Abstract
Using bioassay-guided fractionation, a new steroidal alkaloid, plakinamine M (1), and the known compound, plakinamine L (2), with a unique acyclic side chain, were isolated from the marine sponge Corticium sp. collected from New Britain, Papua New Guinea. The structures were determined on the basis of extensive 1D and 2D NMR and HRESIMS. The two compounds showed inhibition of Mycobacterium tuberculosis with MIC values of 15.8 and 3.6 μg/mL, respectively.
The marine sponge Corticium sp. is well known to produce steroidal alkaloid compounds with a wide range of bioactivities such as cytotoxic, immunomodulatory, anti-HIV, and nucleic acid cleaving and antimicrobial activities.1-3 The majority of these compounds possess a cyclic amine functionality on the side chain as well as an amino group on the A ring of a steroidal framework. As part of our continuing search for bioactive compounds, we examined a specimen of Corticium sp. whose MeOH extracts exhibited specific inhibition of Mycobacterium tuberculosis. Bioassay-guided fractionation of the MeOH extracts led to the isolation of two steroidal alkaloids with acyclic side chains. Herein, we report the isolation, structure elucidation and antimicrobial activities of these two compounds. This represents only the second report of acyclic side chains and the first report of antituberculosis activity for this family of compounds.
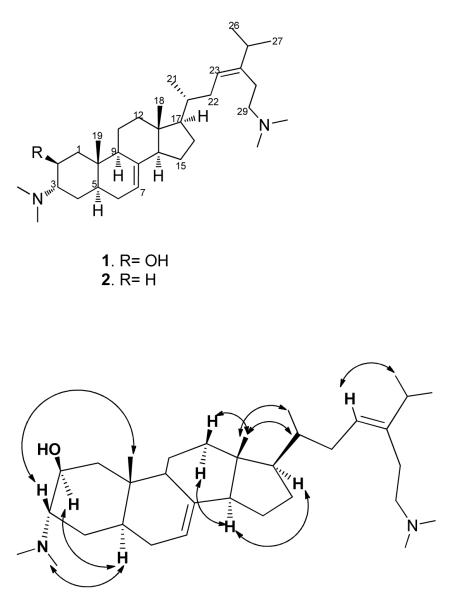
Compound 1 was obtained as colorless oil. The steroidal alkaloid nature of 1 was suggested by the molecular formula C33H58N2O obtained on the basis of HRESIMS and by 1H NMR signals which showed two characteristic angular methyls at δH 0.59 (H3-18, s) and δH 0.91 ppm (H3-19, s). The 1H NMR spectrum also exhibited three doublet methyls at δH 0.95 (H3-21, d, J = 6.6 Hz) and δH 1.05 ppm (H3-26/H3-27, d, J = 6.6 Hz), and four singlet N-methyls at δH 2.83, 2.86 and 2.91 ppm (H3-30, H3-31, and H3-32/H3-33, respectively).The olefinic protons at δH 5.20 (H-7, d, J = 3.6 Hz) and δH 5.36 ppm (H-23, t, J = 7.4 Hz) were assigned to two trisubstituted alkenes. The signals at δH 4.12, 3.26 and 3.05 ppm (H-2, H-3, and H-29), which were correlated by HSQC to carbon signals at δC 64.4, 68.0 and 57.5 ppm, respectively, were assigned as a carbinol, nitrogenated methine, and nitrogenated methylene, respectively. In addition there were several methine and methylene signals apparent in the 1H and 13C NMR spectra of 1. Using COSY correlations it was possible to define two large spin systems, each terminating in a trisubstituted alkene: C=CHCH2CHCH2CH(N(CH3)2)CH(OH)CH2 and C=CHCH2CH(CH3)CHCH2CH2CH, respectively corresponding to C-7, C-6, C-5, C-4, C-3, C-2, C-1 and C-23, C-22, C-20, C-21, C-17, C-16, C-15, C-14. Three smaller spin systems were also identified: C-9, C-11, C-12; C-25, C-26, C-27; and C-28, C-29. Connectivity between fragments was established through analysis of the HMBC spectrum. HMBC correlations from H-7 to C-9 and C-14, from H-1 to C-9, C-10 and C-19 and from H3-19 to C-1, C-9 and C-10 established the presence of fused rings A and B and the position of C-19. HMBC correlations from H-14 to C-9 and C-13, from H-12 to C-9, C-17 and C-18 and from H3-18 to C-12, C-13 and C-17 established rings C and D and the position of C-18. Finally, HMBC correlations from the olefinic proton H-23 to C-25 and C-28, from H2-25 to C-23 and C-28 and from H-28 to C-23 and C-25 established the structure of the side chain attached at C-17. Thus the planar structure of 1 was established, indicating that 1 was a previously undescribed steroidal alkaloid with an acyclic side chain, namely plakinamine M.
