Substantial indirect evidence suggests overlap between bipolar disorder (BIP) and major depressive disorder (MDD). BIP and MDD have in common major depressive episodes with BIP distinguished by the additional presence of manic (bipolar 1) or hypomanic episodes (bipolar 2). Genetic epidemiological (1) and genome-wide linkage studies (2) are also consistent with overlap between genetic risk factors for both disorders. To attempt to identify common genetic risk factors, we conducted a meta-analysis combining data from genome-wide association studies (GWAS) of BIP (4,387 cases and 6,209 controls) (3) and MDD (1,695 cases and 1,761 controls) (4).
Ascertainment, diagnostic assessment, genotyping, quality control, and analysis are detailed elsewhere (3, 4). Both studies were conducted under the appropriate ethical approvals, and all subjects provided written informed consent. Briefly, the BIP results are from a combined analysis of samples from the UK, the US, and Ireland (5, 6) with all subjects genotyped using Affymetrix 500K chips. Most cases met criteria for DSM-IV bipolar 1 (81%) with smaller numbers meeting criteria for bipolar 2 (16%), schizoaffective disorder/manic type (2%), or bipolar NOS (1%). After quality control, 1,769,948 SNPs were analyzed (18.7% directly genotyped and the remainder imputed using HapMap2 CEU) (7, 8). Cases meeting DSM-IV criteria for MDD were ascertained from clinical and community sources, and controls at low liability for MDD were selected from a community sample (9). Genotyping was conducted by Perlegen using a 600K platform. Following quality control (with slightly stricter thresholds to maximize comparability), 1,893,617 SNPs were available (20.4% directly genotyped with the rest imputed using HapMap2 CEU) (10). In both studies, SNPs were dropped for excessive missingness, low minor allele frequencies, and marked deviations from Hardy-Weinberg equilibrium. Subjects were removed for excessive missingness, unusual genome-wide heterozygosity, first- or second-degree relation to any other subject, and if empirical ancestry deviated markedly from other subjects. There was no known subject overlap across studies (BIP subjects were from the US, the UK, and Ireland, and MDD subjects were from The Netherlands).
After merging SNP lists from the BIP and MDD studies (with attention to strand and allele matching), there were 1,472,580 high-quality autosomal SNPs common to both studies (72.3% were imputed in both studies, 5.6% were directly genotyped in both, 11.7% were genotyped in the BIP and imputed in the MDD study, and 10.4% imputed in the BIP and genotyped in the MDD study). Genomic positions were per NCBI Build 36/UCSC hg18.
Fixed-effects meta-analysis was accomplished using a weighted z-score method (11). Figure 1 depicts the results and Table S1 lists the SNPs with p < 10-5 in either primary study or in the meta-analysis. For the combined sample of 6,082 cases and 7,970 controls, λ1000 was 1.019 (i.e., λ scaled to a sample size of 1,000 cases and 1,000 controls and the λ was 1.131) (Figure 1a and 1b).
Figure 1.
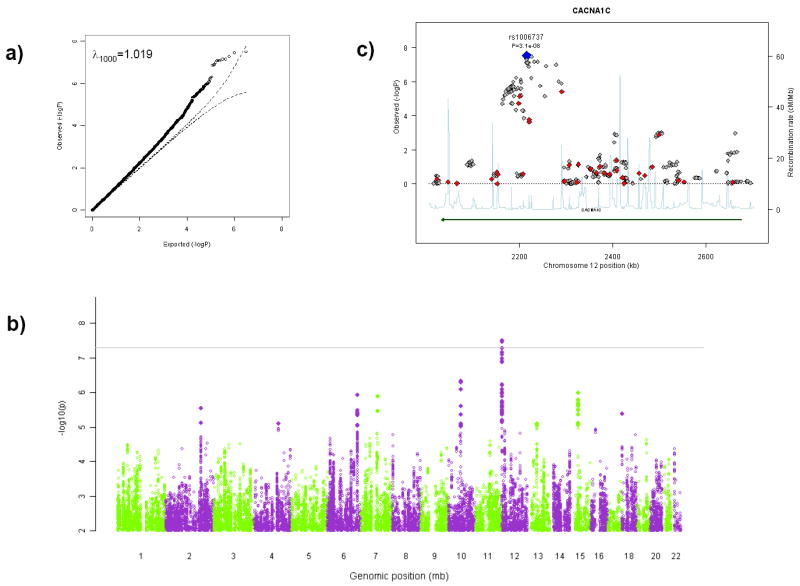
Results of GWAS meta-analysis for BIP and MDD. A) Quantile-quantile plot of the meta-analytic results (observed × expected p-values on −log10 scale). B) Manhattan plot (−log10 of fixed-effects p-value × genomic position). C) The CACNA1C region (red diamonds indicate SNPs genotyped in both studies, gold SNPs genotyped in one study and imputed in the other, and gray SNPs imputed in both studies).
