Abstract
Purpose
To investigate the effects of pirfenidone (PFD) on the migration, differentiation, and proliferation of retinal pigment epithelial (RPE) cells and demonstrate whether the drug induces cytotoxicity.
Methods
Human RPE cells (line D407) were treated with various concentrations of PFD. Cell migration was measured with scratch assay. The protein levels of fibronectin (FN), connective tissue growth factor (CTGF), α-smooth muscle actin (α-SMA), transforming growth factor beta (TGFβS), and Smads were assessed with western blot analyses. Levels of mRNA of TGFβS, FN, and Snail1 were analyzed using reverse transcriptase–polymerase chain reaction. Cell apoptosis was detected with flow cytometry using the Annexin V/PI apoptosis kit, and the percentages of cells labeled in different apoptotic stage were compared. A Trypan Blue assay was used to assess cell viability.
Results
PFD inhibited RPE cell migration. Western blot analyses showed that PFD inhibited the expression of FN, α-SMA, CTGF, TGFβ1, TGFβ2, Smad2/3, and Smad4. Similarly, PFD also downregulated mRNA levels of Snail1, FN, TGFβ1, and TGFβ2. No significant differences in cell apoptosis or viability were observed between the control and PFD-treated groups.
Conclusions
PFD inhibited RPE cell migration, differentiation, and proliferation in vitro and caused no significant cytotoxicity.
Introduction
Proliferative vitreoretinopathy (PVR) is a disease process that follows rhegmatogenous retinal detachment (RRD) secondary to the occurrence and proliferation of ectopic cell sheets in the vitreous and/or periretinal area, causing membrane formation and traction [1]. PVR occurs in 5%–10% of all RRD and is implicated in redetachment after surgery in 75% of cases, which remains a major barrier to successful repair of retinal detachment [2]. Treatment for PVR mostly depends on the surgery, but the outcome is far from satisfactory. Pastor pointed out that only 40%–80% of patients who receive anatomically successful surgeries may regain functional vision (ambulatory vision 5/200 or better) [2]. Thus, further research that aims to improve options for prevention or prophylaxis of PVR with medical treatment is needed. Agents capable of inhibiting inflammation or fibrosis may be of great value. To this end, numerous drugs have been shown to be efficacious in patient studies [3,4], but the potential complications limit the drugs’ clinical application. For example, the antimetabolism drug 5-fluorouracil has garnered much attention because the drug strongly inhibits fibrosis, but some researchers have proposed that 5-fluorouracil is systemically absorbed when added to infusion fluid during vitrectomy. Patient selection is needed to avoid adverse effects on procreativity [5,6]. Thus, oculists have sought to identify a medicine that not only inhibits fibrosis in PVR but also has no serious complications.
Pirfenidone (PFD, 5-cymene-1-phenyl-2-hydrogen-pyridone), used as an antihelminthic or antipyretic, did not garner significant attention as an antifibrotic agent until 1995 when Iyer et al. reported its strong inhibitory effect on bleomycin-induced lung fibrosis in rabbit models [7]. In the same year, Suga et al. showed an antifibrotic effect on sclerosing peritonitis in rats [8]. Since then, research on applying PFD in treating diseases characterized by fibrosis has been increasing [9,10]. Since PFD is a new broad-spectrum anti-inflammation and antiproliferation agent, the safety and effectiveness have been verified through experiments and clinical trials [11,12]. On March 3, 2011, the European Commission (EC) granted marketing authorization for pirfenidone under the trade name Esbriet for treating idiopathic pulmonary fibrosis [13].
Although PFD is frequently applied in various other medical fields, its effects in ocular diseases have seldom been reported. We previously found that PFD prohibited the migration and proliferation of human Tenon’s fibroblasts in vitro [14]. An in vivo study confirmed PFD’s effects on inhibiting the tendency of scar formation after trabeculectomy [15]. We also analyzed the pharmacokinetic properties of PFD when it is topically administered in rabbit eyes [16]. Recently, Choi et al. reported that PFD inhibited the fibroblast-like phenotype of retinal pigment epithelial (RPE) cells induced by transforming growth factor beta 1 (TGFβ1), and the possible mechanism involved blocking TGFβ signaling pathways [17]. This research provided useful information for our aims and supported the application of PFD in treating PVR. However, more research is necessary. For example, since PVR is mediated by many cytokines, it would be useful to study the effect of PFD on the expression of cytokines to clarify the mechanism involved. In addition to investigating the mechanistic effects of PFD, investigating the drug’s safety is also important. Thus, in this article, we explored the effects of PFD on RPE cells in terms of cell motility, differentiation, and proliferation and investigated cytotoxicity.
Methods
Cell culture and treatment
The RPE cell line D407 was obtained from Sun Yat-sen University Zhongshan Ophthalmic Center (State Key Laboratory, GuangZhou, China); cells were cultured in medium essential medium/F12 supplemented with 10% fetal bovine serum (10% FBS-DMEM/F12; Invitrogen-Gibco, Kalsrahe, Germany), 100 U/ml penicillin G, and 100 µg/ml streptomycin (Biochem, Berlin, Germany) and grown in an incubator at 37 °C with 5% CO2. For all experiments except the migration experiment, cells were cultured in complete medium for at least 24 h until they were 25% confluent, at which time, they were subjected to different PFD treatments. PFD (Sigma-Aldrich, St. Louis, MO) was dissolved in sterile distilled water at 60 °C for 30 min. Then, PFD was filtered and stored at −20 °C before being mixed with complete medium during the experiment (the PFD concentrations ranged from 0.3 to 1.0 mg/ml).
