Abstract
NUP98 is among the most promiscuously translocated genes in hematological diseases. Among the 28 known fusion partners, there are two categories: homeobox genes and non-homeobox genes. The homeobox fusion partners are well-studied in animal models, resulting in HoxA cluster overexpression and hematological disease. The non- homeobox fusion partners are less well studied. We created transgenic animal models for three NUP98 fusion genes (one homeobox, two non- homeobox), and show that in this system, the NUP98-homeobox fusion promotes self-renewal and aberrant gene expression to a significantly greater extent. We conclude that homeobox partners create more potent NUP98 fusion oncogenes than do non-homeobox partners.
Keywords: NUP98, homeobox, HOX, leukemia, translocation
Introduction
Chromosomal translocations in human leukemia are a source of novel oncogenes termed fusion genes, which encode fusion proteins combining the properties of two distinct proteins. Some genes are involved in multiple oncogenic fusion proteins with distinct partner genes, indicating that the properties of that gene are generally oncogenic if deregulated, or that there are multiple mechanisms by which oncogenic fusion proteins involving the gene can act. The Nucleoporin 98 (NUP98) gene encodes a protein which is normally involved in nuclear transport, but is also recurrent in many different chromosomal translocations in myelodysplastic syndrome (MDS) and acute leukemia[1]. It is among the most promiscuous known fusion gene partners, having been identified in fusion genes with 28 distinct partner genes. The estimated frequency of NUP98 translocations in human leukemia has been recently revised upward, with modern molecular techniques leading to superior detection of these translocations[2, 3]. Further, the NUP98-JARID1A fusion represents a functionally distinct subclass of AML, suggesting that other NUP98 fusions may do the same [2]. Understanding the leukemic activity of the various NUP98 fusions is therefore of increasing importance.
NUP98 fusion partner genes can be broadly broken into two groups: homeobox genes and non-homeobox genes. Homeobox genes are transcription factors defined by the conserved “homeodomain” DNA-binding domain, and this DNA-binding domain is conserved in the fusion proteins with NUP98, of which there are at least ten [1]. This retention of the DNA-binding domain suggests a transcriptional regulatory mechanism for the NUP98-homeobox fusion proteins. Indeed in the case of the NUP98-HOXA9 (NHA9) fusion, this has been directly shown[4]. For the non-homeobox fusion proteins, of which there are at least eighteen [1], an overarching mechanism has been slower to emerge. Recent work has suggested that regulation of gene expression by chromatin reading and/or writing via indirect DNA binding is the mechanism for at least some non-homeobox fusion partners[1, 5, 6], although not all eighteen have the requisite domains for this activity.
We have previously reported the generation of the NUP98-HOXD13 (NHD13) transgenic mouse[7]. Created using the cloned fusion gene from a human MDS under a transgenic vav promoter[8], the NHD13 mouse develops an MDS by the age of five months (100% penetrance) and an acute leukemia from the age of six months (60% penetrance). The only other reported example of a transgenic NUP98 fusion is also a NUP98-homeobox fusion; the NHA9 transgenic mouse is also reported to develop acute myeloid leukemia (AML), albeit at a lower penetrance and later age (22% by 15 months)[9]. No transgenic models of NUP98-non- homeobox fusion genes have been reported, but there are numerous reports of retrovirally-transduced bone marrow models of these genes, which generally result in myeloproliferative neoplasms or AML [5, 6, 10].
We sought to investigate the relative oncogenic activity of homeobox and non-homeobox NUP98 fusion genes using similar techniques. To this end, we selected three NUP98 fusion genes to study: NUP98-HOXD13 (NHD13) [11], NUP98-TOP1 (NT)[12] and NUP98-RAP1GDS1 (NRG)[13]. All of these fusion genes retain the 5′ portion of NUP98, including the FG repeats responsible for karyopherin docking during nuclear transport[14] (Figure 1A). The 3′ portion of HOXD13 that is retained includes the homeodomain, a DNA binding domain. The 3′ portion of TOP1 (Topoisomerase 1) that is retained includes the core, linker and catalytic domains used in the protein’s normal function of unwinding DNA superstructures [15]. The 3′ portion of the NRG fusion retains the entirety (bar the first methionine) of RAP1GDS1 (RAP1 GTP-GDP dissociation stimulator 1), including the armadillo domain. RAP1GDS1 encodes a protein known as smgGDS, which is involved in guanine nucleotide exchange activity[16]. We created transgenic mouse models expressing NT and NRG from the same vav promoter used in the creation of the NHD13 mouse. We compare phenotype, aberrant gene expression profiles and abnormal self-renewal activity in each of these models to determine the relative oncogenic potency of each of these genes.