We note four findings from the meta-analysis. (A) Two SNPs in a 10.5 kb region of CACNA1C exceeded a genome-wide significance level of 5×10-8 (12): rs1006737 (pfixed=3.1×10-8) and rs7297582 (pfixed=3.4×10-8) (Figure 1c). These SNPs reached genome-wide significance in the initial BIP report and multiple SNPs in this region had p < 0.05 in the MDD study. For rs1006737*A the case frequency/odds ratio estimates were: BIP sample 0.36/1.18, MDD sample 0.32/1.10 (similar to the findings from a different MDD sample, 0.36/1.15) (13). Second, two ANK3 SNPs exceeded genome-wide significance in the initial BIP report but were not supported in the MDD GWAS (rs10994336 and rs10994338 with p-values ~0.9). Third, support for the PCLO SNP of particular interest in the MDD GWAS (rs2522843) was not increased with meta-analysis although several SNPs had p-values < 0.05. Fourth, as shown in Table S1, several areas were of modest significance in each primary GWAS and considerably greater significance in the meta-analysis (although none reached the genome-wide significance level): intergenic regions on chr2:175.95-175.99 Mb and chr13:49.96-49.98 Mb along with SNPs in SYNE1, FAT4, DMTF1, C7orf23, and C15orf53. SYNE1 (a cause of spinocerebellar ataxia) is of immediate interest as it contains a spectrin binding domain suggesting a connection to the function of the BIP susceptibility locus ANK3.
In conclusion, this analysis provides support for a role of CACNA1C risk variants for both bipolar and unipolar major mood disorders. Among the possible explanations, genetic variation in CACNA1C might be common, subtle, and pleomorphic risk factors for mood disorders. Alternatively, the overlap could be due to misclassification – e.g., if some portion of the MDD group were truly “bipolar-like” but misclassified due to diagnostic or nosological error (despite the use of standard and careful methodologies) or if some portion of the BIP group was similarly misclassified (14). In contrast, the bipolar risk locus ANK3 did not find support in this meta-analysis suggesting that its effect may be specific to BIP or that power was insufficient to detect an effect. Finally, our analysis had insufficient power definitively to establish or to exclude the role of several biologically interesting candidate genes (e.g., PCLO and SYNE1), and further insights into their roles in mood disorders await larger-scale mega-analyses (15).
Supplementary Material
Footnotes
Author Contributions
All authors reviewed and approved the final version of the manuscript.
Conflicts of Interest
In the interests of full disclosure, Dr. Sullivan reports receiving unrestricted research funding from Eli Lilly for genetic research in schizophrenia. Dr. Perlis has received speaking or consulting fees from Astra Zeneca, Eli Lilly, GlaxoSmithKline, Pfizer, and Proteus, LLC. Dr. Nolen reports receiving unrestricted research funding and Speaker’s fee from Astra Zeneca, Eli Lilly, GlaxoSmithKline, Pfizer, Servier and Wyeth. The other authors report no conflicts.
Contributor Information
Youfang Liu, Email: youfang@email.unc.edu, University of North Carolina, Chapel Hill.
Douglas H. Blackwood, Email: d.blackwood@ed.ac.uk, University of Edinburgh.
Sian Caesar, Email: e.s.caesar@bham.ac.uk, Birmingham University.
Eco J.C. de Geus, Email: eco@psy.vu.nl, VU University Amsterdam.
Anne Farmer, Email: a.farmer@iop.kcl.ac.uk, Institute of Psychiatry.
Manuel A. R. Ferreira, Email: manuel.ferreira@qimr.edu.au, Queensland Institute of Medical Research.
I. Nicol Ferrier, Email: i.n.ferrier@newcastle.ac.uk, Newcastle University.
Christing Fraser, Email: wpccf2@groupwise.cf.ac.uk, Cardiff University.
Katherine Gordon-Smith, Email: k.m.gordonsmith@bham.ac.uk, Birmingham University.
Elaine K. Green, Email: greenek@cardiff.ac.uk, Cardiff University.
Detelina Grozeva, Email: wmddvg@groupwise.cf.ac.uk, Cardiff University.
Hugh M. Gurling, Email: h.gurling@ucl.ac.uk, University College London.
Marian L. Hamshere, Email: wpcmlh@groupwise.cf.ac.uk, Cardiff University.
Peter Heutink, Email: p.heutink@vumc.nl, VU University Medical Center Amsterdam.
Peter A. Holmans, Email: wpcpah@groupwise.cf.ac.uk, Cardiff University.
Witte J. Hoogendijk, Email: witteh@ggzba.nl, VU University Medical Center Amsterdam.
Jouke Jan Hottenga, Email: jj.hottenga@psy.vu.nl, VU University Amsterdam.
Lisa Jones, Email: l.a.jones@bham.ac.uk, Birmingham University.
