Abstract
Microtia is a rare, congenital malformation of the external ear that in some cases has a genetic etiology. We ascertained a three-generation family with bilateral microtia and hearing loss segregating as an autosomal dominant trait. Exome sequencing of affected family members detected only seven shared, rare, heterozygous, nonsynonymous variants, including one protein truncating variant, a HOXA2 nonsense change (c.703C>T, p.Q235*). The HOXA2 variant was segregated with microtia and hearing loss in the family and was not seen in 6,500 individuals sequenced by the NHLBI Exome Sequencing Project or in 218 control individuals sequenced in this study. HOXA2 has been shown to be critical for outer and middle ear development through mouse models and has previously been associated with autosomal recessive bilateral microtia. Our data extend these conclusions and define HOXA2 haploinsufficiency as the first genetic cause for autosomal-dominant nonsyndromic microtia.
Keywords: microtia, hearing loss, exome sequencing, HOXA2
Microtia is a congenital malformation of the external ear that ranges in severity from mild differences in auricular shape and size to complete absence of the external ear. Microtia is frequently associated with conductive hearing loss due to anomalies of the external ear canal and middle ear ossicles. Prevalence rates of this uncommon anomaly range from 0.83 to 4.34 per 10,000 births in European countries and the United States; however, higher prevalence rates of 12.0–17.4 per 10,000 births have been reported in South America and among Native Americans in the United States [reviewed by Suutarla et al., 2007]. Approximately 80% of microtia cases are unilateral [Canfield et al., 2009; Mastroiacovo et al., 1995; Suutarla et al., 2007], and more than 90% of microtia patients have conductive hearing loss in the affected ears [Suutarla et al., 2007]. Microtia usually appears as an isolated anomaly yet can occur along with other malformations, including hemifacial microsomia, cleft lip and/or palate, limb reduction defects, renal anomalies, vertebral abnormalities, and cardiac defects.
In addition to environmental factors that are hypothesized to cause microtia [Zhang et al., 2009], familial clustering of microtia provides evidence for genetic factors. Estimates of the familial incidence of microtia ranges from 3% to 34%, and both autosomal dominant and autosomal recessive patterns of inheritance have been reported [reviewed by Luquetti et al., 2011]. Several genes have been identified to cause syndromes with a high incidence of microtia, including dominant mutations in EYA1 (branchial-oto-renal; MIM #601653) [Abdelhak et al., 1997], SIX5 (branchial-oto-renal; MIM #600963) [Hoskins et al., 2007], and TCOF1 (TreacherCollins;MIM #606847) [Treacher Collins Syndrome Collaborative Group, 1996], and recessive mutations in HMX1 (oculo-auricular; MIM #142992) [Schorderet et al., 2008], FGF3 (congenital deafness, inner ear agenesis, microtia, microdontia; MIM #164950) [Tekin et al., 2007], and HOXA2 (microtia, hearing impairment, cleft palate; MIM #604685) [Alasti et al., 2008]. In contrast, the genetic etiology of nonsyndromic microtia has remained elusive.