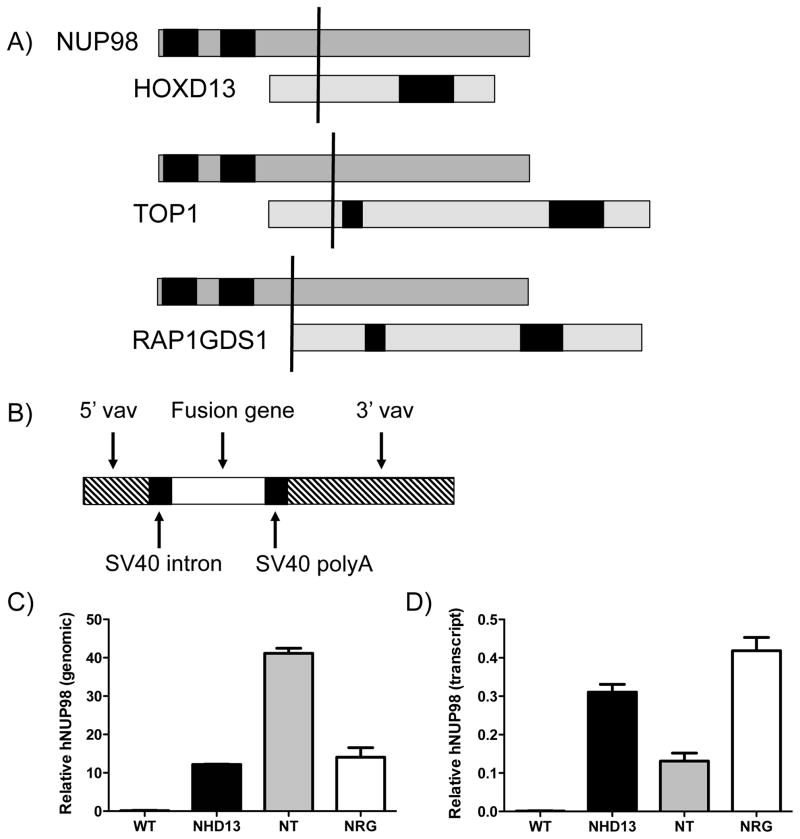
Figure 1. Creation of NT and NRG transgenic mice.
A) Schematic showing the retained domains of each fusion partner gene. In each case the upper gene is NUP98, with the GLFG repeat domains shown in black, and the lower gene is the fusion partner. In all fusions, the 5′ portion of NUP98 is fused to the 3′ portion of the partner. For HoxD13 the black box represents the homeodomain, for Top1 the boxes represent (left to right) the core, linker and catalytic domains, and for Rap1GDS1 the black boxes represent the armadillo domains. The vertical black line represents the fusion point. B) Structure of the vav vector used to generate the transgenic mice. C) Real-time PCR data indicating the relative signal obtained for a transgene-specific NUP98 amplicon from bone marrow obtained from each of the three mouse lines and wild type mice. D) Real- time PCR data indicating the signal (relative to HPRT signal) obtained for a transgene-specific NUP98 amplicon from LK cells obtained from each of the three mouse lines and wild type mice.
Material and methods
Generation of transgenic mice
The NUP98-HOXD13 mice have been described previously (Lin et al 2005). We used the pZVNHD13 vector used in the generation of these mice as the basis for generating the NUP98-TopoisomeraseI (NT) and NUP98-RAP1GDS1 (NRG) mice. Full- length NT and NRG cDNAs were PCR amplified and cloned into the pZVNHD13 vector, replacing the NHD13 sequence in the vector. These resultant vectors (pZVNT and pZVNRG) were sequenced to verify the constructs. The pZVNT and pZVNRG plasmids were digested with PmeI, and the insert containing 5′ and 3′ vav regulatory elements and the respective fusion gene cDNA were purified by agarose gel electrophoresis and Qiagen gel purification, using the manufacturer’s recommendations. The construct was microinjected into zygotes obtained from C57Bl6 mice. Founders were identified using a human NUP98 probe, and offspring were genotyped by PCR amplification of the respective transgene from tail biopsy DNA. Lines were maintained by mating with wild-type C57bl6 mice. All animal experiments were approved by the Animal Care and Use Committee at NCI or the Animal Ethics Committee at Monash University.