Ian R. Jones, Email: wpcirj@groupwise.cf.ac.uk, Cardiff University.
George Kirov, Email: kirov@cardiff.ac.u, Cardiff University.
Danyu Lin, Email: lin@bios.unc.edu, University of North Carolina, Chapel Hill.
Peter McGuffin, Email: p.mcguffin@iop.kcl.ac.uk, Institute of Psychiatry.
Valentina Moskvina, Email: wpcvm@groupwise.cf.ac.uk, Cardiff University.
Willem A. Nolen, Email: w.a.nolen@psy.umcg.nl, University Medical Center Groningen.
Roy H. Perlis, Email: rperlis@chgr.mgh.harvard.edu, Massachusetts General Hospital.
Danielle Posthuma, Email: danielle@psy.vu.nl, VU University Amsterdam.
Edward M. Scolnick, Email: scolnick@broadinstitute.org, Broad Insitute.
August B. Smit, Email: guus.smit@cncr.vu.nl, VU University Amsterdam.
Johannes H. Smit, Email: jh.smit@vumc.nl, VU University Medical Center Amsterdam.
Jordan W. Smoller, Email: jsmoller@hms.harvard.edu, Massachusetts General Hospital.
David St. Clair, Email: d.stclair@abdn.ac.uk, Aberdeen University.
Richard van Dyck, Email: r.van.dyck@ggzba.nl, VU University Medical Center Amsterdam.
Matthijs Verhage, Email: matthijs@cncr.vu.nl, VU University Amsterdam.
Gonneke Willemsen, Email: ahm.willemsen@psy.vu.nl, VU University Amsterdam.
Allan H. Young, Email: allanyoun@gmail.com, UBC, Vancouver, Canada.
Tim Zandbelt, Email: timz@ggzba.nl, VU University Medical Center Amsterdam.
Dorret I. Boomsma, Email: dorret@psy.vu.nl, VU University Amsterdam.
Nick Craddock, Email: craddockn@cardiff.ac.uk, Cardiff University.
Michael C. O’Donovan, Email: odonovanmc@cf.ac.uk, Cardiff University.
Michael J. Owen, Email: owenmj@cardiff.ac.uk, Cardiff University.
Brenda W.J.H. Penninx, Email: b.penninx@vumc.nl, VU University Medical Center Amsterdam.
Shaun Purcell, Email: shaun@pngu.mgh.harvard.edu, Massachusetts General Hospital.
Pamela Sklar, Email: sklar@chgr.mgh.harvard.edu, Massachusetts General Hospital.
Patrick F. Sullivan, Email: pfsulliv@med.unc.edu, University of North Carolina, Chapel Hill.
References
- 1.Tsuang MT, Faraone SV. The Genetics of Mood Disorders. Baltimore: The Johns Hopkins University Press; 1990. [Google Scholar]
- 2.Craddock N, Forty L. Eur J Hum Genet. 2006;14(6):660–8. doi: 10.1038/sj.ejhg.5201549. [DOI] [PubMed] [Google Scholar]
- 3.Ferreira M, O’Donovan M, Meng Y, Jones I, Ruderfer D, Jones L, et al. Nature Genetics. 2008 [Google Scholar]
- 4.Sullivan P, de Geus E, Willemsen G, James MR, Smit JH, Zandbelt T, et al. Molecular Psychiatry. 2009;14:359–75. doi: 10.1038/mp.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WTCCC. Nature. 2007;447(7145):661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sklar P, Smoller JW, Fan J, Ferreira MA, Perlis RH, Chambert K, et al. Mol Psychiatry. 2008 [Google Scholar]
- 7.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira M, Bender D, et al. American Journal of Human Genetics. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, et al. Nature. 2007;449(7164):851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Penninx B, Beekman A, Smit J. International Journal of Methods in Psychiatric Research. 2008;17:121–40. doi: 10.1002/mpr.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang L, Li Y, Singleton AB, Hardy JA, Abecasis G, Rosenberg NA, et al. Am J Hum Genet. 2009;84(2):235–50. doi: 10.1016/j.ajhg.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, Voight BF. Hum Mol Genet. 2008;17(R2):R122–8. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pe’er I, Yelensky R, Altshuler D, Daly MJ. Genet Epidemiol. 2008;32(4):381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 13.Green E, Grozeva D, Jones I, Jones L, Kirov G, Caesar S, et al. Molecular Psychiatry. In press. [Google Scholar]
- 14.Regeer EJ, ten Have M, Rosso ML, Hakkaart-van Roijen L, Vollebergh W, Nolen WA. Acta Psychiatr Scand. 2004;110(5):374–82. doi: 10.1111/j.1600-0447.2004.00363.x. [DOI] [PubMed] [Google Scholar]
- 15.Psychiatric GWAS Consortium. Molecular Psychiatry. 2009;(14):10–7. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