Cell migration
Cells were cultured in serum-free medium for approximately 48 h after they reached confluence. A straight scratch was made in the middle on the monolayer using a sterile pipette. Detached cells were rinsed away with PBS (NaCl 137 mmol/l, KCl 2.7mmol/l, Na2HPO4 10 mmol/l, KH2PO4 2mmol/l, pH 7.4). The scratch gap was recorded and photographed with a light microscope. The distance at the initial time point was set to a value of 100. The cells were then treated with serum-free DMEM/F12 and DMEM/F12 supplemented with 0.3 or 0.5 mg/ml PFD for another 24 h. Finally, the widths of the remaining defect were analyzed and compared using image processing software (Photoshop 7.0, Adobe, San Jose, CA).
Western blot
Samples were divided into three groups. The first group was treated with complete medium, whereas the other two groups were treated with 0.3 mg/ml PFD or 0.5 mg/ml PFD in complete medium, respectively. After incubation for 24 h, cells were lysed with 1 ml RIPA buffer with 10 µl phenylmethanesulfonyl fluoride (PMSF; Shanghai Biocolors, Shanghai, China). Lysates were incubated on ice, and then insoluble material was removed through centrifugation (4 °C, 10,800 × g, 30 min). Thirty micrograms of protein from each group was analyzed. Samples were probed with polyclonal antibodies specific for fibronectin (FN), connective tissue growth factor (CTGF), α-smooth muscle actin (α-SMA), TGFβ1, TGFβ2, Smad2/3, and Smad4 according to the manufacturer’s instructions (Abcam, Cambridge, CA). Glyceraldehyde-3-phosphate dehydrogenase (GADPH) was used to verify equal loading of proteins. The proteins were separated by sodium dodecyl sulfate PAGE (SDS–PAGE) and transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore Co, Billerica, MA). The membranes were blocked with 5% nonfat milk for 2 h at room temperature and incubated with polyclonal antibodies specific for FN, CTGF, α-SMA, TGFβ1, TGFβ2, Smad2/3, and Smad4 according to manufacturer’s instructions (Abcam Ltd, Cambridge, MA) at 4 °C overnight. Glyceraldehyde-3-phosphate dehydrogenase (GADPH) was used to verify equal loading of proteins. Then the membranes were washed and incubated for 1 h at room temperature with horseradish-peroxidase-conjugated secondary antisera (Dako, Hamburg, Germany). The membranes were washed three times with 0.1% Tween-20 in phosphate buffered saline for 10 min before being stained with chemiluminescence western blot detection reagents (Millipore) [14]. X-ray films were scanned on a Model GS-800 imaging densitometer (Bio-Rad Laboratories, Hercules, CA), and the densitometry value of each protein was analyzed with Quantity One software (Bio-Rad Laboratories). Finally, the densitometry values of each protein analyte normalized to GADPH were compared.
Reverse transcriptase–polymerase chain reaction
Cells were divided into three groups: the control group, the 0.3 mg/ml PFD group, and the 0.5 mg/ml PFD group. Twenty-four hours after the beginning of treatment, total RNA was extracted using TRIzol reagent according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA). Synthesis of cDNA and PCR were performed following the manufacturer’s instructions (TaKaRa Biochemicals, Osaka, Japan). The TGFβ1, TGFβ2, Snail1, and FN mRNA expression levels were investigated. Details of the primers are shown in Table 1. PCR amplification was performed with an initial denaturing step at 94 °C for 2 min, followed by cycles of denaturation (94 °C, 30 s), annealing (63 °C and 40 cycles for TGF-β1, 56 °C and 35 cycles for TGF-β2, 55 °C and 35 cycles for Snail1, and 55 °C and 30 s for FN), polymerization (72 °C and 30 s for TGF-β1 and 72 °C and 45 s for TGF-β2, Snail1, and FN), and then terminal polymerization (72 °C and 5 min for TGF-β1 and 72 °C and 6 min for TGF-β2, Snail1, and FN). The results were analyzed with a Gel Genius Bioimaging System (Vilber Lourmat Co, Paris, France), and the densitometry values were read by Quantity One software (Bio-Rad Laboratories). Finally, the densitometry values of each RNA analyte normalized to GADPH were compared.
Table 1. RT–PCR primers.
| Gene | Forward Primer 5′–3′ | Reverse Primer 5′–3′ |
|---|---|---|
| TGFβ1 |
GGGACTATCCACCTGCAAGA |
CCTCCTTGGCGTAGTAGTCG |
| TGFβ2 |
GGAGGTGATTTCCATCTACAAC |
TTCAGGCACTCTGGCTTTT |
| Snail1 |
GAGGCGGTGGCAGACTAGAGT |
CGGGCCCCCAGAATAGTTC |
| FN |
TCGAGGAGGAAATTCCAATG |
CTCTTCATGACGCTTGTGGA |
| GAPDH | CACCACCAACTGCTTAGCAC | CCCTGTTGCTGTAGCCAAAT |
Cell apoptosis and viability
Flow cytometry (FACS Aria; BD Bioscience, Franklin Lakes, NJ) was performed to detect cell apoptosis. The following four groups were under investigation: the control group and the 0.3, 0.5, and 1.0 mg/ml PFD groups. We observed apoptosis rates 24 h after treatment began. In accordance with the Annexin V/PI apoptosis kit (Bio Vision, San Francisco, CA), 5×105 cells were collected in tubes, and to each tube, 1 ml 1× Annexin-binding buffer was added and thoroughly mixed. Then, 5 μl Annexin V- fluorescein isothiocyanate and 10 μl propidium iodide (PI) were added. After intensive mixing, the tube was incubated in the dark at 37 °C for 10 min. For cells in the early apoptotic stage, membrane phosphatidylserine was exposed and combined with Annexin V. The cells were stained with Annexin V with no PI fluorescence and recorded as Annexin V positive (+)/ PI negative (-). For cells in the late apoptotic stage and dead cells, the membranes were permeable to PI. The cells were stained with Annexin V and PI that were recorded as Annexin V (+) PI (+). Finally, Annexin V (+) PI (-) and Annexin V (+) PI (+) cells were detected under flow cytometry (Modifit, BD Biosciences), and the percentages in the total number of cells in each group were compared.