Real time PCR
Total DNA was prepared from tail biopsies using standard techniques. Total RNA was prepared from FACS-sorted LK cells by using the Trizol (Invitrogen) reagent according to the manufacturer’s instructions. In the case of the hNUP98 PCR, RNA was DNAse-treated using the Turbo DNA- free kit (Ambion Life Technologies). cDNA was transcribed from 1 ug of RNA using the Roche Transcriptor kit according to the manufacturer’s instructions. PCR reactions using the Promega GoTaq mastermix were performed on a LightCycler480 (Roche). PCR cycling conditions included an initial denaturation (95°C 60 sec) followed by 95°C 10 sec, 55°C 10 sec and 72°C 30 sec. Data were analyzed using the Roche LightCycler 480 software.
FACS Analysis
BM samples were flushed from femora and tibiae into PBS containing 2% fetal bovine serum (FBS). Antibodies and other reagents for staining were obtained from BD Pharmingen (San Diego, CA): B220 and CD4 as fluorescein isothiocyanate (FITC) conjugates; SCA (E13-161.7) and CD8 (53-6.7) as phycoerythrin (PE) conjugates; CD4 (RM4-5) and c-KIT (2B8) as allo-phycocyanin (APC) conjugates; and biotinylated Mac-1 (M1/70), Gr-1 (RB6-8C5), Ter-119, B220 (RA3-6B2) and CD3 (145-2C11). Second-stage reagents were Streptavidin (SAv) Peridinin Chlorophyll Protein Complex (PerCP). Cell viability was measured by exclusion of propidium iodide (PI; Sigma, St Louis, CA). FACS analysis was performed using a FACSCalibur or LSR-II instrument (BD Biosciences). Cell sorting was performed using a FACSAria instrument (BD Biosciences).
Blood Cell Counts
Blood samples were collected into EDTA-coated tubes and full blood counts were determined on an Advia 120 Automated Hematology Analyzer.
Hematopoietic Progenitor Assays
BM cells were seeded at a density of 5 × 104 cells per 35 mm dish in semi-solid agar for GM colony growth. Cultures were incubated at 37°C in 10% CO2 for 7 days, stained with acetyl-cholinesterase and counted. Replating was performed using unstained plates. Colonies were extracted using a pipette tip, the cells disaggregated in 50 ul of PBS, and each individual colony was then replated in semi- solid agar in the same conditions. For replating assays, BM cells were seeded at a density of 105 cells per 35 mm dish in methylcellulose. Cultures were incubated at 37°C in 10% CO2 for 7 days and colonies counted. Whole plates were harvested and washed, and 105 cells were seeded into fresh methycellulose dishes.
Results
Generation of NT and NRG transgenic mice
We have previously reported the cloning of the NUP98-TOP1 (NT)[12] and NUP98-RAP1GDS1 (NRG)[13] fusion gene cDNAs from patients with therapy-related myelodysplastic syndrome bearing the t(11;20)(p15;q11) and T-cell acute lymphocytic leukemia bearing the t(4;11)(q21;p15), respectively (Figure 1A). We cloned these cDNAs into the identical vav vector used for generation of NHD13 transgenic mice[7] (Figure 1B). Following pronuclear injection, we identified four founder mice for the NT transgene which were able to transmit the transgene through the germline, as determined by PCR genotyping. Three founders were identified for the NRG line which were capable of transmitting the transgene to their offspring. We examined both transgenes to ensure they were expressed, and found them to be expressed exclusively in the hematopoietic compartment (data not shown). To narrow our study, we chose one founder from each of the NT and NRG lines to follow, selecting the one with the highest expression in the marrow.
We examined the relative copy number of each transgene to ensure comparability between the lines. This was achieved by Q-PCR for the shared portion of the human NUP98 gene, using intron-spanning amplicons to ensure that amplification was restricted to the transgenes. To verify this, the endogenous murine Nup98 gene did not amplify from the wild-type mouse. This Q-PCR was normalised using an unrelated endogenous genomic Q-PCR (of the Gata4 locus) The NT line contained approximately four times the number of copies of the transgene as the NRG and NHD13 lines (Figure 1C). To investigate transgene expression level, we performed Q-PCR with primers specific for the retained portion of the human NUP98 common to all three transgenes. We examined expression in Lin- Kit+ (LK) progenitors. Species specificity was confirmed by the lack of amplification from wild type mice. The transgenes in NHD13 and NRG mice were expressed at comparable levels, with the NT mice demonstrating approximately half the level of expression.
Phenotype of NT and NRG mice
NHD13 mice developed hematological phenotype of anemic, lymphopenia and thrombocytopenia by the age of 6 months[7]. In contrast, NT and NRG mice exhibit normal blood counts at six months of age (Figure 2A), and no anemia, lymphopenia or thrombocytopenia emerge as NT and NRG mice are aged out to 600 days (data not shown).
