Abstract
BACKGROUND
Small-cell carcinoma (SCC) of the prostate is an AR-negative variant of prostate cancer found at progression in 10–20% of castrate-resistant disease. Its finding predicts a distinct clinical course and a poor prognosis. Large-cell neuroendocrine carcinoma (LCNEC) is a much rarer variant that behaves similarly to SCC. The biological mechanisms that drive these disease variants are poorly understood.
METHODS
Eight tumor fragments from the salvage pelvic exenteration specimen of a patient with castrate-resistant prostate carcinoma were subcutaneously implanted into 6- to 8-week-old male CB17 SCID mice. Serial tissue sections and tissue microarrays of the resulting MDA PCa 144 xenograft lines were used for histopathologic and immunohistochemical characterization of the xenografts and their tissue of origin. RNA from two representative xenograft sublines was used for gene-expression profiling.
RESULTS
All eight fragments formed tumors: four of the MDA PCa 144 xenograft sublines had morphologic characteristics of SCC and four, of LCNEC. All retained high fidelity to their parent tumor tissue, which remained stable through serial passages. Morphological transitions in the specimen of origin suggested LCNEC represents an intermediate step between adenocarcinoma and SCC. Over 2,500 genes were differentially expressed between the SCC (MDA PCa 144-13) and the LCNEC (MDA PCa 144-4) sublines and enriched in “Nervous System Development” Gene Ontology subtree.
CONCLUSION
The eight xenograft models described represent the spectrum of neuroendocrine carcinomas in prostate cancer and will be valuable preclinical tools to study the pathogenesis of and therapy targets for this increasingly recognized subset of lethal prostate cancer.
Keywords: castrate-resistant, cancer, androgen-independent, neural development, array
INTRODUCTION
Variant forms of prostate cancer account for a substantial proportion of the lethal subset of prostate cancer and are characterized by atypical clinical features distinct from the common bone-homing and bone-forming clinical phenotype of typical prostate cancer. Elucidation of the biology underlying the clinically relevant heterogeneity between or within patients with prostate cancer will lead to the development of individualized treatments. We and others have used tumor xenografts as model systems that reflect the heterogeneity of human prostate cancer to gain insight into their underlying biology.(1,2) In this study, we established and characterized a set of eight morphologically diverse xenografts established from a single salvage pelvic exenteration specimen from a patient with castrate-resistant prostate cancer. The patient’s tumor displayed elements of adenocarcinoma, large-cell neuroendocrine carcinoma (LCNEC), and small-cell carcinoma (SCC).
Transformation to SCC is a well-described phenomenon in prostate cancers subject to androgen deprivation therapy (3,4) and is found in 10–20% of autopsies of patients who die of the disease (5,6). Patients with SCC have a distinct clinical behavior, characterized by rapidly progressive disease with large prostatic or nodal masses, a high frequency of visceral disease, frequently lytic bone metastases, and disproportionately low prostate-specific antigen (PSA) levels for the typically large burden of disease. These cancers respond poorly to hormone therapies (7) but are responsive to chemotherapy, although responses are typically short, and median survival is approximately 1 year (4,8). LCNEC is a rare finding (9,10) that is associated with behavior similar to that of SCC, but has unclear additional significance.
Four of the xenografts we established displayed morphologic features of LCNEC, and the other four, of SCC. Despite these morphologic differences, their immunohistochemical profiles of known markers of prostate cancer prognosis were similar and reflected the molecular characteristics of the donor tumor. Two of the xenograft lines, which represented the extremes of the morphologic spectrum observed in the group, were used for gene expression–array analysis. The differences in gene expression were substantial and interestingly involved a large number of genes involved in development, specifically neural development.
MATERIALS AND METHODS
Donor Patient’s Clinical History
The eight MDA PCa 144 xenografts were obtained from the salvage pelvic exenteration specimen of a 72-year-old man initially diagnosed with a T2cN0M0, Gleason 4+3 prostate adenocarcinoma in December 2002. He was treated with 9 months of androgen deprivation therapy and external beam radiation therapy. His PSA concentration was undetectable(<0.1ng/mL) for 3 years but increased to 4.6ng/mL in April 2006, at which time leuprolide therapy was begun. Despite a decrease in PSA, he developed bladder outlet obstruction and hematuria and, in June 2006, cystoscopy revealed a nodular prostatic mass invading the bladder, which on histologic analysis was consistent with SCC of the prostate.
The patient was enrolled in a phase II study of carboplatin plus docetaxel for patients with anaplastic prostate carcinoma (PCCTC #05-015 [MDA 2006-0097] NCT00514540) and was treated with frontline carboplatin, AUC = 5, and docetaxel, 75 mg/m2, (CD) every 3 weeks. At enrollment, his PSA concentration was 0.4 ng/mL (normal 0.0-4.0ng/mL); prostatic acid phosphatase (PAP), 1 ng/mL (normal 0.5-5.0ng/mL); carcinoembryonic antigen (CEA), 33.8 ng/mL (normal 0.0-3.0ng/mL in non-smokers); and chromogranin A, 144.7 ng/mL (normal ≤ 36.4ng/mL). Computed tomographic imaging revealed an 8.4 × 7 cm prostatic mass involving the rectum, bladder, both distal ureters, and the right obturator internus and puborectalis muscles but no evidence of distant metastatic disease. He experienced a partial response according to the Response Evaluation Criteria in Solid Tumors (RECIST) and a >50% decrease in his CEA concentration (12ng/mL) after two cycles of CD.
However, his disease progressed 5 months after CD was begun and, according to the study protocol, treatment was initiated with etoposide, 120 mg/m2/day, and cisplatin, 25 mg/m2/day, for 3 days every 3 weeks. The prostatic mass decreased from 6.1 × 5.7 cm to 5.7 × 4.2 cm, and his CEA concentration decreased by >50% after four cycles of this regimen from 23.8ng/mL to 7.6ng/mL. Following his fourth cycle, he had a minimal increase in disease and was withdrawn from the study. Nonetheless, his disease remained resectable and a source of substantial morbidity, so he underwent a pelvic exenteration in May 2007. He recovered well from the surgery, experienced no immediate complications, and was discharged after 10 days but died 3 months later.
Xenograft Establishment and Growth
Written informed consent had been obtained from the patient prior to sample acquisition, according to a protocol approved by the Institutional Review Board of The University of Texas M. D. Anderson Cancer Center. Eight small fragments of tumor were obtained from the surgical specimen, placed in cold (4°C), sterile α-MEM (GIBCO–Invitrogen, Life Technologies Corp., Carlsbad, CA and then implanted into subcutaneous pockets in the flanks of 6- to 8-week-old male CB17 SCID mice (Charles River Laboratories International, Inc., Wilmington, MA). Tumor samples taken from locations immediately adjacent to the samples used for implantation were processed for histopathologic analysis.