The toxicity of PFD was further analyzed using a Trypan Blue exclusion assay. After 0.3, 0.5, or 1.0 mg/ml PFD was added to the RPE cells for 24 h, stained (dead) and unstained (viable) cells were imaged under a microscope using a hematocytometer. Cell viability was estimated by comparing the survival rates of cells in each group. The survival rate was calculated according to the following formula: % survival rate=(viable cell count/total cell count) × 100.
Statistical analysis
All experiments were performed in triplicate and repeated at least three times. Data were expressed as the means±standard deviation (SD) of three independent experiments and analyzed using one-way ANOVA, a test of homogeneity of variances, and a post-hoc test (Bonferroni test). P values less than 0.05 were considered significant. In addition, p values less than 0.05 were labeled with a single asterisk and less than 0.01 were designated with double asterisks. The statistical analyses were conducted using Statistical Product and Service Solutions (SPSS) Statistics Software Version 17.0 (SPSS Inc., Chicago, IL).
Results
Pirfenidone-inhibited cell migration
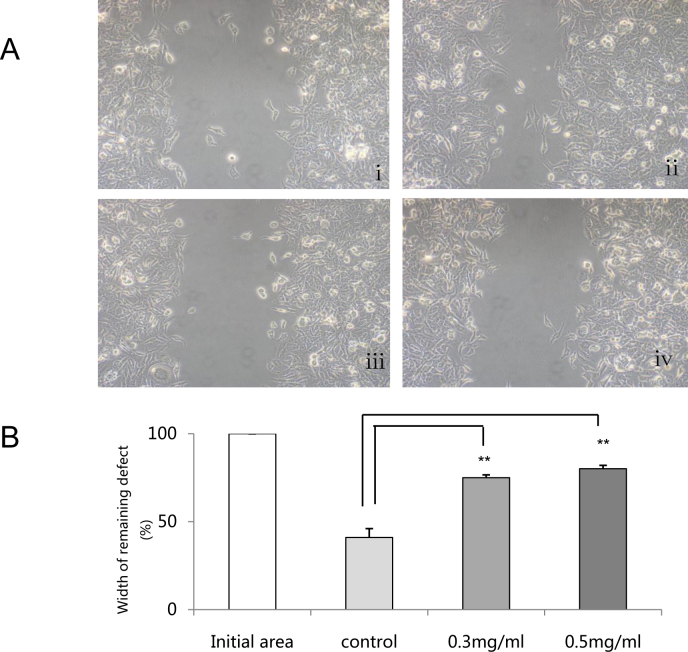
We observed the effects of PFD on cell motility by comparing wound closure in an in vitro wound-healing assay. Values more than the control at 24 h indicated inhibition of migration. As shown in Figure 1A, the width of the wound gap in cells cultured in the absence of PFD was narrower than in cells treated with PFD. The normalized widths of the remaining defects in the control group and the 0.3 and 0.5 mg/ml PFD groups were 41.05%±4.98%, 75%±1.67%, and 80.11%±1.95%, respectively. These results showed that PFD significantly suppressed cell migration (p<0.01; Figure 1B).
Figure 1.
Pirfenidone (PFD) inhibited cell migration. The width of the remaining defect in the control cells was significantly narrower than in cells treated with PFD. A: Light microscope image at the initial time point was taken (i); Light microscope image at 24 h for control cells in serum-free medium was taken (ii); Light microscope image at 24 h for cells treated with 0.3 mg/ml PFD was taken (iii); Light microscope image at 24 h for cells treated with 0.5 mg/ml PFD was taken (iv). B: Summary of three experiments in which the widths of remaining defects were measured in duplicate. The distance between the cell fronts in initial area was set to 100. The widths of the remaining defect after treatment at 24 h were analyzed. Samples significantly different the control sample were indicated with symbols (**n=3, the error bar indicates standard deviation [SD], p<0.01, relative to control).
Pirfenidone-inhibited expression of Snail1, α-smooth muscle actin, and connective tissue growth factor in retinal pigment epithelial cells
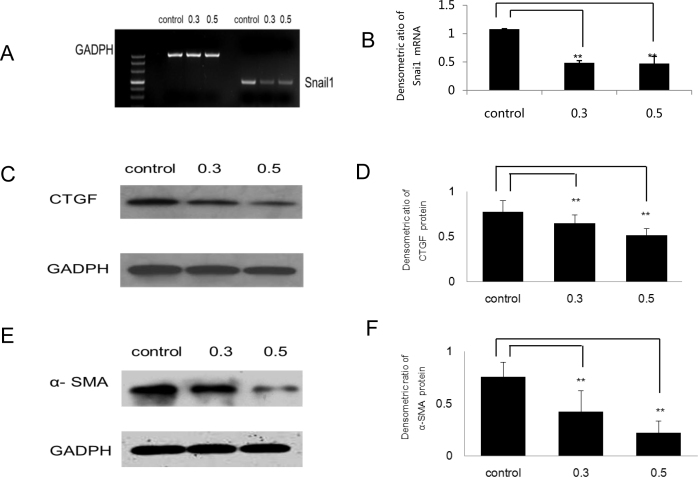
To investigate the effects of PFD on the epithelial-mesenchymal transition (EMT) of RPE cells, we detected the expression of Snail1, α-SMA, and CTGF. After the RPE cells were treated with 0.3 or 0.5 mg/ml PFD for 24 h, the mRNA level of Snail1 was significantly inhibited (p<0.01; Figure 2A,B). From western blot analyses, the results of densitometric analysis showed that PFD significantly downregulated the expression levels of α-SMA (p<0.01) and CTGF (p<0.01; Figure 2C–F).