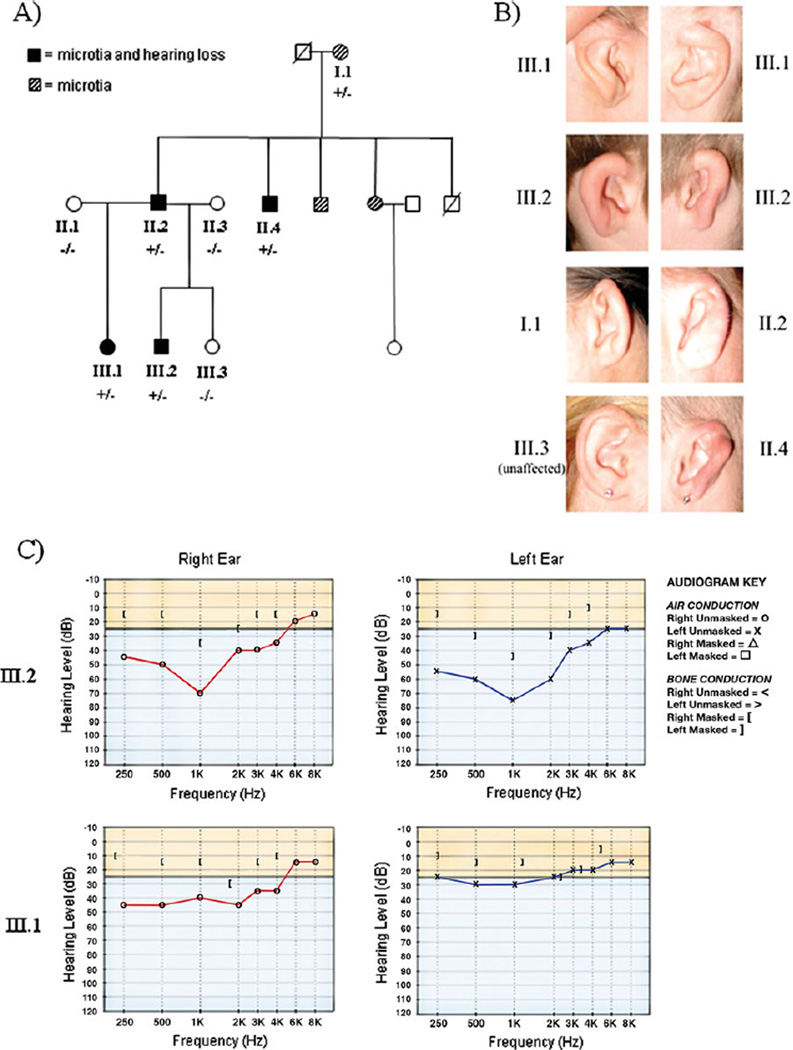
In this study, we harnessed next-generation sequencing technologies to study the exome of affected members of a family of European ancestry with nonsyndromic autosomal dominant bilateral microtia and bilateral mild to severe mixed (conductive and sensorineural) hearing loss. Family history revealed three generations including seven individuals with bilateral microtia with or without hearing loss (Fig. 1A). Written informed consent was obtained for the eight family members (five affected and three unaffected) enrolled in the study, which was performed in accordance with protocols approved by the Institutional Review Boards of Brigham and Women’s Hospital and Geisinger Medical Center. Clinical evaluation and DNA analysis were performed on the enrolled family members. All five affected family members had small, malformed ears with a thickened helix and a superficial postauricular sulcus (Fig. 1B). The external auditory canals were normally formed, and evaluation of other structures derived from the branchial arches revealed no additional anomalies. Notably, the palate was intact in all family members, and renal anatomy was normal in the three family members who had renal imaging. Audiometric testing was performed on three of the affected family members and showed mild to severe bilateral mixed hearing loss (Fig. 1C). Middle ear explorations had been previously performed on individuals II.2 and II.4 and revealed abnormalities of the ossicular chain. In individual II.2, the stapes was noted to have a thickened posterior crus and an absent anterior crus, and the stapedial tendon was absent. Individual II.4 had a rigid ossicular chain.
Figure 1.
Pedigree and clinical features of the family with bilateral microtia and hearing loss. A: Pedigree showing individuals affected with microtia and hearing loss. The labeled individuals (I.1, II.1, II.2, II.3, II.4, III.1, III.2, and III.3) were enrolled in the study. Individuals marked as +/− are heterozygous for the HOXA2 Q235* variant. Individuals marked as −/− do not carry the Q235* variant. B: Pictures of the right and/or left ears of several family members. C: Pure-tone audiometry for family members III.2 and III.1 measuring air and bone conduction of sound. Normal air and bone conduction at different frequencies (Hz) is above the solid horizontal line. Individual III.2 has mixed (diminished bone and air conduction) hearing loss that is moderate in the right ear and severe in the left ear. Individual III.1 has moderate mixed hearing loss in the right ear and mild conductive hearing loss in the left ear.