Mice were monitored weekly for tumor growth. All animal experiments were conducted in accordance with accepted standards of animal care and were approved by the Institutional Animal Care and Use Committee of M. D. Anderson Cancer Center.
Histopathologic and Immunohistochemical Studies
Histopathologic and immunohistochemical studies of the xenografts and patient tissues were performed using the Dako Autostainer Plus (Dako North America, Inc., Carpinteria, CA) on serial 4-μm formalin-fixed paraffin-embedded tissue sections and on tissue microarrays (TMAs) constructed with blocks from the eight xenografts. One to three (median, 3) blocks representing different passages were selected for the TMA. Each xenograft was represented by a median of eight 0.6 mm–diameter cores (range, 3–9; mean ± SD, 7 ± 2 cores). In total, 60 cores were included in the TMA.
Briefly, sections were deparaffinized by incubation at room temperature in xylene for 5 minutes and then rinsed in 100% ethanol. After antigen retrieval, primary antibody (listed in Supplementary Data Table) was applied to each section for 15 minutes and then incubated at room temperature with Dako EnVision labeled polymer (Dako) for 8 minutes. Diaminobenzidine (Dako) was used as the chromogen. Nuclei were counterstained with hematoxylin.
Whole slides of the TMA sections were scanned with the Bliss system (Bacus Laboratories Inc, IL, USA), viewed with WebSlide Browser 4 (Bacus Laboratories Inc, IL, USA) and automatically stored for later retrieval. PAP, P504S, Synaptophysin, Chromogranin, CD56, Cytokeratins, p53, CD20, CD10, CD3, CD30 and bcl-6 were scored as positive when >5% of the cells displayed expression of the marker. For all the other markers the percentage of positive cells (0-100%) was determined for each core and the mean value was calculated for each case. Tissue sections from the donor tumors were viewed with Olympus BX41 Clinical Microscope (Olympus, Center Valley, PA). Areas with the highest intensity of positive cells were selected at low power (x100) magnification and the percentage of positive cells (0-100%) was determined at high power (x400) magnification.
Immunostains were evaluated by two independent pathologists (V.T. and C.C.G). Standard deviation of the scoring between the two observers was <15% and Spearman’s correlation coefficient ranged from 0.457 to 0.963. Discrepancies in scoring between the observers were resolved by review of the slides under a double-headed microscope.
TMPRSS2:ERG Gene Fusion Detection, Gene-Expression Array and Real-Time Reverse-Transcriptase–Polymerase Chain Reaction (RT-PCR)
The TMPRSS2:ERG gene fusion was sought using break-apart fluorescence in situ hybridization as previously described.(11) Briefly, tissue was pretreated with the Paraffin Pretreatment Kit I (Vysis, Des Plaines, IL, USA) and hybridized and washed with Vysis hybridization reagents, as per manufacturer’s recommendations. The rhodamine-labeled 5′-ERG probe (BAC RP11-95I21) and the fluorescein isothiocyanate-labeled 3′-ERG probe (BAC RP11-476D17) were obtained from the Children’s Hospital of Oakland Research Institute (Oakland, CA, USA). The metaphase spread of normal peripheral lymphocytes was used as control. One hundred 100 cells per tumor focus were evaluated. Cells without TMPRSS2-ERG gene fusion contained two pairs of co-localized green and red signals while cells with the TMPRSS2-ERG gene fusion contained only one pair of co-localized green and red signals, with the other broken into one green and one red signal, or only one green signal if 3’ERG was deleted resulting in loss of the red signal.
Total RNA was extracted by using an RNeasy mini kit (Qiagen, Inc., Valencia, CA) from four fresh MDA PCa 144-13 (passages 1, 2, and 3) and five MDA PCa 144-4 (passages 1 and 3) tumors grown in three different mice. All RNA samples were submitted to the M. D. Anderson Genomics Core Facility and converted to cDNA, labeled and hybridized to an Affymetrix U133A 2.0 Plus Array (Affymetrix, Inc., Santa Clara, CA). The DNA-Chip Analyzer 2004 version was used for quantification and normalization of the array data. Gene-expression measurements were logarithm transformed (base 2) for analysis. Quality control included an evaluation of the percentage of the signal present, the distribution of background signals, and estimates of the level of RNA degradation. The percentage of signals present was no less than 43%, which is within the Affymetrix recommendation of 30–60%. We found that average, minimum, and maximum backgrounds were similar across arrays. Scaling factors used to bring arrays into similar scale did not differ by more than threefold, as recommended by Affymetrix. Probes to both ends and the middle of the beta-actin and GAPDH genes were used. We observed no significant RNA degradation on the basis of intensity ratios.
The relative mRNA level for each gene was quantified by using real-time RT-PCR with SYBR Green (Applied Biosystems, Inc., Foster City, CA). Primer sequences are listed in Supplemental Table 2. mRNA levels for each gene were calculated from the values in the linear range of the PCR cycles and were normalized with the values of the mRNA of GAPDH and β-actin (‘reference’) as follows: ΔCT = average CT of reference– average CT of the specific gene, in which CT is the threshold cycle. The relative level of expression of each gene with respect to the reference gene (GAPDH or β-actin) was then calculated as 2ΔCT. For each gene, data from three independent measurements were used to calculate the means and standard deviations.
Statistical Analysis
Standard statistical analyses were used to describe the immunohistochemical test results.
The Robust Multichip Analysis algorithm(12) was used for quantification of the data. We used one-way analysis of variance to identify genes expressed differently and to adjust for multiple testing; the resulting p-values were analyzed using a beta-uniform mixture model (13). To adjust for multiple testing, we computed Tukey’s HSD (honestly significant differences) with a false discovery rate of 1e-5. If the mean between samples 144-4 and 144-13 was larger than the Tukey’s HSD cutoff, we called that probe differently expressed between the two samples.
The GOTM (Gene Ontology Tree Machine) is a Web-based tool for data analysis and data visualization for gene sets using Gene Ontology (GO) hierarchies (14). We used the GOTM to identify GO categories that were enriched by genes expressed differently between 144-13 and 144-4.