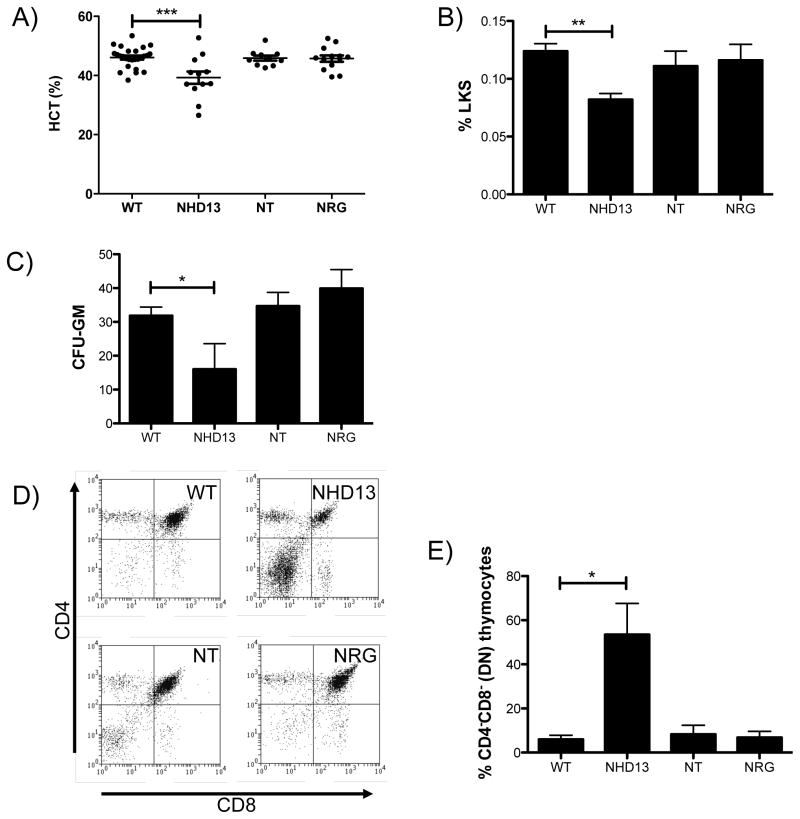
Figure 2. Pre-leukemic phenotype of NT and NRG mice.
A) Hematocrit of mice of each genotype. B) Percentage of Lin- Kit+ Sca+ cells in the bone marrow of mice of each genotype (n=5). C) Colonies formed in CFU-GM assays per 5 × 104 cells seeded (n=6). D) Representative CD4 vs CD8 FACS plot from thymus of mice of each genotype. E) Quantitation of CD4− CD8− cells from thymus of mice of each genotype (n=3). * indicates p<0.05; ** indicates p<0.01; *** indicates p<0.001. All assays performed on mice at 5 months of age.
NHD13 mice have decreased progenitor cells as defined either by FACS staining for the LKS population or functionally by colony assay (CFU-GM) due to increased apoptosis [17]. Consistent with the normal blood counts, numbers of LKS (Figure 2B) and CFU-GM (Figure 2C) in the NT and NRG mice were normal. Examination of mature lineages and progenitors at 12 months of age also revealed no differences between NT or NRG mice and wild type littermates (data not shown).
The thymic CD4/CD8 profiles of NT and NRG mice were normal (Figure 2D&E), indicating that differentiation is not impaired in these cells, in contrast to the NHD13 phenotype which shows an increase in CD4/CD8 double negative (DN) thymocytes, consistent with previous results [18].
