitness each displayed significant year by genotype interactions. In 2008, H alleles were favored in the dry but not the wet microsite (Table 1). In 2008, we observed higher pollinator visitation in dry microsite, and this combined with the higher reproductive capacity of larger flowers, could explain the fitness advantage of H alleles in the dry microsite. However, this spatial pattern of selection was not temporally consistent. Selection did not clearly favor H alleles in the dry microsite in either 2009 or 2010.
In 2010, there was microsite dependent epistasis for survival. QTL specific effects changed with genetic background. In the dry environment, the L allele was superior to the H allele at QTL 1 if QTL 8 was L. With H at QTL 8, selection reversed at QTL 1 (H > L; Table 1). This kind of sign epistasis (Weinreich et al. 2005) was not evident in the wet microsite. Brock et al. (2010) also noted a strong environmental dependence of QTL by QTL interactions in Brassica rapa. Korves and colleagues (2007) argued that the combination of variable selection and epistasis maintain geographical variation at the FRIGIDA locus of Arabidopsis. Given that we could estimate epistasis in only one year of our study (2010), it is premature to draw conclusions about whether interactions among QTL facilitate the maintenance of floral variation in Mimulus. However, we have recently demonstrated that QTL of M. guttatus routinely exhibit epistasis in their effects on floral morphology (Kelly & Mojica 2011).
Conclusions
We performed a three-year field transplant experiment measuring selection on two intra-population flower size QTL. We found antagonistic pleiotropy with H alleles (alleles that increase flower size) generally increasing fecundity but reducing survivorship to flowering relative to L alleles. The direction and magnitude of selection fluctuated yearly and at the spatial scale of meters. Considering the point estimates for total fitness (mean number of seeds produced per transplant across microsites), H genotypes were superior to L genotypes in 2008 while the reverse was true for fitness in 2009. In 2010, the H allele had higher fitness than L at QTL1, but L was higher than H at QTL8.
In classical population genetics, balancing selection is indicated by a ‘protected polymorphism’ wherein selection acts to prevent either allele at a locus from going extinct (Christiansen 1974). Given only three years of data and ignorance of some important genetic details (e.g. dominance relations between H/L at each locus, the possible existence of other alleles at these loci, effects of genetic background, etc.), it is premature to claim that these loci are protected polymorphisms.
However, these data clearly provide direct evidence for at least three distinct balancing selection mechanisms: antagonistic pleiotropy, spatial variation in fitness, and temporal variation in fitness. If these selective processes are simultaneously operating with the recurrent introduction of variation by mutation and gene flow, variation may persist at elevated levels regardless of genetic details specific to particular loci.
Supplementary Material
Fig. 4.
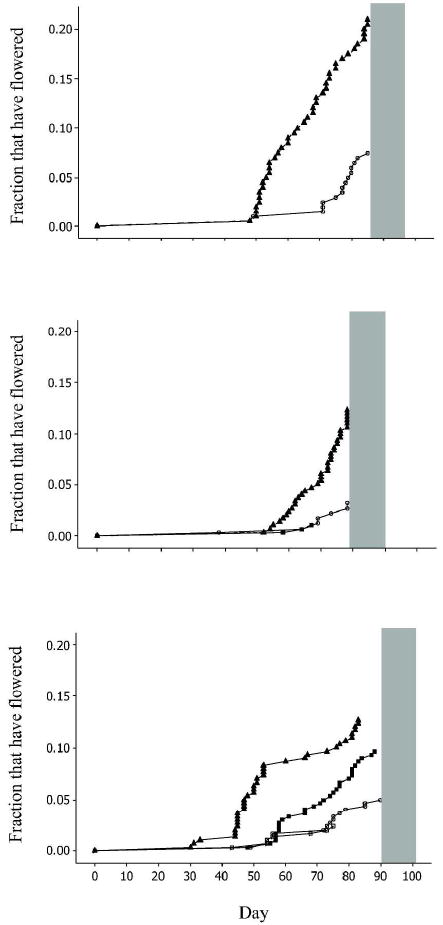
Cumulative proportion of flowered HH (open circle), LL (shaded triangle), HL (shaded square), and LH (open square) Mimulus guttatus genotypes per day across three years: (top panel) 2008; (middle panel) 2009; and (bottom panel) 2010. Grey bar indicates the final drought phase in each year.