Figure 2.
Pirfenidone (PFD) inhibited expression of Snail1, α-smooth muscle actin, and connective tissue growth factor in retinal pigment epithelial cells. Retinal pigment epithelial (RPE) cells were incubated for 24 h in complete medium (control), 0.3 mg/ml PFD, or 0.5 mg/ml PFD. A: The expression of Snail1 was assayed with reverse transcriptase (RT)–PCR. C and E: Cells were harvested and subjected to western blot analysis with antibodies against connective tissue growth factor (CTGF) and α-smooth muscle actin (α-SMA). The experiment was repeated at least three times. B, D, and F: The densitometry ratio data are presented. The densitometric ratio is the gene/protein of interest band density divided by the glyceraldehyde-3-phosphate dehydrogenase (GADPH) band density. PFD inhibited expression of Snail1, α-SMA, and CTGF in RPE cells. Samples significantly different from the control sample were indicated with symbols (**n=3, the error bar indicates standard deviation [SD], p<0.01, relative to control).
Pirfenidone-inhibited expression of fibronectin, transforming growth factor beta 1, transforming growth factor beta 2, Smad2/3, and Smad4 in retinal pigment epithelial cells
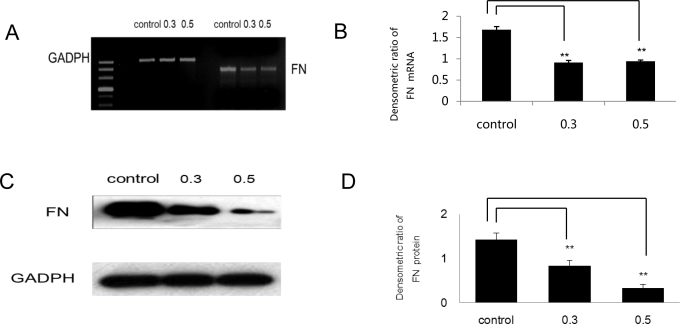
Because FN is an important glycoprotein of the extracellular matrix (ECM) [18], we investigated the expression of FN with reverse transcriptase (RT)–PCR and western blot. Figure 3A,B showed that the mRNA level of FN was significantly inhibited by PFD treatment (p<0.01). The quantitative effect of PFD on FN was shown with densitometry analysis of three independent experiments; at PFD levels of 0.3 mg/ml and 0.5 mg/ml, the protein levels were significantly inhibited (p<0.01; Figure 3C,D).
Figure 3.
Pirfenidone (PFD) inhibited expression of fibronectin in retinal pigment epithelial cells. Retinal pigment epithelial (RPE) cells were treated in the complete medium (control) or 0.3 mg/ml or 0.5 mg/ml PFD for 24 h. Glyceraldehyde-3-phosphate dehydrogenase (GADPH) was used to verify equal loading of genes and proteins. All experiments were repeated in triplicate, and the results were reproducible. A and C: The expression of fibronectin (FN) were assayed using reverse transcriptase (RT)–PCR and western blot analyses. B and D: Compared with cells cultured in the complete medium, the densitometric ratios of FN mRNA and protein (normalized to GADPH) in cells treated with PFD were significantly decreased (**n=3, the error bar indicates standard deviation [SD], p<0.01, relative to control).
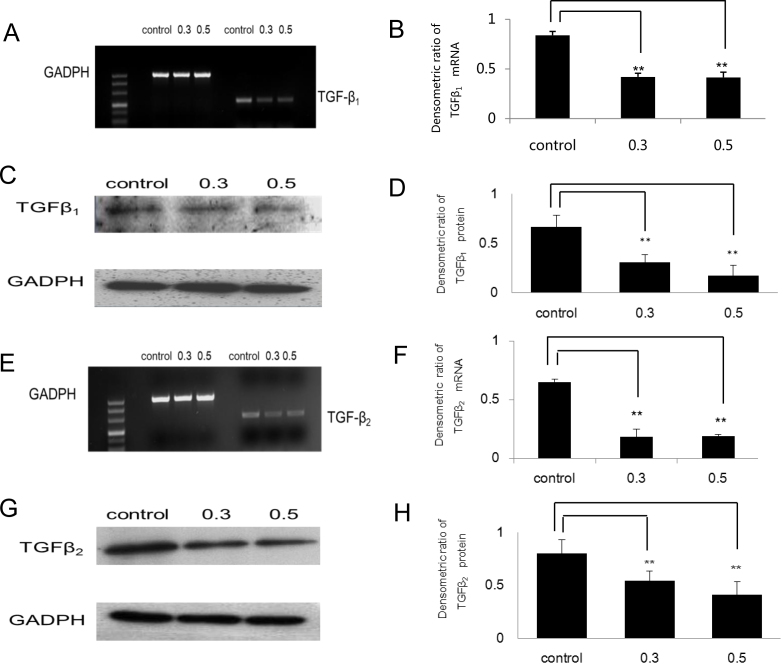
Many reports indicate that diseases characterized by fibrosis can be attributed, at least in part, to increased levels of TGFβ [19,20]. Thus, we examined the effects of PFD on the expression levels of TGFβ1 and TGFβ2. As shown in Figure 4A,B, in part of the TGFβ1 gene, PFD significantly suppressed mRNA expression at the concentrations tested (0.3 mg/ml and 0.5 mg/ml; p<0.01). As for TGFβ2 gene, PFD also significantly downregulated transcription at the 0.3 mg/ml and 0.5 mg/ml doses (p<0.01; Figure 4E,F). Similar results were obtained for protein expression. Densitometry analysis of three independent western blot analyses showed the quantitative inhibition of PFD on the protein levels of TGFβ1 (p<0.01; Figure 4C,D) and TGFβ2 (p<0.01; Figure 4G,H).