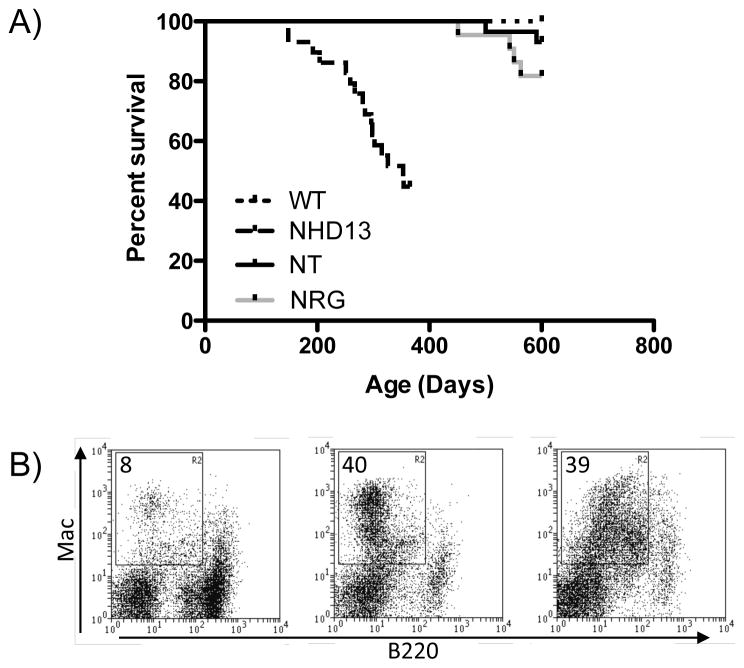
Aged cohorts of each strain were monitored for the development of acute leukemia. As previously reported, NHD13 mice developed leukemia beginning prior to 6 months of age, with 62% (13/21) having established leukemia by the age of 12 months. Of these, nine (43%) leukemias had features of AML and four (19%) had features of T-ALL. The remaining eight mice that survived to 12 months were severely cytopenic but without apparent acute disease, in keeping with the known phenotype of NHD13 mice. In contrast, by 600 days of age only 2/29 (7%) NT and 4/22 (18%) NRG mice developed AML (Figure 3A), as determined by infiltration of spleen and marrow with Mac-1+ cells (Figure 3B). This disease onset late, with the earliest occurring at 451 days of age. All wild type mice remained healthy through 600 days. These data indicate that NT and NRG have oncogenic potential, but are weaker oncogenes than NHD13.
Figure 3. Leukemic phenotype of NT and NRG mice.
A) Survival curve for cohorts of wild type (n=25), NHD13 (n=21), NT (n=29) and NRG (n=22) mice. NHD13 mice were not monitored beyond 12 months of age. Other cohorts were monitored for 600 days. B) Representative FACS profiles of leukemias shows Mac vs B220 profile of spleen. Image on the left is a representative wild-type mouse, middle image is an NT leukemia, and image on the right is an NRG leukemia.
NT and NRG fusion proteins are less potent inducers of self- renewal and homeobox gene expression
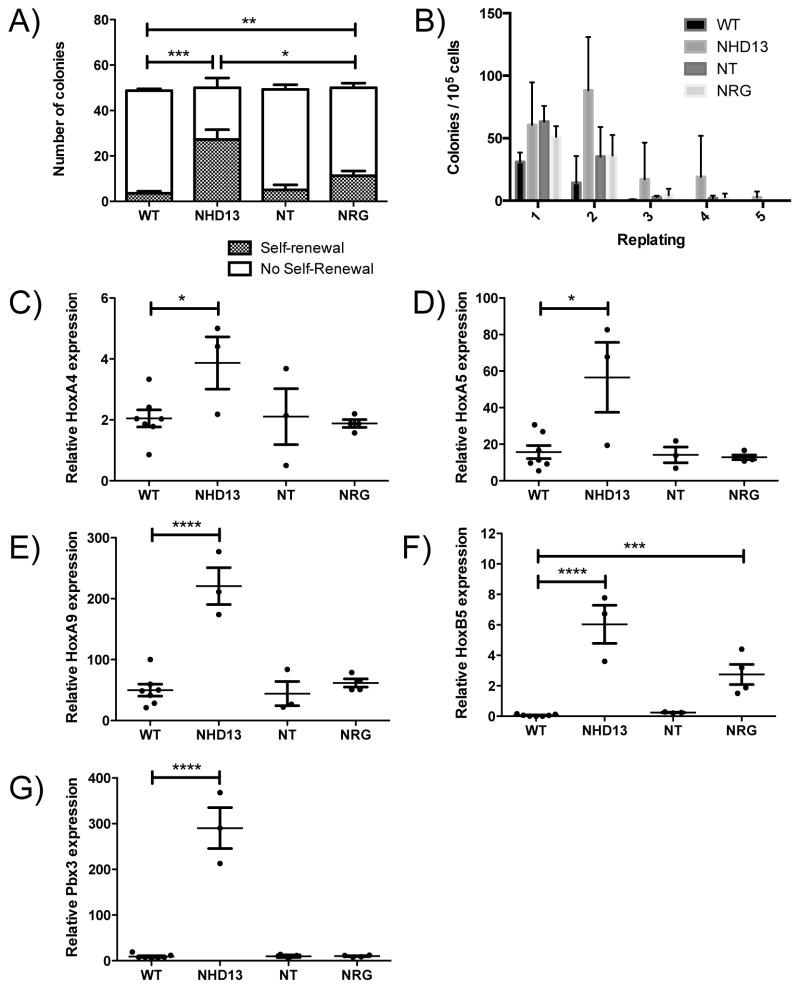
To investigate the self-renewal properties of these progenitor cells, we performed serial replating of individual CFU-GM colonies. NHD13 CFU-GM colonies were able to form secondary colonies with a frequency of ~ 50% while wild type CFU-GMs replated with a frequency of ~ 2%, indicating a clear increase in self-renewal in NHD13 CFU-GMs in this assay. NRG CFU-GMs had a replating efficiency of 10%, while NT CFU-GM colonies showed no increased replating ability above wild type (Figure 4A). We also conducted standard replating experiments in which 105 cells were replated from each plate. In this assay, NHD13 colonies replated indefinitely as previously shown [7], both NT and NRG colonies replated on the fourth but not the fifth replating, while wild type cells were not able to produce colonies after two replatings (Figure 4B).