Acknowledgments
We thank M. Rausher, T. Mitchell-Olds, M. Trenary, K. Koch, S. Easter, C. Wallace, S. Bodbyl-Roels, V. Koelling, L., S. Macdonald, and T. Lubin for assistance with field work and/or comments on the paper. Thorough reviews were given by three anonymous referees, Bob Latta, and Aurélie Bonin. This project was supported by grants NIH GM073990 (to JKK and JHW), and NSF grant DEB-0910321 (to JPM), and by funding from the University of Kansas Botany Endowment Funds.
Footnotes
Data accessibility. Phenotype and fitness data: Dryad, entry doi:10.5061/dryad.s0n20
References
- Anderson J, Lee C, Mitchell-Olds T. Life history QTLs and natural selection on flowering time in Boechera stricta, a perennial relative of Arabidopsis. Evolution. 2011;65:771–787. doi: 10.1111/j.1558-5646.2010.01175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH. Pleiotropic models of quantitative trait variation. Genetics. 1990;124:773–782. doi: 10.1093/genetics/124.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw HD, Schemske DW. Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature. 2003;426:176–178. doi: 10.1038/nature02106. [DOI] [PubMed] [Google Scholar]
- Brock MT, Dechaine JM, Iniguez-Luy FL, et al. Floral genetic architecture: An examination of QTL architecture underlying floral covariation across environments. Genetics. 2010;186:1451–U1596. doi: 10.1534/genetics.110.119982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero A, Keightley PD. A pleiotropic nonadditive model of variation in quantitative traits. Genetics. 1994;138:883–900. doi: 10.1093/genetics/138.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen FB. Sufficient conditions for protected polymorphism in a subdivided population. American Naturalist. 1974;108:157–166. [Google Scholar]
- Clausen J, Keck DD, Hiesey WM. Experimental studies on the nature of species. I. The effect of varied environments on western American plants. Carnegie institute; Washington: 1940. p. 452. [Google Scholar]
- Colosimo PF, Hosemann KE, Balabhadra S, et al. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science. 2005;307:1928–1933. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- Coop G, Pickrell JK, Novembre J, et al. The role of geography in human adaptation. PLoS Genet. 2009;5:e1000500. doi: 10.1371/journal.pgen.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky TH, Levene H. Genetics of natural populations. XVII. Proof of operation of natural selection in wild populations of Drosophila pseudoobscura. Genetics. 1948;33:537–547. doi: 10.1093/genetics/33.6.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford EB. Ecological genetics. 3. Chapman and Hall; London: 1971. [Google Scholar]
- Fry JD. The “General Vigor” problem: Can antagonistic pleiotropy be detected when genetic covariances are positive? Evolution. 1993;47:327–333. doi: 10.1111/j.1558-5646.1993.tb01224.x. [DOI] [PubMed] [Google Scholar]
- Gardner KM, Latta RG. Identifying loci under selection across contrasting environments in Avena barbata using quantitative trait locus mapping. Molecular Ecology. 2006;15:1321–1333. doi: 10.1111/j.1365-294X.2005.02835.x. [DOI] [PubMed] [Google Scholar]
- Gillespie JH, Turelli M. Genotype-environment interactions and the maintenance of polygenic variation. Genetics. 1989;121:129–138. doi: 10.1093/genetics/121.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves JH, Redfern R, Ayres PB, Gill JE. Warfarin resistance-balanced polymorphism in Norway rat. Genetical Research. 1977;30:257–263. doi: 10.1017/s0016672300017663. [DOI] [PubMed] [Google Scholar]
- Hall MC, Lowry DB, Willis JH. Is local adaptation in Mimulus guttatus caused by tradeoffs at individual loci? Molecular Ecology. 2010;19:2739–2753. doi: 10.1111/j.1365-294X.2010.04680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MC, Willis JH. Divergent selection on flowering time contributes to local adaptation in Mimulus guttatus populations. Evolution. 2006;60:2466–2477. [PubMed] [Google Scholar]