Figure 4.
Pirfenidone (PFD) inhibited expression of transforming growth factor beta 1 and transforming growth factor beta 2 in retinal pigment epithelial cells. Retinal pigment epithelial (RPE) cells were treated in the absence or presence of PFD (control, 0.3 mg/ml or 0.5 mg/ml PFD) for 24 h. A and C: Reverse transcriptase (RT)–PCR and western blot analyses for transforming growth factor beta 1 (TGFβ1) were performed. E and G: RT–PCR and western blot results for detecting transforming growth factor beta 2 (TGFβ2) are shown. B, D, G, and H: The densitometry ratio data are presented. The densitometric ratio is the gene/protein of interest band density divided by the glyceraldehyde-3-phosphate dehydrogenase (GADPH) band density. PFD inhibited the mRNA and protein levels of TGFβ1 and TGFβ2 in RPE cells (**n=3, the error bar indicates standard deviation [SD], p<0.01, relative to control).
TGFβ regulates the expression of target genes through several signal pathways, among which the Smad family is an important target [21]. Therefore, western blots were conducted to detect whether PFD interfered with the expression of Smad2/3 and Smad4. A decrease in the expression of Smad2/3 was seen with the application of 0.3 mg/ml and 0.5 mg/ml PFD. Inhibition was statistically significant when evaluated with densitometric analysis (p<0.01; Figure 5A,B). The expression level of Smad4 as measured by western blot was also reduced at doses of 0.3 mg/ml and 0.5 mg/ml PFD (p<0.01; Figure 5C,D).
Figure 5.
Pirfenidone (PFD) inhibited expression of Smad2/3 and Smad4 in retinal pigment epithelial cells. Retinal pigment epithelial (RPE) cells were treated in the complete medium (control) or 0.3 mg/ml or 0.5 mg/ml PFD for 24 h. A and C: The protein expression of Smad2/3 and Smad4 were investigated using immunoblot analysis. B and D: Densitometry ratio data are presented. The densitometric ratio is the protein of interest band density divided by the glyceraldehyde-3-phosphate dehydrogenase (GADPH) band density. PFD inhibited protein levels of Smad2/3 and Smad4 in RPE cells. Samples significantly different from the control sample are indicated with symbols (**n=3, the error bar indicates standard deviation [SD], p<0.01, relative to control).
Pirfenidone had no effect on cell apoptosis or viability
To confirm that the effect of PFD was not a result of cytotoxicity, cells were exposed to PFD concentrations of 0, 0.3, 0.5, and 1.0 mg/ml. Cell apoptosis and survival rate were then examined.
Through Annexin V/PI staining, cells in the apoptotic stages were labeled and detected with flow cytometry. As shown in Figure 6A, most cells were normal that were negative in staining (shown in the Q3 area). Cells in the early apoptotic stage were stained with Annexin V labeled as Annexin V (+) PI (-; shown in the Q4 area). Cells in the late apoptotic stage and dead cells were stained with Annexin V and PI labeled as Annexin V (+) PI (+; shown in the Q2 area). The percentage of cells labeled as Annexin V (+) PI (-) was 1.33%±1.02%, 1.57%±0.93%, 2.63%±1.08%, and 1.90%±1.13% in cells untreated and cells treated with 0.3, 0.5, and 1.0 mg/ml PFD, respectively. No significant differences were found between the cells (p=0.701, p>0.05). The percentages of cells labeled as Annexin V (+) PI (+) were 2.17%±1.46%, 3.77%±2.14%, 4.46%±1.10%, and 4.65%±0.35% in the control group and the 0.3, 0.5, and 1.0 mg/ml PFD-treated groups. There was a slight trend toward more late apoptosis with PFD treatment, but this did not reach statistical significance (p=0.283, p>0.05; Figure 6B).
Figure 6.
Pirfenidone (PFD) had no effect on cell apoptosis. Retinal pigment epithelial (RPE) cells were incubated for 24 h with complete medium (control), 0.3 mg/ml pirfenidone, 0.5 mg/ml PFD, or 1.0 mg/ml PFD. A: Cells labeled with Annexin V (-) PI (+) were shown in Q1 area; cells labeled with Annexin V (+) PI (+) were shown in Q2 area; cells labeled with Annexin V (-) PI (-) were shown in Q3 area; cells labeled with Annexin V (+) PI (-) were shown in Q4 area. B: The percentages of cells labeled as Annexin V (+) PI (-) and Annexin V (+) PI (+) were investigated using flow cytometry. There were no significant differences between the groups (n=4, the error bar indicates standard deviation [SD], p>0.05).