Figure 4. The self-renewal ability and Hox gene expression signature of NT and NRG mice.
A) Number of colonies that were successfully re-plated, from the 50 colonies that were picked from each plate (n=6). B) Number of colonies obtained from successive replating of 105 cells (n=3). C–F) Expression of HoxA4, HoxA5, HoxA9, HoxB5 and Pbx3, shown relative to the expression of Hprt, is shown for mice of each genotype. * indicates p<0.05; ** indicates p<0.01; *** indicates p<0.001 ; **** indicates p<0.0001. All assays performed on mice at 5 months of age.
Gene expression profiling of NHD13 progenitor (Lin- Kit+; LK) cells indicated that NHD13 induces high levels of expression of numerous Hox genes[17]. We examined the relative expression in NT and NRG LK cells of four Hox genes that are overexpressed in NHD13 LK cells (Hoxa4, Hoxa5, Hoxa9 and Hoxb5), to determine if failure to induce Hox gene expression could be a molecular explanation for the weak oncogenic potential of NT and NRG. Indeed, only Hoxb5 was significantly overexpressed in NRG LK cells, and none of the four genes were overexpressed in NT LK cells (Figure 4B–E). In addition, expression of the homeobox gene Pbx3 [19], an important co-factor for the Hox genes[20] and which is also overexpressed in NHD13 LK cells, was not increased in NT or NRG LK cells (Figure 4F).
Discussion
NUP98 forms fusion genes with 28 known partner genes in human MDS and leukemia. These partner genes fall into two clear categories; ten are homeobox transcription factors, and the remainder comprise a set of unrelated genes, none of which are known transcription factors[1]. The only noted commonality they share is the presence of at least one coiled-coil domain[21].
In the present study, we insert two non-homeobox NUP98 fusion genes, NT and NRG, into a transgenic system identical to that previously used to generate the NHD13 mouse model, and find that the two non-homeobox fusions are substantially less potent in this system. While oncogenic potential in transgenic mice can affected by the type of promoter, mouse strain and transgene copy number, these factors are not likely to be resulting in the phenotypic differences seen in this study because all strains were generated using the same vav promoter construct, copy number was similar or higher in the two non- homeobox models, expression at the RNA level was comparable in all three models, and all strains were generated and maintained on a C57BL/6J background.
Phenotypic disease was much less penetrant in both the NT and NRG mice. There was no evidence of a “pre- malignant” phenotype as there is in the NHD13 mice, with peripheral blood counts remaining normal throughout life in the majority of animals. In late age, 2 of 29 NT mice and 4 of 22 NRG mice developed acute leukemia. This penetrance was greatly reduced in comparison to the 62% of acute leukemia at 365 days of age in the NHD13 cohort. Onset of the disease was also much later in the NT and NRG mice, with no disease apparent prior to 400 days of age. Taken together, we conclude that NT and NRG are less potent oncogenes in this assay than is NHD13.