- Hancock AM, Witonsky DB, Ehler E, et al. Human adaptations to diet, subsistence, and ecoregion are due to subtle shifts in allele frequency. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:8924–8930. doi: 10.1073/pnas.0914625107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick PW. Antagonistic pleiotropy and genetic polymorphism: a perspective. Heredity. 1999;82:126–133. [Google Scholar]
- Hereford J. A Quantitative Survey of Local Adaptation and Fitness Trade-Offs. American Naturalist. 2009;173:579–588. doi: 10.1086/597611. [DOI] [PubMed] [Google Scholar]
- Hill WG. Selection with recurrent backcrossing to develop congenic lines for quantitative trait loci analysis. Genetics. 1998;148:1341–1352. doi: 10.1093/genetics/148.3.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecology Letters. 2004;7:1225–1241. [Google Scholar]
- Keightley PD, Hill WG. Variation maintained in quantitative traits with mutation selection balance - pleiotropic side-effects on fitness traits. Proceedings Of the Royal Society Of London Series B-Biological Sciences. 1990;242:95–100. [Google Scholar]
- Kelly JK. Deleterious mutations and the genetic variance of male fitness components in Mimulus guttatus. Genetics. 2003;164:1071–1085. doi: 10.1093/genetics/164.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JK. Geographical variation in selection, from phenotypes to molecules. American naturalist. 2006;167:481–495. doi: 10.1086/501167. [DOI] [PubMed] [Google Scholar]
- Kelly JK. Testing the rare-alleles model of quantitative variation by artificial selection. Genetica. 2008;132:187–198. doi: 10.1007/s10709-007-9163-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JK, Mojica JP. Interactions among flower-size QTL of Mimulus guttatus are abundant but highly variable in nature. Genetics. 2011;189:1461–+. doi: 10.1534/genetics.111.132423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JK, Willis JH. Deleterious mutations and genetic variation for flower size in Mimulus guttatus. Evolution. 2001;55:937–942. doi: 10.1554/0014-3820(2001)055[0937:dmagvf]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Kingsolver JG, Hoekstra HE, Hoekstra JM, et al. The strength of phenotypic selection in natural populations. American Naturalist. 2001;157:245–261. doi: 10.1086/319193. [DOI] [PubMed] [Google Scholar]
- Kondrashov AS, Turelli M. Deleterious mutations, apparent stabilizing selection and the maintenance of quantitative variation. Genetics. 1992;132:603–618. doi: 10.1093/genetics/132.2.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korves TM, Schmid KJ, Caicedo AL, et al. Fitness effects associated with the major flowering time gene FRIGIDA in Arabidopsis thaliana in the field. American Naturalist. 2007;169:E141–E157. doi: 10.1086/513111. [DOI] [PubMed] [Google Scholar]
- Lande R. The maintenance of genetic variability by mutation in a polygenic character with linked loci. Genetic Researches on Population Ecology. 1975;26:221–235. doi: 10.1017/s0016672300016037. [DOI] [PubMed] [Google Scholar]
- Lee YW. Genetic analysis of standing variation for floral morphology and fitness components in a natural population of Mimulus guttatus (common monkeyflower) PhD. Duke University; 2009. [Google Scholar]
- Lexer C, Welch ME, Durphy JL, Rieseberg LH. Natural selection for salt tolerance quantitative trait loci (QTLs) in wild sunflower hybrids: Implications for the origin of Helianthus paradoxus, a diploid hybrid species. Molecular Ecology. 2003;12:1225–1235. doi: 10.1046/j.1365-294x.2003.01803.x. [DOI] [PubMed] [Google Scholar]
- Li ZK, Yu SB, Lafitte HR, et al. QTL x environment interactions in rice. I. Heading date and plant height. Theoretical and Applied Genetics. 2003;108:141–153. doi: 10.1007/s00122-003-1401-2. [DOI] [PubMed] [Google Scholar]
- Lowry DB, Hall MC, Salt DE, Willis JH. Genetic and physiological basis of adaptive salt tolerance divergence between coastal and inland Mimulus guttatus. New Phytologist. 2009;183:776–788. doi: 10.1111/j.1469-8137.2009.02901.x. [DOI] [PubMed] [Google Scholar]
- Mackay TFC, Lyman R, Jackson MS. Effects of P elements on quantitative traits in Drosophila melanogaster. Genetics. 1992;130:315–332. doi: 10.1093/genetics/130.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauricio R. Costs of resistance to natural enemies in field populations of the annual plant Arabidopsis thaliana. American Naturalist. 1998;151:20–28. doi: 10.1086/286099. [DOI] [PubMed] [Google Scholar]