The relative configuration of 1 was determined from measurement of 1H-1H scalar coupling interactions and NOESY correlations (Figure 1). The large coupling interaction between H-2 and H-3 (J = 13.7 Hz) indicated axial orientations for both. H-2 displayed a NOESY correlation with H-5, whereas H-3 showed a correlation with H3-19 indicating a trans juncture for ring A/B. Similarly, H3-18 showed correlations with the axially located H-12b (δH 2.09, br d, J = 13.2 Hz), H-20 and H3-21, whereas H-14 showed correlations with H-12a and H-17 indicating a trans juncture for ring C/D. The C-23/C-24 double bond was assigned a Z geometry on the basis of NOESY correlations from H-23 to H-25, H3-26 and H3-27.
Figure 1.
Key NOESY interactions of 1.
The molecular formula of compound 2 was determined to be C33H58N2 based on an observed protonated pseudomolecular ion at m/z = 483.4691 Da by HRESIMS and the 13C NMR data. Analysis of the 1D and 2D NMR spectra revealed 2 to be the known compound plakinamine L.4
The inhibition of M. tuberculosis by the two compounds was evaluated. Plakinamine M (1) inhibited the growth of M. tuberculosis5,6 with a minimal inhibitory concentration of 15.8 μg/mL, whereas the MIC for plakinamine L (2) was 3.6 μg/mL. The presence of a hydroxy group at C-2 led to a decrease in potency of approximately five-fold, suggesting that the hydroxy group is somewhat deleterious for activity. Also, the inhibition activities of 1 and 2 against Candida albicans (fungus), Staphylococcus aureus (Gram-positive bacterium), Bacillus subtilis (Gram-positive bacterium) and Escherichia coli (Gram-negative bacterium) were assessed.7 The assay results showed that 1 and 2 were weakly active against C. albicans8 with partial inhibition at 50 μg/mL and displayed no activity against the bacteria (data not shown). This is the first report of M. tuberculosis inhibition for this family of compounds.
Experimental Section
General Experimental Procedures
Optical rotations were measured on a JASCO DIP-370 digital polarimeter. UV spectra were acquired in spectroscopy grade MeOH using a Hewlett-Packard 8452A diode array spectrophotometer. NMR data were collected using a Varian INOVA 500 (1H 500 MHz, 13C 125 MHz) NMR spectrometer with a 3 mm Nalorac MDBG probe with a z-axis gradient and utilized residual solvent signals for referencing (δH 3.30 ppm, δC 49.0 ppm for CD3OD). High-resolution ESIMS analyses were obtained using a Q-Tof Micro mass spectrometer (Waters, Inc). Analytical and semipreparative HPLC was accomplished utilizing a Beckman System Gold 126 solvent module equipped with a 168 photodiode array detector.
Sponge Material
Corticium sp., was collected by SCUBA at New Britain, Papua New Guinea (S 04° 08.967’ E 151° 33.832’) and frozen immediately after collection. PNG07-12-171 is an undescribed species of Corticium (Homosclerophorida, Plakinidae). This Corticium sp. is encrusting, with a black exterior and slightly lighter interior. Candelabra form a surface layer and line the aquiferous system; the choanosome is densely packed with large tetracts. There appear to be several species of Corticium in the Indo-Pacific that do not match published species. The present undescribed species of Corticium differs from other undescribed species in our own collections by having abundant large tetracts in the choanosome and different chemical composition. The sample was identified by M.K.H. A voucher specimen is maintained at the University of Utah under accession number PNG07-12-171.