Exome sequencing was performed on four affected family members (I.1, II.4, III.1, and III.2). For each individual, a barcoded whole-genome library was generated from DNA extracted from a blood sample. The libraries were pooled and the exomes were captured using the NimbleGen SeqCap EZ Human Exome Library v2.0 (Roche). Sequencing of the captured exomes was done on an Illumina HiSeq2000 producing 50 basepair paired end reads. Reads were aligned to the human reference genome (hg19) using Novoalign (Novocraft.com), quality measures were assessed using Picard HsMetrics (picard.sourceforge.net), and variants were called using GATK Unified Genotyper [Depristo et al., 2011].We obtained an average of 49 million unique reads per individual, resulting in about 20-fold coverage over all exons with 96% of regions covered at least twice and 70%of regions covered at least 10 times. As analysis of this initial sequencing data revealed a variant of interest, additional runs to increase depth of coverage were not performed.
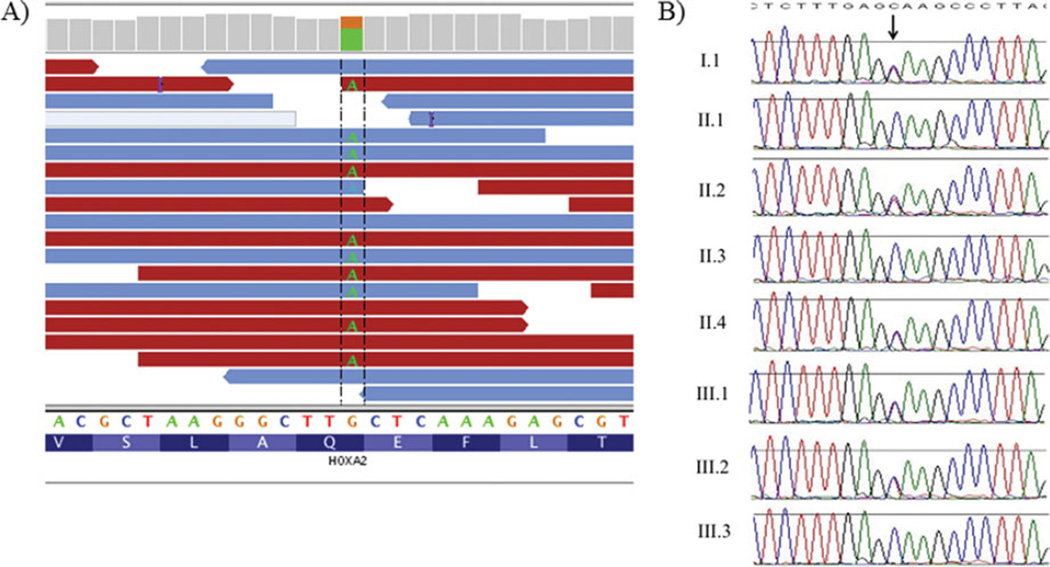
After applying standard quality metrics [Herman et al., 2012], we employed the following filters to prioritize heterozygous nonsynonymous variants for further study: (1) variants were shared by each of the four samples studied by exome sequencing; (2) variants altered conserved residues in protein-encoding sequences or consensus splice sequences; (3) variants had a minor allele frequency ≤0.001 in the 1000 Genomes Project (Phase I data) and the NHLBI Exome Sequencing Project, and were absent from other exomes studied in our laboratory. After filtering by these criteria, only seven potential candidate variants remained, including one nonsense variant in HOXA2, five missense variants (one each in TTN, FN1, NPAT, HMBS, and NOTUM), and one in-frame deletion of two amino acids in SUSD2 (Supp. Table S1). The HOXA2 change, c.703C>T (NM_006735.3), p.Q235* (Fig. 2A), is predicted to result in truncation of 142 amino acids from the C-terminal end of the HOXA2 protein. Given that the HOXA2 variant is predicted to truncate the encoded protein and that a recessive mutation in HOXA2 was previously identified in a consanguineous family with syndromicmicrotia [Alasti et al., 2008], we investigated this variant further.
Figure 2.
Exome sequencing and Sanger sequencing results for the HOXA2 variant. A: Illustration, using the Integrative Genomics Viewer (www.broadinstitute.org/igv/), of the heterozygous HOXA2 variant (c.703C>T, p.Q235*) detected by exome sequencing in family member II.4. B: Sanger sequencing traces of the HOXA2 variant showing segregation in affected family members. The HOXA2 variant was submitted to a public LSDB (http://www.lovd.nl/HOXA2).