- Mojica JP, Kelly JK. Viability selection prior to trait expression is an essential component of natural selection. Proceedings of the Royal Society B-Biological Sciences. 2010;277:2945–2950. doi: 10.1098/rspb.2010.0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennings PS, Hermisson J. Soft sweeps III: The signature of positive selection from recurrent mutation. PLoS Genet. 2006;2:e186. doi: 10.1371/journal.pgen.0020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Di Rienzo A. Adaptation - not by sweeps alone. Nature Reviews Genetics. 2010;11:665–667. doi: 10.1038/nrg2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Pickrell JK, Coop G. The genetics of human adaptation: Hard sweeps, soft sweeps, and polygenic adaptation. Current Biology. 2010;20:R208–R215. doi: 10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radwan J. Maintenance of genetic variation in sexual ornaments: a review of the mechanisms. Genetica. 2008;134:113–127. doi: 10.1007/s10709-007-9203-0. [DOI] [PubMed] [Google Scholar]
- Rausher MD. The measurement of selection on quantitative traits: biases due to the environmental covariances between traits and fitness. Evolution. 1992;46:616–626. doi: 10.1111/j.1558-5646.1992.tb02070.x. [DOI] [PubMed] [Google Scholar]
- Rose MR. Antagonistic pleiotropy, dominance, and genetic-variation. Heredity. 1982;48:63–78. [Google Scholar]
- Sabeti PC, Varilly P, Fry B, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoville AG, Lee YW, Willis JH, Kelly JK. Explaining the heritability of an ecologically significant trait in terms of individual QTLs. Biology letters. 2011;7:896–898. doi: 10.1098/rsbl.2011.0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonson TS, Yang YZ, Huff CD, et al. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329:72–75. doi: 10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
- Stewart SC, Schoen DJ. PATTERN OF PHENOTYPIC VIABILITY AND FECUNDITY SELECTION IN A NATURAL-POPULATION OF IMPATIENS-PALLIDA. Evolution. 1987;41:1290–1301. doi: 10.1111/j.1558-5646.1987.tb02467.x. [DOI] [PubMed] [Google Scholar]
- Strauss SY, Rudgers JA, Lau JA, Irwin RE. Direct and ecological costs of resistance to herbivory. Trends in Ecology & Evolution. 2002;17:278–285. [Google Scholar]
- Subramaniam B, Rausher MD. Balancing selection on a floral polymorphism. Evolution. 2000;54:691–695. doi: 10.1111/j.0014-3820.2000.tb00070.x. [DOI] [PubMed] [Google Scholar]
- Teshima KM, Coop G, Przeworski M. How reliable are empirical genomic scans for selective sweeps? Genome research. 2006;16:702–712. doi: 10.1101/gr.5105206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Ranciaro A, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M. Heritable genetic variation via mutation -selection balance: Lerch’s zeta meets the abdominal bristle. Theoretical population biology. 1984;25:138–193. doi: 10.1016/0040-5809(84)90017-0. [DOI] [PubMed] [Google Scholar]
- Verhoeven KJF, Vanhala TK, Biere A, Nevo E, Van Damme JMM. The genetic basis of adaptive population differentiation: A quantitative trait locus analysis of fitness traits in two wild barley populations from contrasting habitats. Evolution. 2004;58:270–283. [PubMed] [Google Scholar]
- Weinig C, Stinchcombe JR, Schmitt J. QTL architecture of resistance and tolerance traits in Arabidopsis thaliana in natural environments. Molecular Ecology. 2003;12:1153–1163. doi: 10.1046/j.1365-294x.2003.01787.x. [DOI] [PubMed] [Google Scholar]
- Weinreich DM, Watson RA, Chao L. Perspective: Sign epistasis and genetic constraint on evolutionary trajectories. Evolution. 2005;59:1165–1174. [PubMed] [Google Scholar]
- Willis JH. Measures of phenotypic selection are biased by partial inbreeding. Evolution. 1996;50:1501–1511. doi: 10.1111/j.1558-5646.1996.tb03923.x. [DOI] [PubMed] [Google Scholar]
- Wright S. The genetics of quantitative variability. In: Reeve ECR, Waddington CH, editors. Quantitative inheritance. Her Majesty’s Stationary Office; London, UK: 1952. pp. 5–41. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