RESULTS
Histopathologic and Immunohistochemical Description
MDA PCa 144 human specimen
On gross examination, the prostate gland was distorted and diffusely infiltrated by a whitish firm neoplasm that extended into the rectal wall, the urinary bladder wall, and the periprostatic and perivesical soft tissue. On microscopic examination, various morphologic patterns were noted (Fig. 1). Most of the intraprostatic mass displayed closely packed fused and cribriform glands, consistent with conventional acinar adenocarcinoma of Gleason score 8 (4+4). However, focally in the intraprostatic mass and diffusely in the extraprostatic mass were areas displaying the whole range of neuroendocrine (NE) differentiation. These areas consisted mostly of small cells with hyperchromatic nuclei (with salt-and-pepper stippled chromatin and inconspicuous nucleoli) and scanty cytoplasm, characteristic of SCC. The cells were either arranged in a diffuse fashion, forming large sheets and irregular nodules, or infiltrated the tissues as single cells in an “Indian file” pattern. In other areas, the typical features of LCNEC were noted: nodules, nests, and rosettes of cells with large nuclei, vesicular chromatin, easily discernible nucleoli, and moderate amounts of amphophilic cytoplasm. A gradual transition among the three components was noted in the interface areas, where the cells were of a transitional type, not easily assignable to any of the three components. Additionally, in various foci within the tumor mass, cells of the three components were intermingled.
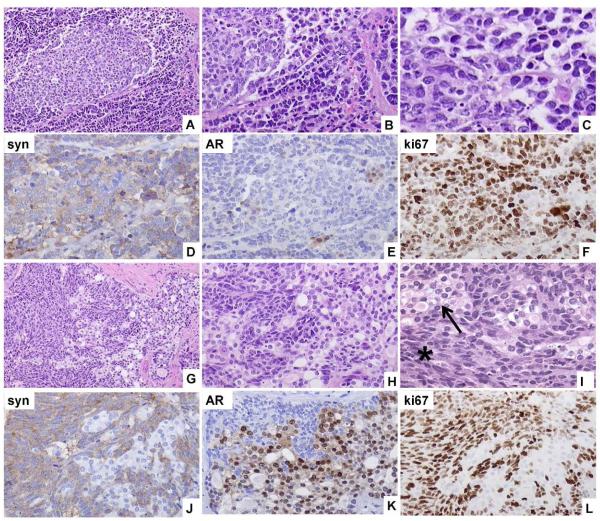
Fig. 1.
Representative images of the human specimen of origin of the MDA PCa 144 xenografts. A and B photomicrographs show a gradual transition from small-cell carcinoma (SCC) to large-cell neuroendocrine carcinoma (LCNEC). C is a high magnification photomicrograph of the small cell component. Synaptophysin (syn) is expressed diffusely in SCC and LCNEC cells (D) whereas androgen receptor (AR) is expressed only rarely (E). Ki67 is highly expressed in the cells of the neuroendocrine components of the tumor (F). Gradual transition of adenocarcinoma to LCNEC is shown in G, H and I. In I a higher magnification of the adenocarcinoma (arrow) and the LCNEC (star) cells is shown. LCNEC cells express synaptophysin (J), are negative for AR (K), and have a high Ki67 index (L), whereas the opposite is true for the adenocarcinoma cells shown in the same photomicrographs. Original magnification of A, G, × 100; B, D–F, H, J-L, × 200, C, I × 400.
Immunohistochemically, the cells of the SCC component displayed focal paranuclear dot-like staining with anti-cytokeratin antibodies and intense staining for chromogranin, synaptophysin, and CD56, whereas staining for various lymphocytic markers (L26, CD10, CD3, CD30, bcl-6), androgen receptor (AR), PSA, PAP and P504S (alpha-methylacyl-CoA racemase, AMACR) was negative. Ki67 expression was noted in the majority of cells (>90%). The cells of the LCNEC component also stained positively for chromogranin, synaptophysin, CD56, and cytokeratin (both membranous and cytoplasmic expression) and stained negatively for AR, PSA, PAP and P504S (alpha-methylacyl-CoA racemase, AMACR). As was the case with the SCC component of the tumor, Ki67 expression was noted in >90% of the cells. In the adenocarcinoma component of the tumor, intense membranous and cytoplasmic expression of cytokeratin cocktail and intense nuclear expression of AR were noted. PAP, PSA and P504S stains were positive, whereas staining for NE markers (chromogranin A, synaptophysin, CD56) was negative, and Ki67 was expressed in 30% of the tumor cells. Nuclear expression of p53 was noted in all three components of the tumor. The immunohistochemical results and the histopathological features of the different components of the donor tumor are summarized in Tables I and II, respectively.
TABLE I.
Results of Immunohistochemical Analyses in the Eight MDA PCa 144 Xenografts and the Pelvic Exenteration Specimen of Origin*
| # Blocks | # Cores |
% AR + Nuclear |
% PSA + Cytoplasmic |
Chr A + Cytoplasmic |
Syn + Cytoplasmic |
% Ki-67 + Nuclear |
p53 + Nuclear |
% PTEN + Cytoplasmic |
% pAKT + Cytoplasmic |
% bcl-2 + Cytoplasmic |
% Sox9 Nuclear |
% PTPRZ Cytoplasmic |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PATIENT DONOR TUMOR TISSUE COMPONENTS | |||||||||||||
| ADENO | n/a | n/a | 90 | 40 | − | − | 30 | + | 10 | 20 | 20 | 80 | 10 |
| LCNEC | n/a | n/a | <1 | 5 | + | + | 95 | + | 0 | 60 | 70 | 60 | 60 |
| SCC | n/a | n/a | <1 | <1 | + | + | 95 | + | 0 | 10 | 40 | 15 | 70 |
| XENOGRAFTS | |||||||||||||
| 144-13 | 3 | 9 |
0.3±0.5 0(0-1) |
0±0 | + | + |
85±12 90(60-95) |
+ | 10±0 |
91±11 95(70-100) |
0±0 |
12±10 10 (0-40) |
99±2 100(95-100) |
| 144-11 | 2 | 6 |
0.8±2 0(0-5) |
0±0 | + | + | 40±0 | + |
1±0.5 1(0-1) |
75±19 75(50-100) |
8±13 0 (0-30) |
13±8 10(10-30) |
72±20 70(40-90) |
| 144-20 | 3 | 9 | 0±0 | 0±0 | + | + |
45±31 50(10-80) |
+ | 1±0 |
89±8 90(80-100) |
2±8 0 (0-30) |
0±0 |
75±20 70 (60-100) |
| 144-23 | 2 | 6 | 0±0 | 0±0 | + | + |
31±27 25(5-70) |
+ | 1±0 |
72±10 75(60-80) |
0±0 | 0±0 |
95±5 95 (90-100) |
| 144-4 | 3 | 9 | 0±0 | 0±0 | + | + |
95±0 95(95-95) |
+ |
11±14 5(0-30) |
93±14 100(60-100) |
41±21 40 (0-70) |
98±4 100(90-100) |
77±13 80(60-90) |
| 144-2 | 3 | 9 | 0±0 | 0±0 | + | + |
14±2 15(10-15) |
+ | 0±0 |
85±17 90(60-100) |
100±0 | 100±0 |
93±10 95(80-100) |
| 144-6 | 3 | 9 |
0.7±2 0(0-5) |
0±0 | + | + |
87.5±8.5 90(70-95) |
+ |
8±8 5(1-20) |
100±0 |
72±17 70(40-100) |
n/a |
88±14 90(60-100) |
| 144-9 | 1 | 3 |
5±5 0(0-10) |
0±0 | + | + |
43±15 40(30-60) |
+ |
4±5 1(1-10) |
100±0 |
2±2 1(1-5) |
83±8 85(70-90) |
75±7 75(70-80) |
Abbreviations: AR, androgen receptor; PSA, prostate-specific antigen; Chr A, chromogranin A; Syn, synaptophysin; Adeno, adenocarcinoma; LCNEC, large-cell neuroendocrine carcinoma; SCC, small-cell carcinoma.