Sanger sequencing for the HOXA2 variant was performed in all enrolled family members. These studies confirmed the exome sequencing data and revealed that HOXA2 c.703C>T (p.Q235*) was present in all affected family members and absent in unaffected family members (Fig. 2B). Additionally, Sanger sequencing confirmed that no family member had an additional nonsynonymous variant in either HOXA2 allele.
The cosegregation of the HOXA2 nonsense variant with dominant microtia/hearing loss in this family yielded a LOD score of 1.51 (odds = 1/32; θ = 0). The HOXA2 c.703C>T (p.Q235*) variant was not detected in over 6,500 individuals studied in the NHLBI Exome Sequencing Project. Furthermore, as no nonsense variants are reported at any position in the HOXA2 gene by the NHLBI Exome Sequencing Project or any other source, we suggest that haploinsufficiency of HOXA2 protein is deleterious. Taken together, we conclude that HOXA2 c.703C>T is the likely cause of microtia and hearing loss in this family.
To extend these findings, we sequencedHOXA2 in a cohort of 119 microtia subjects recruited from Colombia and Ecuador (regions with the highest prevalence of microtia) and 218 ethnically matched controls. This cohort included 100 individuals with nonsyndromic, unilateral microtia, and 19 individuals with nonsyndromic bilateral microtia.
Barcoded genomic libraries were produced for each case and control, HOXA2 sequences were captured by a filter-based hybridization method [Herman et al., 2009], and the enriched libraries were sequenced on an Illumina HiSeq2000. Regions of HOXA2 with insufficient depth of coverage in the patient samples were filled in using Sanger sequencing. In addition, HOXA2 was also fully sequenced in two additional bilateral microtia patients using Sanger sequencing. The c.703C>T (p.Q235*) variant was not detected in any of the case or control samples. Other heterozygous nonsynonymous HOXA2 variants were detected in two microtia subjects. One subject with nonsyndromic unilateral microtia from Ecuador had a missense variant that changes a leucine to a phenylalanine (c.766G>A, p.L256F), and a second nonsyndromic unilateral microtia subject from Colombia had a missense variant that changes a leucine to an isoleucine (c.124G>T, p.L42I). Both of these nonsynonymous changes affect leucine residues with high evolutionary conservation; however, each of these variants was also found in one control, indicating that either these variants are benign or that these variants in combination with another undetected HOXA2 variant (possibly in a noncoding portion of the gene) are responsible for microtia in these patients. Neither of these variants has been reported by the NHLBI Exome Sequencing Project, suggesting that they are specific to these South American populations, where the incidence of microtia is high. We conclude that HOXA2 mutations are an uncommon cause of nonsyndromic unilateral microtia in South American populations.
HOXA2 is a member of the homeobox gene family, which encodes highly conserved transcription factors that control body patterning and morphogenesis during embryonic development. All homeobox genes encode a helix-turn-helix homeodomain, which facilitates site-specific DNA binding. HOXA2 is strongly expressed in the second branchial arch, and it appears to specify cell fate, steering cells of the second arch to develop into second arch derivatives, including the external ear and the stapes [Gendron-Maguire et al., 1993; Rijli et al., 1993].HOXA2 may prevent the formation of structures of the first arch, as lack of HOXA2 in the second arch causes duplication of first arch structures [Gendron-Maguire et al., 1993; Rijli et al., 1993], and overexpression of HOXA2 in the first arch causes duplication of second arch structures [Grammatopoulos et al., 2000].
Hoxa2 knockout mice die shortly after birth due to feeding difficulties. The knockout mice are easily distinguished from their littermates as they lack the pinna of the external ear and have a wide cleft of the secondary palate and a cleft of the root of the tongue [Gendron-Maguire et al., 1993; Rijli et al., 1993]. Hoxa2-null mice also lacked second arch derivatives of the middle ear, including the stapes, the styloid bone, the lesser horn of the hyoid bone, and the stapedius muscle. Additionally, the malleus and the incus, first arch structures, were duplicated.