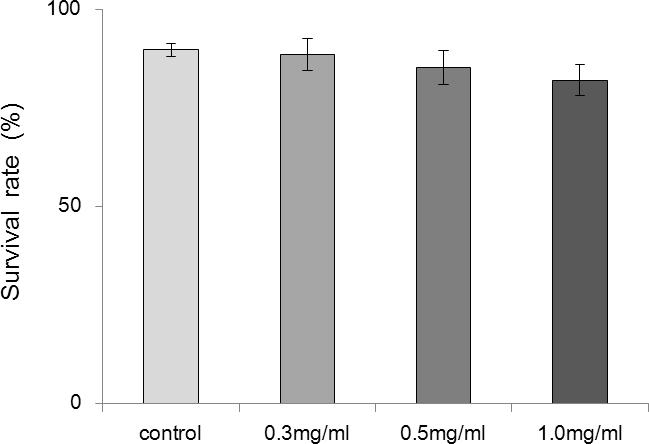
A Trypan Blue assay was also used to detect RPE cell viability. The survival rates of untreated cells and cells treated with 0.3, 0.5, or 1.0 mg/ml PFD were 89.75%±1.71%, 88.50%±4.04%, 85.25%±4.27%, and 82%±3.92%, respectively. These differences were insignificant (p=0.113, p>0.05), further confirming the lack of toxicity of PFD on RPE cells at concentrations below 1.0 mg/ml (Figure 7).
Figure 7.
Pirfenidone had no effect on cell viability. Retinal pigment epithelial (RPE) cells were incubated for 24 h with complete medium (control), 0.3 mg/ml pirfenidone (PFD), 0.5 mg/ml PFD, or 1.0 mg/ml PFD. Cell viability was estimated using a Trypan Blue assay. The survival rates were compared. There were no significant differences between the groups (n=4, the error bar indicates standard deviation [SD], p>0.05).
Discussion
RPE cells have been regarded as a predominant cell type in the pathogenesis of PVR [22]. Thus, agents capable of interfering with biologic behaviors of RPE cells might mitigate the development of PVR. PFD exhibits its antifibrotic effect on a host of cell types in vitro. On this basis, we investigated PFD’s effects on the migration, differentiation, and proliferation of RPE cells cultured in vitro. Our results demonstrated that PFD significantly suppressed cell migration and mRNA expression of Snail1, FN, TGFβ1, and TGFβ2. We also found that PFD inhibited protein levels of α-SMA, CTGF, FN, TGFβ1, TGFβ2, Smad2/3, and Smad4. As far as we know, no receptor for PFD has been found. PFD takes effect mainly through regulation of several signal pathways involved in inflammation and proliferation. In a vitro study on cultured hepatic stellate cells, PFD inhibits platelet derived growth factor (PDGF)-induced activation of the Na+/K+ exchanger and protein kinase C, but the inhibition is irrelevant with PDGF-receptor autophosphorylation. PFD also inhibits collagen accumulation induced by TGFβ1 [23]. In a mouse model of left ventricular remodeling, PFD attenuated expression of interleukin-1β, and interleukin-1β induced inflammatory and profibrotic responses [24].
Some researchers have observed RPE cells can lose their intrinsic epithelial characteristics and acquire mesenchymal features, which implies that RPE cells undergo EMT [25]. Choi et al. added exogenous TGFβ1 to induce RPE cell transformation and observed typical morphological changes such as elongated and spindle-like morphology, which was suppressed by pretreatment with PFD. He also found that TGFβ1 induced the reorganization of F-actin and that cells failed to mature in the presence of PFD. We investigated whether PFD was able to interfere with the EMT in RPE cells in another way. Snail1 is a transcription repressor often considered a marker of the EMT [26]. Olmeda et al. showed that if the transcription of Snail1 in renal tubular epithelial cells was blocked, the EMT would stop entirely [27]. In our experiments, 0.3 mg/ml and 0.5 mg/ml PFD both suppressed Snail1 mRNA significantly. α-SMA is an ultrastructure of the cytoskeleton and is the most widely used marker to identify fibroblasts [28]. CTGF is abundantly expressed in human connective tissues and organs [29]. We demonstrated that PFD inhibited the protein levels of α-SMA and CTGF significantly. The densitometry values in cells treated with PFD were significantly decreased compared to the untreated cells. Because these proteins are often associated with the EMT, our results imply that PFD might interfere with the EMT in RPE cells.
TGFβs are among the most important cytokines involved in regulating cell proliferation [30]. Current evidence suggests that TGFβs and molecules downstream of TGFβ are key therapeutic targets in treating fibrotic disorders in the eye [31,32]. In addition, TGFβs not only affect cell proliferation but may also be therapeutic targets for inhibiting the EMT by disrupting TGFβ-associated signaling and/or suppressing Smad activation [28]. According to our results, TGFβ1 and TGFβ2 were downregulated at the mRNA and protein levels. Densitometric analysis with protein/RNA analyte showed significant inhibition in the PFD group when compared with the control group implying that PFD significantly suppresses TGFβ expression.
Choi et al. showed that PFD had no effect on the expression of p-Smad2/3 induced by TGFβ1, but instead inhibited the translocation of active Smad2/3 into the nucleus [17]. We examined total Smad2/3 and Smad4 expression levels. The observation that PFD suppressed total Smad2/3 and Smad4 synthesis showed that PFD disrupted Smad signaling. Choi et al. investigated only the effects over 6 h, but we prolonged the observation time to 24 h. Moreover, Choi et al. mainly focused on 0.5 or 1.0 mg/ml concentrations of PFD over various time periods up to 6 h. We investigated one time point under different concentrations of PFD and demonstrated that beginning at a dose of 0.3 mg/ml, PFD inhibited the migration, differentiation, and proliferation of RPE cells. As shown in the migration assay, RT–PCR, and western blot analyses, inhibition was statistically significant. These data add to our knowledge of the dose and effect response of PFD.