Data are expressed for tissue microarray results as means ± standard deviations (SD) of positive staining. When SD>0, medians with ranges in parentheses are given below. For 4-μm tissue sections, results are given as positive (+) or negative (−).
TABLE II.
Cytoarchitectural Features of the MDA PCa 144-4 and MDA PCa 144-13 Xenografts and Their Tissue of Origin
| Feature | Adenocarcinoma | LCNEC | MDA 144-4 | SCC |
MDA 144-
13 |
|---|---|---|---|---|---|
| Architecture | Irregular fused glands/cribriform |
Diffuse pattern, nodules and solid sheets with occasional |
Diffuse
pattern, nodules and solid sheets with rosettes |
Diffuse pattern |
Diffuse
focally nested pattern |
| Cell size | Medium to large | Large | Large | Medium | Medium |
| Nuclear size | Small to medium | Large | Large | Medium | Medium |
|
Nuclear
shape |
Round | Oval or elongated |
Oval or
elongated |
Round | Round |
| Cytoplasm | Abundant, clear | Abundant, amphopihilc |
Abundant,
amphopihilc |
Scanty | Scanty |
| Chromatin | Vesicular | Vesicular | Vesicular | Salt-and- pepper stippled |
Salt-and-
pepper stippled |
| Nucleoli | Prominent; 1 centrally located |
Prominent; 1 or 2 centrally or peripherally |
Prominent;
centrally located |
Indistinct | Indistinct |
|
Mitotic
index |
1 or 2 per 10 HPF | 15 per 10 HPF |
>20 per 10 HPF |
15 per 10 HPF |
>20 per 10 HPF |
| Apoptosis | Absent | Present (abundant) |
Present
(abundant) |
Present (abundant) |
Present
(abundant) |
| Necrosis | Absent | Present | Present | Present | Present |
|
Percentage
of tumor mass |
20 | 10 | 70 |
Abbreviations: LCNEC, large-cell neuroendocrine carcinoma; SCC, small-cell carcinoma; HPF, high-power microscope field.
MDA Pca 144 xenografts
All eight fragments obtained from the pelvic exenteration specimen resulted in tumor formation in the mice. The MDA Pca cell sublines 144-13 and 144-11 (both from the bladder neck) and 144-20 and 144-23 (both from the rectal wall) were first passaged 42 days after implantation, whereas sublines 144-4 and 144-9 (both from the right prostate) and 144-2 and 144-6 (both from the left prostate) were first passaged 84 days after implantation. The salient immunohistochemical features of the three tissue components of the tumor of origin and of all eight 144 sublines are compared in Table I.
Sublines 144-13 and 144-4 were deemed the most representative of the SCC and LCNEC components of the tumor, respectively (Table II); both were selected for more detailed studies and propagated for more than five passages (Fig. 2).
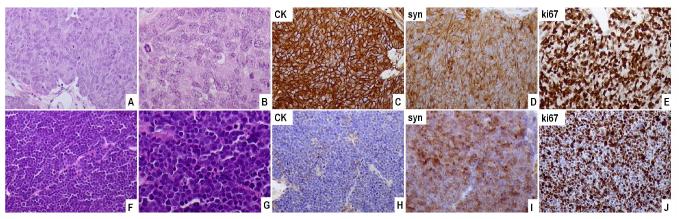
Fig. 2.
Representative images of the MDA PCa 144 xenograft. (A–E) MDA PCa 144-4 xenograft. The tumor is composed of large cells with a nested arrangement (A,B). The tumor cells express cytokeratin (CK) (C), synaptophysin (syn) (D), and Ki67 diffusely (E). (F–J) MDA PCa 144-13 xenograft. The tumor is composed of small hyperchromatic cells (F, G) and expresses cytokeratin in a dot-like pattern (H). High expression of synaptophysin (I) and Ki67 (J) is noted (original magnification, A, C-F, H-J, × 200, B, G, × 400.
Subline 144-13 (SCC)
The 144-13 tumor subline is composed of medium-sized cells arranged in solid sheets with a focally nested pattern owing to the presence of thin-walled vessels and scanty amounts of connective tissue. The tumor cells display round, hyperchromatic nuclei with indistinct nucleoli and scanty cytoplasm. Scattered small clusters of apoptotic cells are visible. Mitotic activity is high (> 20 mitotic figures per 10 high-power microscope fields). Additionally, there are areas with coagulative necrosis.
Immunohistochemically, 144-13 cells exhibit focal dot-like positive staining for cytokeratins and positive staining for synaptophysin, chromogranin, and CD56. Ki67 expression is noted in the majority of the cells (85%), whereas AR, PAP, PSA, and AMACR are not expressed. The morphologic characteristics of the tumor and the immunohistochemical results are consistent with a diagnosis of SCC; the characteristics of sublines 144-11, 144-20, and 144-23 are consistent with those of 144-13.
Subline 144-4 (LCNEC)
The 144-4 tumor subline is composed of large cells arranged in a diffuse or nodular/nested pattern with focal peripheral palisading and frequent rosette formation. The tumor cells display large oval nuclei with vesicular chromatin and medium-sized centrally located nucleoli. The cytoplasm is abundant and the cell borders indistinct. Numerous mitotic figures (> 20 mitotic figures per 10 high-power microscope fields), apoptotic cells (scattered and in small clusters), and areas of coagulative necrosis are visible. Immunohistochemically, 144-4 cells display intense cytoplasmic and membranous expression of cytokeratins, cytoplasmic expression of chromogranin and synaptophysin, and focal membranous expression of CD56. Ki67 is expressed in 95% of the cells, whereas < 5% of the cells express AR and PAP. AMACR and PSA immunostaining are negative. The morphologic and immunohistochemical features of the 144-4 tumors are consistent with a diagnosis of LCNEC; the characteristics of sublines 144-2, 144-6, and 144-9 are consistent with those of 144-4. The immunohistochemical and histopathological features of the different sublines are summarized in Tables I and II, respectively.