A study of mice carrying a Hoxa2 hypomorphic allele demonstrated that these phenotypes are dosage sensitive, with outer and middle ear structures being most sensitive to decreased Hoxa2 levels [Ohnemus et al., 2001]. While most mice heterozygous for one wild-type allele and one null allele appear phenotypically normal, some had a malformed stapes, abnormally located styloid process, and absent processus brevis. All mice homozygous for the hypomorphic allele had malformed stapes and pinnae that appeared to be slightly smaller than normal. Compound mice, heterozygous for a hypomorphic allele and a null allele, had an overtly malformed pinna and more severe middle ear defects, including absence of the stapes.
In humans, HOXA2 has previously been associated with an autosomal recessive syndromic microtia. Using genome-wide linkage analysis and candidate gene sequencing, a highly conserved homozygous HOXA2 missense variant c.558C>A (p.Q186K) was found to segregate with bilateral microtia, mixed (conductive and sensorineural) hearing loss, and cleft palate in one consanguineous family from Iran [Alasti et al., 2008]. The missense variant is located within the homeodomain and is predicted by computational modeling to affect DNA-binding activity. The affected individuals from the Iranian family have malformed ears with thickened helices bilaterally, short and severely narrowed auditory canals, malformed ossicular chains that appeared to be fixated, severe to profound mixed hearing impairment affecting all frequencies, and partial cleft palate.
While the external and middle ear features of the family studied here are similar to those of the Iranian family, we did not identify abnormalities of the auditory canal, profound hearing loss with a significant sensorineural component, or the cleft palate phenotype found in the Iranian family. These differences in clinical severity are not surprising, given the dosage sensitivity observed inHoxa2 mouse models. Since only one allele contained the Q235* variant, affected subjects are predicted to have about a 50% reduction in normal HOXA2 activity. In contrast, the Q186K variant could completely abrogate the DNA-binding activity of HOXA2, so homozygous subjects may have little to no physiologic HOXA2 activity. The smaller reduction in HOXA2 activity predicted for our family members is consistent with their milder clinical features.
We observed no increased burden of rare and deleterious variants in HOXA2 in microtia cases from South America. This cohort included primarily cases of unilateral microtia (83%), whereas affected individuals with recessive or dominant HOXA2 mutations all had bilateral microtia. As deleterious mutations in HOXA2 were also absent in the 21 cases with bilateral microtia, we predict that mutations in HOXA2 may account for less than 5% of bilateral microtia cases. Asmost of the cases in our cohort had sporadic microtia (no first degree relatives with microtia), it is possible that among familial microtia cases or in microtia patients froma different ethnic background HOXA2 mutations may be more common.
In summary, we have identified a nonsense mutation (Q235*) in HOXA2 that segregates with bilateral nonsyndromic microtia and hearing loss through three generations of a family in an autosomal dominant pattern. Based on the absence of this mutation in over 6,500 control samples, and evidence for Hoxa2 dosage sensitivity in mice, we conclude that HOXA2 haploinsufficiency causes microtia and hearing loss. This idea is strengthened by the known role of HOXA2 in the development of the outer and middle ear, the malformations seen in null and hypomorphic Hoxa2 mouse models, and the similar features of a previously reported family in whom microtia segregates with a homozygous HOXA2 missense variant. This study is the first to identify a genetic etiology for autosomal dominant nonsyndromic microtia and implies that HOXA2 and its downstream targets may be responsible for microtia in other affected individuals.