Thus far, no life-threatening effects of PFD have been reported. In vivo and in vitro experiments on the application of PFD have verified its safety at the appropriate concentrations [33,34]. Clinical trials have also verified the safety of PFD in oral usage up to 1,800 mg daily [10]. Our previous studies also showed that PFD below 1.0 mg/ml caused no cytotoxicity to human Tenon’s fibroblasts. Furthermore, 0.5% PFD eye drops did not harm ocular tissues [14,15]. Consistent with these findings, we also found that PFD did not cause toxicity at concentrations up to 1.0 mg/ml in RPE cells. However, we just verified no cytotoxicity of PFD to RPE cells. Evaluating the toxicity in other retinal cells such as retinal neurosensorial cells through experiments in vitro would be meaningful. Moreover, since an in vitro experiment to assess the effect and toxicity of PFD may be insufficient, our group is conducting more experiments in vivo to investigate the effect of PFD.
In summary, PFD treatment can suppress certain behaviors of RPE cells, including migration, differentiation, and proliferation. These effects were not attributed to cytotoxicity for concentrations up to 1.0 mg/ml. These findings might contribute to further therapeutic solutions for PVR.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (Grant No. 81,170,848, Minbin Yu) and the Doctoral Fund of Ministry of Education of China (Grant No. 20120171130013, Minbin Yu).
References
- 1.Sadaka A, Giuliari GP. Proliferative vitreoretinopathy: current and emerging treatments. Clin Ophthalmol. 2012;6:1325–33. doi: 10.2147/OPTH.S27896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pastor JC. Proliferative vitreoretinopathy: An Overview. Surv Ophthalmol. 1998;43:3–18. doi: 10.1016/s0039-6257(98)00023-x. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Chen H, Hou P, Fok A, Hu Y, Lam DS. Midterm results of low-dose intravitreal triamcinolone as adjunctive treatment for proliferative vitreoretinopathy. Retina. 2011;31:1137–42. doi: 10.1097/IAE.0b013e3181fe5427. [DOI] [PubMed] [Google Scholar]
- 4.Blumenkranz M, Hernadez E, Ophir A, Norton EW. 5-fluorouracil: new applications in complicated retinal detachment for an established antimetabolite. Ophthalmology. 1984;91:122–30. doi: 10.1016/s0161-6420(84)34318-4. [DOI] [PubMed] [Google Scholar]
- 5.Wickham L, Bunce C, Wong D, McGurn D, Charteris DG. A randomized controlled trial of combined 5-fluorouracil and low-molecular-weight heparin in the management of unselected rhegmatogenous retinal detachments undergoing primary vitrectomy. Ophthalmology. 2007;114:698–704. doi: 10.1016/j.ophtha.2006.08.042. [DOI] [PubMed] [Google Scholar]
- 6.Creten O, Spileers W, Stalmans P. Systemic resportion of 5-fluorouracil used in infusion fliud during vitrectomy. Bull Soc Belge Ophtalmol. 2007;303:37–41. [PubMed] [Google Scholar]
- 7.Iyer SN, Gurujeyalakshmi G, Giri SN. Effects of pirfenidone on procollagen gene expression at the transcriptional level in bleomycin hamster model of lung fibrosis. Pharmacol Exp Ther. 1999;289:211–8. [PubMed] [Google Scholar]
- 8.Suga H, Teraoka S, Ota K, Komemushi S, Furutani S, Yamauchi S. Margolin s. Preventive effect of pirfenidone against experimental sclerosing peritonitis in rats. Exp Toxicol Pathol. 1995;47:287–91. doi: 10.1016/s0940-2993(11)80261-7. [DOI] [PubMed] [Google Scholar]
- 9.Miric G, Dallemagne C, Endre Z, Margolin S, Taylor SM, Brown L. Reversal of cardiac and renal fibrosis by pirfenidone and spironolactone in streptozotocin-diabetic rats. Br J Pharmacol. 2001;133:687–94. doi: 10.1038/sj.bjp.0704131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker JE, Margolin SB. Pirfenidone for chronic progressive multiple sclerosis. Mult Scler. 2001;7:305–12. doi: 10.1177/135245850100700506. [DOI] [PubMed] [Google Scholar]
- 11.Azuma A, Nukiwa T, Tsuboi E, Suga M, Nakata K, Taguchi Y, Nagai S, Itoh H, Ochi M, Sato A, Kudoh S. Double-blind, placebo-controlled trial of pirfenidone in patients with idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2005;171:1040–7. doi: 10.1164/rccm.200404-571OC. [DOI] [PubMed] [Google Scholar]
- 12.Azuma A, Nukiwa T, Tsuboi E, Suga M, Abe S, Nakata K, Taguchi Y, Nagai S, Itoh H, Ohi M, Sato A, Kudoh S. Double-blind, placebo-controlled trial of pirfenidone in patients with idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2005;171:1040–7. doi: 10.1164/rccm.200404-571OC. [DOI] [PubMed] [Google Scholar]
- 13.Available at http:// www. intermune. com/ Pirfenidone. Accessed January, 5th, 2013.