TMPRSS2:ERG Gene Fusion and ERG Transcript
The TMPRSS2:ERG gene fusion was detected by FISH in the adenocarcinoma, LCNEC and SCC areas of the patient’s tumor, as well as in the SCC (144-13) and LCNEC (144-4) xenografts. However, neither TMPRSS2:ERG nor ERG transcript were detected by quantitative RT-PCR in either xenograft (data not shown), which is consistent with an absence of AR signaling.
Gene-Expression Array, Subline 144-13 vs 144-4
The gradual transition from adenocarcinoma to LCNEC and from LCNEC to SCC observed in the histopathologic evaluation of the human tumor specimen (Fig. 1) suggested a progression from the adenocarcinoma to the SCC components, with LCNEC as an intermediate step. We reasoned that understanding the differences between the SCC (144-13 xenograft) and the LCNEC (144-4 xenograft) components of the tumor could help us discern whether LCNEC is truly a separate entity and gain insight into the mechanisms underlying this proposed progression.
In total, we found 3,890 probes (7.1%), representing 2,618 genes, that were significantly different between 144-4 and 144-13. The 1,394 genes that were up-regulated and the 1,224 genes that were down-regulated in 144-13 relative to their expression in 144-4 were then ranked according to mean absolute log ratios. The top 500 genes (limit imposed by the software) were used as the test gene list to query the GO database. By comparing the observed number of genes in a particular GO category with the expected number of genes in that category, we calculated enrichment P values. We found that 66 GO categories were enriched (P < 0.01) within the LCNEC up-regulated genes and 44, within the SCC up-regulated genes (Table III).
Table III.
Enriched Gene Ontology Categories in Genes Expressed Differently in LCNEC (MDA PCa 144-4) and SCC (MDA PCa 144-13) cells
| GO CATEGORY | LCNEC(144-4) >SCC(144-13) | SCC(144-13) > LCNEC(144-4) | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| I. Biological process | O | E | R | P | O | E | R | P |
|
| ||||||||
| Development | 83 | 45.8 | 1.8 | 1.90E-08 | 73 | 43.8 | 1.7 | 4.42E-06 |
|
|
||||||||
| ■ Anatomic structure development |
63 | 32.3 | 1.9 | 1.25E-07 | 52 | 30.9 | 1.7 | 1.15E-04 |
|
|
||||||||
| ▫ System development | 25 | 11.6 | 2.2 | 2.25E-04 | 27 | 11.1 | 2.4 | 1.85E-05 |
|
|
||||||||
| ▪ Nervous system development |
25 | 11.5 | 2.2 | 2.25E-04 | 27 | 11.1 | 2.5 | 1.59E-05 |
|
|
||||||||
| ▫ Organ development | 33 | 13.8 | 2.4 | 2.97E-06 | 23 | 13.2 | 1.7 | 6.95E-03 |
|
|
||||||||
| ▪ Skeletal development |
11 | 3.3 | 3.3 | 5.09E-04 | ||||
|
|
||||||||
| ▫ Morphogenesis | 26 | 14.8 | 1.7 | 3.87E-03 | ||||
|
|
||||||||
| ■ Cell differentiation | 27 | 13.4 | 2.0 | 4.35E-04 | ||||
|
| ||||||||
| II. Molecular function | O | E | R | P | O | E | R | P |
|
| ||||||||
| Transcription regulator activity | 52 | 32.5 | 1.6 | 4.2E-04 | ||||
|
|
||||||||
| ■ Transcription factor activity | 42 | 22.4 | 1.9 | 6.25E-05 | ||||
|
|
||||||||
| Enzyme regulator activity | 28 | 16.5 | 1.7 | 4.29E-03 | 25 | 14.9 | 1.7 | 8.25E-03 |
|
| ||||||||
| III. Cellular component | O | E | R | P | O | E | R | P |
|
| ||||||||
| Membrane | 181 | 139. 9 |
1.3 | 2.46E-06 | ||||
|
|
||||||||
| Neurofilament | 2 | 0.1 | 14.3 | 7.41E-03 | ||||
Abbreviations: LCNEC, large-cell neuroendocrine carcinoma; SCC, small-cell carcinoma; GO, Gene Ontology; O, observed; E, expected; R, ratio; P, P value.
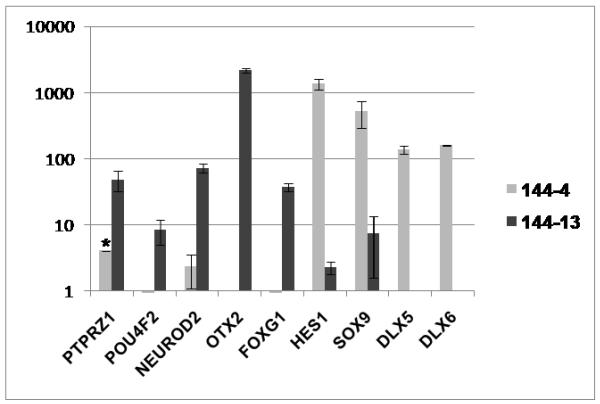
The top enriched “Biological Process” tree for both sets was “Development” and, within it, the subtree “Nervous System Development” was also enriched in both sets. On the basis of these and previously published findings (2,15-17) implicating the neural features of SCC in its biologic behavior, we selected genes within the “Development” category that encoded for proteins involved in neural differentiation to validate our gene-expression results (Fig. 3). We selected the transcription factors POU4F2, NEUROD2, OTX2, FOXG1, HES1, SOX9, DLX5, and DLX6 as potential “master regulators” of the disease progression and the protein tyrosine phosphatase receptor type Z (PTPRZ1) as a potential target for therapy (18). We also obtained antibodies for proteins derived from two of these genes, PTPRZ1 and SOX9, and confirmed their overexpression in the xenograft TMA (Table I) (Fig.4).
Fig. 3.
Reverse-transcriptase polymerase chain reaction analysis validation of gene-expression array results for MDA PCa 144-13 and MDA PCa 144-4 sublines. Data for the listed transcription factors and protein tyrosine phosphatase receptor type Z (PTPRZ1), a potential target for therapy, are expressed as mean relative mRNA level to GAPDH (× 10−4) ± standard deviation. Similar results were obtained using β-actin gene as a reference (data not shown). (*Standard deviation of PTPRZ1 in 144-4 is greater than mean value.)