The cellular effect of most transcription factor fusion proteins is to block differentiation and increase self-renewal[22]. A standard assay for hematopoietic progenitor cell self-renewal is agar colony replating. We use a colony recloning assay as well as the more standard replating assay. Both assays showed that NRG myeloid progenitors have increased self-renewal capacity, but significantly less than those from NHD13 mice. Cells from NT mice showed aberrant self-renewal ability in the standard assay but not in the colony recloning assay, suggesting a mild self-renewal advantage. We conclude that both NT and NRG cells have an increased renewal capability compared to wild type cells, but not to the same extent as do NHD13 cells. NUP98-homeobox gene fusions have been widely demonstrated to upregulate expression of the HOXA cluster [1, 19], in a manner similar to many MLL fusion genes [23]. Recently, evidence has been presented that NUP98-NSD1[5], -PHF23 and -JARID1A[6] cause HOXA cluster overexpression by induction of histone modification changes. Here, we show that two other non- homeobox fusion partners of NUP98 (RAP1GDS and TOP1) have much weaker oncogenic potential than the homeobox fusion partner HOXD13. We propose that this is due to an inability of these two fusions to activate the HOXA gene cluster required for enhanced self-renewal of progenitors. HOX genes are expressed at high levels in hematopoietic precursors, and expression is gradually extinguished as cells mature (reviewed in [24]). Increased expression of HOX genes is common in both MDS and AML, and is a driving factor in aberrant self-renewal in these diseases[25]. The measurably increased self-renewal induced by NRG compared to NT is consistent with the increased expression of at least one Hox gene (Hoxb5) in NRG but not NT progenitors.
Our results differ from those of a previous study which used bone marrow transduction to enforce expression of NT [10]. The mice in this study developed AML with 100% penetrance and a median survival of 233.5 days. Possible explanations for the enhanced oncogenic potential in the retroviral system compared to our transgenic system include higher expression of the fusion gene. We have made every effort to ensure that the transgenes are expressed at similar levels, but can not rule out the possibility that different levels of each fusion protein may be present. A more attractive hypothesis to explain the difference between the transgenic and retroviral systems is that cooperative oncogenic hits arise from retroviral insertion effects. This latter hypothesis is supported by the mono- or oligoclonal basis of disease in the original study, and that at leastone such cooperative hit resulting from retroviral insertion was identified [10].
With a lesser increase in self-renewal than that of NHD13 mice, NT and NRG mice have a decreased propensity to develop leukemia. The lower penetrance and increased latency of disease suggests a greater requirement for additional mutations, which may combine with NT or NRG to increase self- renewal, alongside mutations required to overcome other barriers to transformation. Alternatively, the greater amount of aberrant self-renewal in the NHD13 mice may create an environment in which these latter mutations occur with higher frequency, while the environment in NT or NRG mice may promote these mutations at a lesser rate. While the precise oncogenic mechanism remains to be determined, we have shown that, in this transgenic system, there are different levels of oncogenic potency among NUP98 fusion oncogenes.
Acknowledgments
We thank Lionel Feigenbaum of the NCI Transgenic core facility for the generation of the mice, animal facility staff for animal care, and Geza Paukovics, Michael Thomson and Jeanne Le Massurier for flow cytometric sorting. This research was supported by the National Health and Medical Research Council of Australia, the Cancer Council of Victoria and the Intramural Research Program of the National Institutes of Health, National Cancer Institute.
Footnotes
Conflict of interest statement
CIS and PDA receive royalites from the National Institutes of Health Technology Transfer Office for the invention of the NUP98-HOXD13 mice. The remaining authors declare no competing financial interests.
Authors’ Contribution
JS and CS performed experiments and prepared the first draft. DJC, PDA and CS conceived and designed the study. DJH and AD provided critical reagents and revised the article. CS prepared the final draft of the manuscript. All authors edited and approved the final manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gough SM, Slape CI, Aplan PD. NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood. 2011;118:6247–57. doi: 10.1182/blood-2011-07-328880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Rooij JD, Hollink IH, Arentsen-Peters ST, van Galen JF, Berna Beverloo H, Baruchel A, et al. NUP98/JARID1A is a novel recurrent abnormality in pediatric acute megakaryoblastic leukemia with a distinct HOX gene expression pattern. Leukemia. 2013 doi: 10.1038/leu.2013.87. [DOI] [PubMed] [Google Scholar]