- 14.Lin X, Yu M, Wu K, Yuan H, Zhong H. Effects of pirfenidone on proliferation, migration, and collagen contraction of human Tenon's fibroblasts in vitro. Invest Ophthalmol Vis Sci. 2009;50:3763–70. doi: 10.1167/iovs.08-2815. [DOI] [PubMed] [Google Scholar]
- 15.Zhong H, Sun G, Lin X, Wu K, Yu M. Evaluation of pirfenidone as a new postoperative antiscarring agent in experimental glaucoma surgery. Invest Ophthalmol Vis Sci. 2011;52:3136–42. doi: 10.1167/iovs.10-6240. [DOI] [PubMed] [Google Scholar]
- 16.Sun G, Lin X, Zhong H, Yang Y, Qiu X, Ye C, Wu K, Yu M. Pharmacokinetics of pirfenidone after topical administration in rabbit eye. Mol Vis. 2011;17:2191–6. [PMC free article] [PubMed] [Google Scholar]
- 17.Choi K, Lee K, Ryu S-W, Im M, Kook KH, Choi C. Pirfenidone inhibits transforming growth factor-β1-induced fibrogenesis by blocking nuclear translocation of Smads in human retinal pigment epithelial cell line ARPE-19. Mol Vis. 2012;18:1010–20. [PMC free article] [PubMed] [Google Scholar]
- 18.Fromme HG, Voss B, Pfautsch M, Grote M, von Figura K, Beeck H. Immunoelectron-microscopic study on the location of fibronectin in human fibroblast cultures. Ultrastruct Res. 1982;80:264–9. doi: 10.1016/s0022-5320(82)80039-7. [DOI] [PubMed] [Google Scholar]
- 19.Saika S. TGF beta pathobiology in the eye. Lab Invest. 2006;86:106–15. doi: 10.1038/labinvest.3700375. [DOI] [PubMed] [Google Scholar]
- 20.Saika S, Yamanaka O, Okada Y, Tanaka S, Miyamoto T, Sumioka T, Kitano A, Shirai K, Ikeda K. TGF beta in fibroproliferative diseases in the eye. Front Biosci. 2009;1:376–90. doi: 10.2741/S32. Schol Ed. [DOI] [PubMed] [Google Scholar]
- 21.Pennison M, Pasche B. Targeting transforming growth factor beta signaling. Curr Opin Oncol. 2007;19:579–85. doi: 10.1097/CCO.0b013e3282f0ad0e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pastor JC, de la Rúa ER, Martín F. Proliferative vitreoretinopathy: risk factors and pathobiology. Prog Retin Eye Res. 2002;21:127–44. doi: 10.1016/s1350-9462(01)00023-4. [DOI] [PubMed] [Google Scholar]
- 23.Di Sario A, Bendia E, Baroni GS, Ridolfi F, Casini A, Ceni E, Saccomanno S, Marzioni M, Trozzi L, Sterpetti P, Taffetani S, Benedetti A. Effect of pirfenidone on rat hepatic stellate cell proliferation and collagen production. J Hepatol. 2002;37:584–91. doi: 10.1016/s0168-8278(02)00245-3. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Wu Y, Chen J, Zhan S, Li H. Pirfenidone Attenuates Cardiac Fibrosis in a Mouse Model of TAC-Induced Left Ventricular Remodeling by Suppressing NLRP3 Inflammasome Formation. Cardiology. 2013;126:1–11. doi: 10.1159/000351179. [DOI] [PubMed] [Google Scholar]
- 25.Laqua H, Machemer R. Clinical-pathological correlation in massive periretinal proliferation. Am J Ophthalmol. 1975;80:913–29. doi: 10.1016/0002-9394(75)90289-5. [DOI] [PubMed] [Google Scholar]
- 26.Kirchhof B, Kirchhof E, Ryan SJ, Sorgente N. Vitreous modulation of migration and proliferation of retinal pigment epithelial cells in vitro. Invest Ophthalmol Vis Sci. 1989;30:1951–7. [PubMed] [Google Scholar]
- 27.Olmeda D, Jordá M, Peinado H, Fabra A, Cano A. Snail silencing effectively suppresses tumour growth and invasiveness. Oncogene. 2007;26:1862–74. doi: 10.1038/sj.onc.1209997. [DOI] [PubMed] [Google Scholar]
- 28.Hinton DR, He S, Jin ML, Barron E, Ryan SJ. Novel growth factors involved in the pathogenesis of proliferative vitreoretinopathy. Eye (Lond) 2002;16:422–8. doi: 10.1038/sj.eye.6700190. [DOI] [PubMed] [Google Scholar]
- 29.Wang Q, Usinger W, Nichols B, Gray J, Xu L, Seeley TW, Brenner M, Guo G, Zhang W, Oliver N, Lin A, Yeowell D. Cooperative interaction of CTGF and TGF-beta in animal models of fibrotic disease. Fibrogenesis Tissue Repair. 2011;4:4. doi: 10.1186/1755-1536-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pennison M, Pasche B. Targeting transforming growth factor beta signaling. Curr Opin Oncol. 2007;19:579–85. doi: 10.1097/CCO.0b013e3282f0ad0e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parapuram SK, Chang B, Li L, Hartung RA, Chalam KV, Nair-Menon JU, Hunt DM, Hunt RC. Differential Effects of TGFβ and Vitreous on the Transformation of Retinal Pigment Epithelial Cells. IVOS. 2009;50:5965–74. doi: 10.1167/iovs.09-3621. [DOI] [PubMed] [Google Scholar]
- 32.Kita T, Hata Y, Arita R, Kawahara S, Miura M, Nakao S, Mochizuki Y, Enaida H, Goto Y, Shimokawa H, Hafezi-Moghadam A, Ishibashi T. Role of TGF-β in proliferative vitreoretinal diseases and ROCK as a therapeutic target. Proc Natl Acad Sci USA. 2008;105:17504–9. doi: 10.1073/pnas.0804054105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tada S, Nakamuta M, Enjoji M, Sugimoto R, Iwamoto H, Kato M, Nakashima Y, Nawata H. Pirfenidone inhibits dimethylnitrosamine induced hepatic fibrosis in rat. Clin Exp Pharmacol Physiol. 2001;28:522–7. doi: 10.1046/j.1440-1681.2001.03481.x. [DOI] [PubMed] [Google Scholar]
- 34.Kaibori M, Yanagida H, Uchida Y, Yokoigawa N, Kwon AH, Okumura T, Kamiyama Y. Pirfenidone protects endotoxin-induced liver injury after hepatic ischemia in rats. Transplant Proc. 2004;36:1973–4. doi: 10.1016/j.transproceed.2004.08.061. [DOI] [PubMed] [Google Scholar]