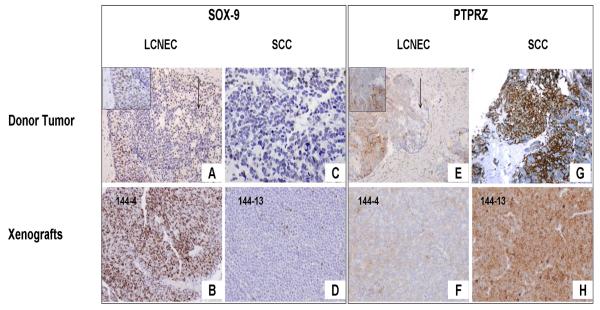
Fig. 4.
Photomicrographs illustrate immunohistochemical validation of gene-expression array results for the transcription factor SOX-9 (A-D) and the protein tyrosine phosphatase receptor type Z (PTPRZ1) (E-H), a potential target for therapy. High expression of SOX-9 is noted in the large-cell neuroendocrine carcinoma (LCNEC) (A, inset in A) and the adenocarcinoma (arrow in A) components of the donor tumor and the MDA PCa 144-4 xenograft (B), whereas low expression is noted in the small-cell carcinoma (SCC) component of both the donor tumor (C) and the MDA PCa 144-13 xenograft (D). PTPRZ expression is low in the adenocarcinoma component (arrow in E) and moderate in the LCNEC component of the donor tumor (E, inset in E) and the 144-4 xenograft (F) but is high in the SCC component of the donor tumor (G) and the 144-13 xenograft (H). (Original magnification of A, E, × 100; inset boxes in A and E, BD, F-H × 200).
DISCUSSION
Castrate-resistant prostate cancer is a heterogeneous disease. Emerging data suggest that a significant proportion of castrate-resistant prostate cancers are still driven by AR signaling (19-21). However, several studies (5,6,22) have shown that 10–20% of the lethal prostate cancers display small cell carcinoma (SCC) elements which are typically AR negative and portend a very poor prognosis, with only 6-month median survival (3,23). It is reasonable to postulate that this subset is not driven by AR signaling but that alternative driving mechanisms are at play. To gain insight into the biology underlying this subset of castrate-resistant prostate cancer and into developing therapeutic strategies specific to it, we generated the MDA PCa 144 models, which represent the spectrum of NE carcinoma in prostate cancer. The results we report here show that these 144 models retain a high degree of fidelity to the human specimen of origin and display morphologic and immunohistochemical features that are found in advanced castrate-resistant prostate tumors. Furthermore, these features are maintained after serial passages of the xenografts; thus, these models will be valuable preclinical tools for analyzing the heterogeneity of prostate cancer and developing specific treatment strategies that will improve the outcome of the disease.
Previous prostatic SCC xenografts have been reported in the literature, all from patients undergoing treatment with androgen-deprivation therapy for previously diagnosed prostate adenocarcinoma: UCRU-PR-2 (24) and WISH-PC2 (25) were from specimens obtained during transurethral resection, and LuCaP 49 was obtained from an omental metastasis (26). Like our 144 xenografts, those three lines are all negative for AR, PSA, and PAP and express NE markers to various degrees. However, to the best of our knowledge, ours is the first report of prostatic LCNEC xenograft models. LCNEC of the prostate is rare, and most of the literature on it is in the form of single case reports and small series (9,10). Similarly to SCC of the prostate, LCNEC reportedly develops most frequently after long-term treatment with androgen-ablation therapy, is usually found in combination with conventional adenocarcinoma, and pursues an aggressive clinical course. Based on the criteria applied in lung neuroendocrine tumors, LCNEC is characterized by large sized cells with coarse chromatin, frequent nucleoli, high mitotic rate (>10 mitotic figures/10 high power microscope fields) and immunohistochemical or ultrastructural evidence of neuroendocrine differentiation.(27) In contrast, SCC is characterized by medium sized cells with scant cytoplasm, hyperchromatic nuclei and indiscernible nucleoli, albeit a continuous spectrum of some of the morphologic characteristics between the two extremes (SCC and LCNEC) has been reported.(27) Despite the morphologic differences, it remains unclear whether LCNEC of the prostate is a separate entity; in fact, the 2004 WHO classification of prostate tumors does not recognize it as such (28). Our study revealed no significant differences between the growth pattern and Ki-67 expression in the SCC and the LCNEC xenograft models. The expression of p53, PTEN, and pAKT was also similar between the xenografts, but not bcl-2 expression which was lost in the xenografts displaying SCC morphologic characteristics. These results, combined with the presence of focal transitions and intermediate cell types in the human specimen of origin suggest that LCNEC and SCC are points on a biologic continuum, not distinct entities. Similar findings have been reported in neuroendocrine tumors of the lung, with minor or no differences in terms of prognosis and gene expression profiles being noted between LCNEC and SCC.(29-31)
Gene-expression profiling showed a number of genes that were expressed differently between the SCC and the LCNEC xenografts; most of those genes are associated with development, specifically neural development. Given the number of biologic replicates in each group and the fact that all of our specimens were derived from a single patient, we believe that our findings are robust, although their significance is also limited by the fact that they are derived from only one patient’s tumor. Various genes associated with neural development were differently expressed in the 144-13 and 144-4 sublines. For example, the mRNA levels of the proneural transcription factors OTX2, FOXG1, POU4F2, and NEUROD2, were higher in 144-13 (SCC) than in 144-4 (LCNEC). This is consistent with previous gene-expression profiles of a prostate SCC xenograft (2) and a transgenic model (16) in which relatively increased levels of mRNA associated with neural differentiation, such as ASCL1, were identified. It is interesting that the expression of both OTX2 and FOXG1 has been correlated with an anaplastic phenotype of medulloblastoma (32,33). In contrast, the 144-4 (LCNEC) xenograft expressed higher mRNA levels than 144-13 (SCC) of HES1 (a repressor of ASCL1), DLX5, DLX6, and SOX9 (involved in skeletal development). Notably, SOX-9 expression at the protein level was higher in the LCNEC xenografts than in the SCC xenografts and in the LCNEC and adenocarcinoma components of the donor tumor than in the SCC component of the tumor. This again suggests that LCNEC is indeed an intermediate step between the adenocarcinoma and the SCC components and that adaptation of a neural development program by the malignant cells may be implicated in the evolution of the phenotype. Further studies in additional patients’ tumors is necessary to strengthen these hypotheses.