- 3.Hollink IH, van den Heuvel-Eibrink MM, Arentsen-Peters ST, Pratcorona M, Abbas S, Kuipers JE, et al. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood. 2011;118:3645–56. doi: 10.1182/blood-2011-04-346643. [DOI] [PubMed] [Google Scholar]
- 4.Kasper LH, Brindle PK, Schnabel CA, Pritchard CE, Cleary ML, van Deursen JM. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol Cell Biol. 1999;19:764–76. doi: 10.1128/mcb.19.1.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9:804–12. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]
- 6.Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, et al. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459:847–51. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin YW, Slape C, Zhang Z, Aplan PD. NUP98-HOXD13 transgenic mice develop a highly penetrant, severe myelodysplastic syndrome that progresses to acute leukemia. Blood. 2005;106:287–95. doi: 10.1182/blood-2004-12-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogilvy S, Metcalf D, Print CG, Bath ML, Harris AW, Adams JM. Constitutive Bcl-2 expression throughout the hematopoietic compartment affects multiple lineages and enhances progenitor cell survival. Proc Natl Acad Sci U S A. 1999;96:14943–8. doi: 10.1073/pnas.96.26.14943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwasaki M, Kuwata T, Yamazaki Y, Jenkins NA, Copeland NG, Osato M, et al. Identification of cooperative genes for NUP98-HOXA9 in myeloid leukemogenesis using a mouse model. Blood. 2005;105:784–93. doi: 10.1182/blood-2004-04-1508. [DOI] [PubMed] [Google Scholar]
- 10.Gurevich RM, Rosten PM, Schwieger M, Stocking C, Humphries RK. Retroviral integration site analysis identifies ICSBP as a collaborating tumor suppressor gene in NUP98-TOP1-induced leukemia. Experimental hematology. 2006;34:1192–201. doi: 10.1016/j.exphem.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 11.Raza-Egilmez SZ, Jani-Sait SN, Grossi M, Higgins MJ, Shows TB, Aplan PD. NUP98-HOXD13 gene fusion in therapy-related acute myelogenous leukemia. Cancer Res. 1998;58:4269–7. [PubMed] [Google Scholar]
- 12.Ahuja HG, Felix CA, Aplan PD. The t(11;20)(p15;q11) chromosomal translocation associated with therapy-related myelodysplastic syndrome results in an NUP98-TOP1 fusion. Blood. 1999;94:3258–61. [PubMed] [Google Scholar]
- 13.Hussey DJ, Nicola M, Moore S, Peters GB, Dobrovic A. The (4;11)(q21;p15) translocation fuses the NUP98 and RAP1GDS1 genes and is recurrent in T-cell acute lymphocytic leukemia. Blood. 1999;94:2072–9. [PubMed] [Google Scholar]
- 14.Griffis ER, Altan N, Lippincott-Schwartz J, Powers MA. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Molecular biology of the cell. 2002;13:1282–97. doi: 10.1091/mbc.01-11-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang JC. DNA topoisomerases. Annual review of biochemistry. 1996;65:635–92. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 16.Mizuno T, Kaibuchi K, Yamamoto T, Kawamura M, Sakoda T, Fujioka H, et al. A stimulatory GDP/GTP exchange protein for smg p21 is active on the post-translationally processed form of c-Ki-ras p21 and rhoA p21. Proc Natl Acad Sci U S A. 1991;88:6442–6. doi: 10.1073/pnas.88.15.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slape CI, Saw J, Jowett JB, Aplan PD, Strasser A, Jane SM, et al. Inhibition of apoptosis by BCL2 prevents leukemic transformation of a murine myelodysplastic syndrome. Blood. 2012;120:2475–83. doi: 10.1182/blood-2012-05-430736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi CW, Chung YJ, Slape C, Aplan PD. A NUP98-HOXD13 fusion gene impairs differentiation of B and T lymphocytes and leads to expansion of thymocytes with partial TCRB gene rearrangement. J Immunol. 2009;183:6227–35. doi: 10.4049/jimmunol.0901121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novak RL, Harper DP, Caudell D, Slape C, Beachy SH, Aplan PD. Gene expression profiling and candidate gene resequencing identifies pathways and mutations important for malignant transformation caused by leukemogenic fusion genes. Experimental hematology. 2012;40:1016–27. doi: 10.1016/j.exphem.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Zhang Z, Li Y, Arnovitz S, Chen P, Huang H, et al. PBX3 is an important cofactor of HOXA9 in leukemogenesis. Blood. 2012 doi: 10.1182/blood-2012-07-442004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hussey DJ, Dobrovic A. Recurrent coiled-coil motifs in NUP98 fusion partners provide a clue to leukemogenesis. Blood. 2002;99:1097–8. doi: 10.1182/blood.v99.3.1097. [DOI] [PubMed] [Google Scholar]
- 22.Gilliland DG, Tallman MS. Focus on acute leukemias. Cancer Cell. 2002;1:417–20. doi: 10.1016/s1535-6108(02)00081-8. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen AT, Taranova O, He J, Zhang Y. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9- mediated leukemogenesis. Blood. 2011;117:6912–22. doi: 10.1182/blood-2011-02-334359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26:6766–76. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 25.Thorsteinsdottir U, Mamo A, Kroon E, Jerome L, Bijl J, Lawrence HJ, et al. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood. 2002;99:121–9. doi: 10.1182/blood.v99.1.121. [DOI] [PubMed] [Google Scholar]