Supplementary Material
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, Weil D, Cruaud C, Sahly I, Leibovici M, Bitner-Glindzicz M, Francis M, et al. A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat Genet. 1997;15:157–164. doi: 10.1038/ng0297-157. [DOI] [PubMed] [Google Scholar]
- Alasti F, Sadeghi A, Sanati MH, Farhadi M, Stollar E, Somers T, Van Camp G. A mutation in HOXA2 is responsible for autosomal-recessive microtia in an Iranian family. Am J Hum Genet. 2008;82:982–991. doi: 10.1016/j.ajhg.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canfield MA, Langlois PH, Nguyen LM, Scheuerle AE. Epidemiologic features and clinical subgroups of anotia/microtia in Texas. Birth Defects Res A Clin Mol Teratol. 2009;85:905–913. doi: 10.1002/bdra.20626. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron-Maguire M, Mallo M, Zhang M, Gridley T. Hoxa-2 mutant mice exhibit homeotic transformation of skeletal elements derived from cranial neural crest. Cell. 1993;75:1317–1331. doi: 10.1016/0092-8674(93)90619-2. [DOI] [PubMed] [Google Scholar]
- Grammatopoulos GA, Bell E, Toole L, Lumsden A, Tucker AS. Homeotic transformation of branchial arch identity after Hoxa2 overexpression. Development. 2000;127:5355–5365. doi: 10.1242/dev.127.24.5355. [DOI] [PubMed] [Google Scholar]
- Herman DS, Hovingh GK, Iartchouk O, Rehm HL, Kucherlapati R, Seidman JG, Seidman CE. Filter-based hybridization capture of subgenomes enables resequencing and copy-number detection. Nat Methods. 2009;6:507–510. doi: 10.1038/nmeth.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman DS, Lam L, Taylor MR, Wang L, Teekakirikul P, Christodoulou D, Conner L, DePalma SR, McDonough B, Sparks E, Teodorescu DL, Cirino AL, et al. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366:619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins BE, Cramer CH, II, Silvius D, Zou D, Raymond RM, Jr, Orten DJ, Kimberling WJ, Smith RJH, Weil D, Petit C, Otto EA, Xu PX, et al. Transcription factor SIX5 is mutated in patients with branchio-oto-renal syndrome. Am J Hum Genet. 2007;80:800–804. doi: 10.1086/513322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luquetti DV, Heike CL, Hing AV, Cunningham ML, Cox TC. Microtia: epidemiology and genetics. Am J Med Genet A. 2011;158A:124–139. doi: 10.1002/ajmg.a.34352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroiacovo P, Corchia C, Botto LD, Lanni R, Zampino G, Fusco D. Epidemiology and genetics of microtia-anotia: a registry based study on over one million births. J Med Genet. 1995;32:453–457. doi: 10.1136/jmg.32.6.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnemus S, Bobola N, Kanzler B, Mallo M. Different levels of Hoxa2 are required for particular developmental processes. Mech Dev. 2001;108:135–147. doi: 10.1016/s0925-4773(01)00502-0. [DOI] [PubMed] [Google Scholar]
- Rijli FM, Mark M, Lakkaraju S, Dierich A, Dollé P, Chambon P. Ahomeotic transformation is generated in the rostral branchial region of the head by disruption of Hoxa-2, which acts as a selector gene. Cell. 1993;75:1333–1349. doi: 10.1016/0092-8674(93)90620-6. [DOI] [PubMed] [Google Scholar]
- Schorderet DF, Nichini O, Boisset G, Polok B, Tiab L, Mayeur H, Raji B, de la Houssaye G, Abitbol MM, Munier FL. Mutation in the human homeobox gene NKX5–3 causes an oculo-auricular syndrome. Am J Hum Genet. 2008;82:1178–1184. doi: 10.1016/j.ajhg.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suutarla S, Rautio J, Ritvanen A, Ala-Mello S, Jero J, Klockars T. Microtia in Finland: comparison of characteristics in different populations. Int J Pediatr Otorhinolaryngol. 2007;71:1211–1217. doi: 10.1016/j.ijporl.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Tekin M, Hişmi BO, Fitoz S, Ozdağ H, Cengiz FB, Sirmaci A, Aslan I, Inceoğlu B, Yüksel-Konuk EB, Yilmaz ST, Yasun O, Akar N. Homozygous mutations in fibroblast growth factor 3 are associated with a new form of syndromic deafness characterized by inner ear agenesis, microtia, and microdontia. Am J Hum Genet. 2007;80:338–344. doi: 10.1086/510920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treacher Collins Syndrome Collaborative Group. Positional cloning of a gene involved in the pathogenesis of Treacher Collins syndrome. Nat Genet. 1996;12:130–136. doi: 10.1038/ng0296-130. [DOI] [PubMed] [Google Scholar]
- Zhang QG, Zhang J, Yu P, Shen H. Environmental and genetic factors associated with congenital microtia: a case-control study in Jiangsu, China, 2004 to 2007. Plast Reconstr Surg. 2009;124:1157–1164. doi: 10.1097/PRS.0b013e3181b454d8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.