In conclusion, the eight new subcutaneous xenograft models of prostate carcinoma we developed in SCID mice reflect the spectrum of NE carcinomas in prostate cancer. They will be valuable preclinical tools for elucidating the pathogenesis of NE carcinoma of the prostate and evaluating the efficacy of therapies in this increasingly recognized variant of prostate carcinoma.
Supplementary Material
Supplementary Table I. Antigen retrieval method, dilution and source of the antibodies used.
Supplementary Table II. Quantitative RT-PCR Primer Sequences.
ACKNOWLEDGMENTS
The authors wish to acknowledge the important contributions that the following individuals made to the work presented in this manuscript: Jun Yang; Jing-Fang Lu; Brittany Kleb; Eleni Efstathiou, MD PhD; John W.Davis, MD; Jiexin Zhang; Ivan P.Gorlov, PhD; Anh G. Hoang; Karen Phillips.
Grant sponsor: This research is supported in part by the National Institutes of Health through the Prostate Cancer SPORE Career Development Award Number P50 CA140388 from the National Cancer Institute, the M. D. Anderson’s Cancer Center Support Grant, CA016672 and the Prostate Cancer Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institute of Health.
REFERENCES
- 1.Li ZG, Mathew P, Yang J, Starbuck MW, Zurita AJ, Liu J, Sikes C, Multani AS, Efstathiou E, Lopez A, Wang J, Fanning TV, Prieto VG, Kundra V, Vazquez ES, Troncoso P, Raymond AK, Logothetis CJ, Lin SH, Maity S, Navone NM. Androgen receptor-negative human prostate cancer cells induce osteogenesis in mice through FGF9-mediated mechanisms. J Clin Invest. 2008;118(8):2697–2710. doi: 10.1172/JCI33093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clegg N, Ferguson C, True LD, Arnold H, Moorman A, Quinn JE, Vessella RL, Nelson PS. Molecular characterization of prostatic small-cell neuroendocrine carcinoma. Prostate. 2003;55(1):55–64. doi: 10.1002/pros.10217. [DOI] [PubMed] [Google Scholar]
- 3.Tetu B, Ro JY, Ayala AG, Johnson DE, Logothetis CJ, Ordonez NG. Small cell carcinoma of the prostate. Part I. A clinicopathologic study of 20 cases. Cancer. 1987;59(10):1803–1809. doi: 10.1002/1097-0142(19870515)59:10<1803::aid-cncr2820591019>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 4.Papandreou CN, Daliani DD, Thall PF, Tu SM, Wang X, Reyes A, Troncoso P, Logothetis CJ. Results of a phase II study with doxorubicin, etoposide, and cisplatin in patients with fully characterized small-cell carcinoma of the prostate. J Clin Oncol. 2002;20(14):3072–3080. doi: 10.1200/JCO.2002.12.065. [DOI] [PubMed] [Google Scholar]
- 5.Turbat-Herrera EA, Herrera GA, Gore I, Lott RL, Grizzle WE, Bonnin JM. Neuroendocrine differentiation in prostatic carcinomas. A retrospective autopsy study. Arch Pathol Lab Med. 1988;112(11):1100–1105. [PubMed] [Google Scholar]
- 6.Tanaka M, Suzuki Y, Takaoka K, Suzuki N, Murakami S, Matsuzaki O, Shimazaki J. Progression of prostate cancer to neuroendocrine cell tumor. Int J Urol. 2001;8(8):431–436. doi: 10.1046/j.1442-2042.2001.00347.x. discussion 437. [DOI] [PubMed] [Google Scholar]
- 7.Moore SR, Reinberg Y, Zhang G. Small cell carcinoma of prostate: effectiveness of hormonal versus chemotherapy. Urology. 1992;39(5):411–416. doi: 10.1016/0090-4295(92)90235-o. [DOI] [PubMed] [Google Scholar]
- 8.Amato RJ, Logothetis CJ, Hallinan R, Ro JY, Sella A, Dexeus FH. Chemotherapy for small cell carcinoma of prostatic origin. J Urol. 1992;147(3 Pt 2):935–937. doi: 10.1016/s0022-5347(17)37427-x. [DOI] [PubMed] [Google Scholar]
- 9.Wynn SS, Nagabundi S, Koo J, Chin NW. Recurrent prostate carcinoma presenting as omental large cell carcinoma with neuroendocrine differentiation and resulting in bowel obstruction. Arch Pathol Lab Med. 2000;124(7):1074–1076. doi: 10.5858/2000-124-1074-RPCPAO. [DOI] [PubMed] [Google Scholar]
- 10.Evans AJ, Humphrey PA, Belani J, van der Kwast TH, Srigley JR. Large cell neuroendocrine carcinoma of prostate: a clinicopathologic summary of 7 cases of a rare manifestation of advanced prostate cancer. Am J Surg Pathol. 2006;30(6):684–693. doi: 10.1097/00000478-200606000-00003. [DOI] [PubMed] [Google Scholar]
- 11.Guo CC, Zuo G, Cao D, Troncoso P, Czerniak BA. Prostate cancer of transition zone origin lacks TMPRSS2-ERG gene fusion. Mod Pathol. 2009;22(7):866–871. doi: 10.1038/modpathol.2009.57. [DOI] [PubMed] [Google Scholar]
- 12.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 13.Pounds S, Morris SW. Estimating the occurrence of false positives and false negatives in microarray studies by approximating and partitioning the empirical distribution of p-values. Bioinformatics. 2003;19(10):1236–1242. doi: 10.1093/bioinformatics/btg148. [DOI] [PubMed] [Google Scholar]
- 14.Zhang B, Schmoyer D, Kirov S, Snoddy J. GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics. 2004;5:16. doi: 10.1186/1471-2105-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vias M, Massie CE, East P, Scott H, Warren A, Zhou Z, Nikitin AY, Neal DE, Mills IG. Pro-neural transcription factors as cancer markers. BMC Med Genomics. 2008;1:17. doi: 10.1186/1755-8794-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Y, Ippolito JE, Garabedian EM, Humphrey PA, Gordon JI. Molecular characterization of a metastatic neuroendocrine cell cancer arising in the prostates of transgenic mice. J Biol Chem. 2002;277(46):44462–44474. doi: 10.1074/jbc.M205784200. [DOI] [PubMed] [Google Scholar]