Extraction and Isolation
The frozen sponge (190 g wet wt) was exhaustively extracted with MeOH to yield 1.1 g of extract that was then separated on HP20SS resin using a gradient of H2O to IPA in 25% steps, and a final wash of 100% MeOH, yielding five fractions. The third fraction (1:1 H2O/IPA) was active and further fractionationed by C18 flash chromatography using a stepped gradient from 0-100% aqueous MeOH to give four fractions after combination. The active fraction eluted with 100% MeOH (0.1% TFA) and was further chromatographed by HPLC using a Luna C18 column (250 × 10 mm; 5 mm; Phenomenex, Inc.) employing a linear gradient from 5% CH3CN/95% H2O (+0.1% TFA) to 100% CH3CN over 45 min at 4 mL/min to yield compounds 1 (5.0 mg; tR = 25 min) and 2 (3.3 mg; tR = 28 min).
Plakinamine M (1)
colorless oil; [a]20D −40 (c 0.1, MeOH); UV (MeOH) λmax (log ε) 206 (4.30) nm; 1H and 13C NMR data Table 1; HRESIMS m/z 499.4626 [M+H]+ (calcd for C33H59N2O, 499.4622).
Table 1.
NMR Spectroscopic Data for Plakinamine M (1) in CD3ODa
| position | δC, type | δH (J in Hz) | COSY | HMBCb |
|---|---|---|---|---|
| 1 | 43.4, CH2 | 1.46, m; 1.91, m | H-2 | 2, 3, 5, 9, 10, 19 |
| 2 | 64.4, CH | 4.12, ddd (5.5, 8.5, 13.7) |
H-1, H-3 | 1, 3, 4, 10 |
| 3 | 68.0, CH | 3.26, ddd (5.5, 8.3, 13.7) |
H-2, H-4 | 1, 2, 4, 3-NCH3 |
| 4 | 24.1, CH2 | 1.80, m; 1.91, m | H-3, H-5 | 2, 3, 6, 10 |
| 5 | 36.1, CH | 1.67, m | H-4, H-6 | 4, 6, 9, 10, 19 |
| 6 | 29.7, CH2 | 1.79, m; 1.91, m | H-5, H-7 | 4, 10 |
| 7 | 118.2, CH | 5.20, br d (3.6) | H-6 | 5, 6, 9, 14 |
| 8 | 140.8, C | |||
| 9 | 50.8, CH | 1.89, m | H-11 | 10, 11, 12 |
| 10 | 35.3, C | |||
| 11 | 22.4, CH2 | 1.55, m; 1.62, m | H-9, H-12 | 9, 10, 12, 13 |
| 12 | 40.5, CH2 | 1.29, m; 2.09, br d (13.2) |
H-11 | 9, 11, 13, 14, 17, 18 |
| 13 | 44.4, C | |||
| 14 | 56.0, CH | 1.88, m | H-15 | 9, 13 |
| 15 | 23.7, CH2 | 1.48, m; 1.53, m | H-14, H-16 | 8, 14, 16 |
| 16 | 28.8, CH2 | 1.35, m; 1.95, m | H-15, H-17 | 13, 15, 17 |
| 17 | 56.9, CH | 1.31, m | H-16, H-20 | 12, 13, 16, 20, 21 |
| 18 | 12.1, CH3 | 0.59, s | 12, 13, 17 | |
| 19 | 17.4, CH3 | 0.91, s | 1, 5, 9, 10 | |
| 20 | 38.2, CH | 1.48, m | H-17, H-21, H-22 |
16, 17 |
| 21 | 19.1, CH3 | 0.95, d (6.6) | H-20 | 17, 20, 22 |
| 22 | 34.9, CH2 | 1.82, m; 2.15, m | H-20, H-23 | 17, 20, 21, 23 |
| 23 | 126.1, CH | 5.36, br t (7.4) | H22 | 20, 22, 25, 28, 29 |
| 24 | 141.1, C | |||
| 25 | 35.5, CH | 2.27, q (6.6) | H-26, H-27 | 23, 24, 26, 27, 28 |
| 26/27 | 22.2, CH3 | 1.05, d (6.6) | H-25 | 24, 25, 27/26 |
| 28 | 25.6, CH2 | 2.45, m | H-29 | 23, 24, 25, 29 |
| 29 | 57.5, CH2 | 3.05, t (9.0) | H-28 | 24, 28, 29-NCH3 |
| 29-NCH3 | 43.0, CH3 | 2.91, s | 29, 29-NCH3 | |
| 29-NCH3 | 43.1, CH3 | 2.91, s | 29, 29-NCH3 | |
| 3-NCH3 | 37.9, CH3 | 2.83, s | 3, 3-NCH3 | |
| 3-NCH3 | 41.9, CH3 | 2.86, s | 3, 3-NCH3 |
Spectra were recorded at 500 MHz for 1H and 125 MHz for 13C.