- 17.Rapa I, Ceppi P, Bollito E, Rosas R, Cappia S, Bacillo E, Porpiglia F, Berruti A, Papotti M, Volante M. Human ASH1 expression in prostate cancer with neuroendocrine differentiation. Mod Pathol. 2008;21(6):700–707. doi: 10.1038/modpathol.2008.39. [DOI] [PubMed] [Google Scholar]
- 18.Foehr ED, Lorente G, Kuo J, Ram R, Nikolich K, Urfer R. Targeting of the receptor protein tyrosine phosphatase beta with a monoclonal antibody delays tumor growth in a glioblastoma model. Cancer Res. 2006;66(4):2271–2278. doi: 10.1158/0008-5472.CAN-05-1221. [DOI] [PubMed] [Google Scholar]
- 19.Scher HI, Sawyers CL. Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis. J Clin Oncol. 2005;23(32):8253–8261. doi: 10.1200/JCO.2005.03.4777. [DOI] [PubMed] [Google Scholar]
- 20.Attar RM, Takimoto CH, Gottardis MM. Castration-resistant prostate cancer: locking up the molecular escape routes. Clin Cancer Res. 2009;15(10):3251–3255. doi: 10.1158/1078-0432.CCR-08-1171. [DOI] [PubMed] [Google Scholar]
- 21.Attard G, Reid AH, A’Hern R, Parker C, Oommen NB, Folkerd E, Messiou C, Molife LR, Maier G, Thompson E, Olmos D, Sinha R, Lee G, Dowsett M, Kaye SB, Dearnaley D, Kheoh T, Molina A, de Bono JS. Selective inhibition of CYP17 with abiraterone acetate is highly active in the treatment of castration-resistant prostate cancer. J Clin Oncol. 2009;27(23):3742–3748. doi: 10.1200/JCO.2008.20.0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shah RB, Mehra R, Chinnaiyan AM, Shen R, Ghosh D, Zhou M, Macvicar GR, Varambally S, Harwood J, Bismar TA, Kim R, Rubin MA, Pienta KJ. Androgen-independent prostate cancer is a heterogeneous group of diseases: lessons from a rapid autopsy program. Cancer Res. 2004;64(24):9209–9216. doi: 10.1158/0008-5472.CAN-04-2442. [DOI] [PubMed] [Google Scholar]
- 23.Oesterling JE, Hauzeur CG, Farrow GM. Small cell anaplastic carcinoma of the prostate: a clinical, pathological and immunohistological study of 27 patients. J Urol. 1992;147(3 Pt 2):804–807. doi: 10.1016/s0022-5347(17)37390-1. [DOI] [PubMed] [Google Scholar]
- 24.van Haaften-Day C, Raghavan D, Russell P, Wills EJ, Gregory P, Tilley W, Horsfall DJ. Xenografted small cell undifferentiated cancer of prostate: possible common origin with prostatic adenocarcinoma. Prostate. 1987;11(3):271–279. doi: 10.1002/pros.2990110307. [DOI] [PubMed] [Google Scholar]
- 25.Pinthus JH, Waks T, Schindler DG, Harmelin A, Said JW, Belldegrun A, Ramon J, Eshhar Z. WISH-PC2: a unique xenograft model of human prostatic small cell carcinoma. Cancer Res. 2000;60(23):6563–6567. [PubMed] [Google Scholar]
- 26.True LD, Buhler K, Quinn J, Williams E, Nelson PS, Clegg N, Macoska JA, Norwood T, Liu A, Ellis W, Lange P, Vessella R. A neuroendocrine/small cell prostate carcinoma xenograft-LuCaP 49. Am J Pathol. 2002;161(2):705–715. doi: 10.1016/S0002-9440(10)64226-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Travis WD, Linnoila RI, Tsokos MG, Hitchcock CL, Cutler GB, Jr., Nieman L, Chrousos G, Pass H, Doppman J. Neuroendocrine tumors of the lung with proposed criteria for large-cell neuroendocrine carcinoma. An ultrastructural, immunohistochemical, and flow cytometric study of 35 cases. Am J Surg Pathol. 1991;15(6):529–553. doi: 10.1097/00000478-199106000-00003. [DOI] [PubMed] [Google Scholar]
- 28.di Sant’Agnese PAE L, Epstein JI, Helpap B, Humphrey PA, Montironi R, Rubin MA, Sakr WA, Tan PH. Neuroendocrine Tumors. In: Eble JNSG, Epstein JI, Sesterhenn IA, editors. World Health Organization Classification of Tumors Pathology and Genetics of Tumours of the Urinary System and Male Genital Organs. International Agency for Research on Cancer (IARC); Lyon: 2004. p. 207. [Google Scholar]
- 29.Jones MH, Virtanen C, Honjoh D, Miyoshi T, Satoh Y, Okumura S, Nakagawa K, Nomura H, Ishikawa Y. Two prognostically significant subtypes of high-grade lung neuroendocrine tumours independent of small-cell and large-cell neuroendocrine carcinomas identified by gene expression profiles. Lancet. 2004;363(9411):775–781. doi: 10.1016/S0140-6736(04)15693-6. [DOI] [PubMed] [Google Scholar]
- 30.Hiroshima K, Iyoda A, Shida T, Shibuya K, Iizasa T, Kishi H, Tanizawa T, Fujisawa T, Nakatani Y. Distinction of pulmonary large cell neuroendocrine carcinoma from small cell lung carcinoma: a morphological, immunohistochemical, and molecular analysis. Mod Pathol. 2006;19(10):1358–1368. doi: 10.1038/modpathol.3800659. [DOI] [PubMed] [Google Scholar]
- 31.Sun L, Sakurai S, Sano T, Hironaka M, Kawashima O, Nakajima T. High-grade neuroendocrine carcinoma of the lung: comparative clinicopathological study of large cell neuroendocrine carcinoma and small cell lung carcinoma. Pathol Int. 2009;59(8):522–529. doi: 10.1111/j.1440-1827.2009.02402.x. [DOI] [PubMed] [Google Scholar]
- 32.Adesina AM, Nguyen Y, Mehta V, Takei H, Stangeby P, Crabtree S, Chintagumpala M, Gumerlock MK. FOXG1 dysregulation is a frequent event in medulloblastoma. J Neurooncol. 2007;85(2):111–122. doi: 10.1007/s11060-007-9394-3. [DOI] [PubMed] [Google Scholar]
- 33.Adamson DC, Shi Q, Wortham M, Northcott PA, Di C, Duncan CG, Li J, McLendon RE, Bigner DD, Taylor MD, Yan H. OTX2 is critical for the maintenance and progression of Shh-independent medulloblastomas. Cancer Res. 70(1):181–191. doi: 10.1158/0008-5472.CAN-09-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table I. Antigen retrieval method, dilution and source of the antibodies used.
Supplementary Table II. Quantitative RT-PCR Primer Sequences.