HMBC correlations are from proton(s) stated to the indicated carbon.
Antimicrobial Bioassays
Mycobacterium tuberculosis strain H37Ra (ATCC 25177) was obtained from American Type Culture Collection (ATCC). Mycobacterium tuberculosis was grown in 7H9 media (Becton Dickinson) with albumin/dextrose/catalase (ADC) enrichment (Remel). Rifampicin (Sigma Aldrich) served as positive control. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was obtained from Thermo Fisher (Thermo Fisher Scientific Inc.). The inoculum for test plates came from a premade frozen bacterial pellet of which a 100 μL aliquot was mixed with 20 mL fresh media for inoculation of test plates resulting in a final concentration of 5×105 CFU/mL. The bacterial suspension was then dispensed into 96-well plates containing 1 μL of test compound in quadruplicate wells, as well as control wells containing DMSO only (negative control) or rifampicin (positive control 250 ng/mL). After 4 days of incubation at 37 °C 11 μL of MTT4 was added followed by further incubation for 12 h. After resuspension of the formazan precipitate with a solution containing 50% H2O / 45% DMF / 5% SDS absorbance at 570 nm was measured using a Biorad Model 450 microtiter plate reader (Biorad). After subtraction of uninoculated media-only wells from all data percent inhibition was derived using the following relationship:
Supplementary Material
Acknowledgment
This work was supported by NIH grant 5U01TW006671-08. Funding for the Varian INOVA 500 MHz NMR spectrometer was provided through NIH grant RR06262. We gratefully acknowledge Dr. Z. Lin (Medicinal Chemistry Department University of Utah) for his assistance in the preparation of this manuscript.
Footnotes
Supporting Information Relevant NMR spectra used in the identification of plakinamine M and the collection picture of the Corticium sp. sponge are presented in the Supporting information. This material is available free of charge via the Internet at http://www.acs.org.
References and Notes
- (1).Jurek J, Scheuer PJ, Kelly-Borges M. J. Nat. Prod. 1994;57:1004–1007. doi: 10.1021/np50109a022. [DOI] [PubMed] [Google Scholar]
- (2).De Marino S, Iorizzi M, Zollo F, Roussakis C, Debitus C. Eur. J. Org. Chem. 1999;3:697–701. [Google Scholar]
- (3).Lee HS, Seo Y, Rho JR, Shin J, Paul V. J. Nat. Prod. 2001;64:1474–1476. doi: 10.1021/np0101649. [DOI] [PubMed] [Google Scholar]
- (4).Aknin M, Rudi A, Kashman Y, Vacelet J, Gaydou EM. Nat. Prod. Commun. 2010;5:33–34. [PubMed] [Google Scholar]
- (5).Koch M, Bugni TS, Pond CD, Sondossi M, Dindi M, Piskaut P, Ireland CM, Barrows LR. Planta Med. 2009;75:1326–1330. doi: 10.1055/s-0029-1185687. [DOI] [PubMed] [Google Scholar]
- (6).Foongladda S, Roengsanthia D, Arjrattanakool W, Chuchottaworn C, Chaiprasert A, Franzblau SG. Int J. Tuberc. Lung Dis. 2002;6:1118–1122. [PubMed] [Google Scholar]
- (7).Ferraro MJ, Craig WA, Dudley MN, Eliopoulos GM, Hecht DW, Hindler J, Reller LB, Sheldon AT, Swenson JM, Tenovar FC, Testa RT, Weinstein MP, Winkler MA. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard. 5th Edition NCCLS Press; Wayne: 2000. [Google Scholar]
- (8).Galgiani JN, Bartlett MS, Mahmoud A. Ghannoum MA, Espinel-Ingroff A, Lancaster MV, Odds FC, Pfaller MA, Rex JH, Rinaldi MG, Walsh TJ. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standard. NCCLS Press; Wayne: 1997. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.