Abstract
Although dedifferentiation, transformation of differentiated cells into progenitor cells, is a critical step in the regeneration of amphibians and fish, the molecular mechanisms underlying this process, including epigenetic changes, remain unclear. Dot blot assays and immunohistochemical analyses revealed that, during regeneration of zebrafish fin, the levels of 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) are transiently reduced in blastema cells and cells adjacent to the amputation plane at 30 h post-amputation (hpa), and the level of 5mC, but not 5hmC, is almost restored by 72 hpa. We observed that the dedifferentiated cells showed reduced levels of 5mC and 5hmC independent of cell proliferation by 24 hpa. Furthermore, expressions of the proposed demethylation- and DNA repair-related genes were detected during fin regeneration. Taken together, our findings illustrate that the transient reduction of 5mC and 5hmC in dedifferentiated cells is associated with active demethylation during regeneration of zebrafish fin.
Keywords: zebrafish, fin regeneration, 5-methylcytosine, 5-hydroxymethylcytosine, active DNA demethylation
Introduction
In animals, cytosine methylation (5-methylcytosine; 5mC) of genomic DNA is an epigenetic mechanism that is implicated in many biological processes, such as regulation of gene expression, cellular proliferation, differentiation, pluripotency, oncogenesis, and genomic imprinting.1-3 The methylation pattern of genomic DNA is established and maintained by the activity of de novo and maintenance methyltransferases.4,5 Conversely, 5mC is reverted to cytosine through active demethylation, which is well studied in the reprogramming of the paternal pronuclei in fertilized mouse zygotes and in primordial germ cells during mouse embryonic development.1,3 However, the mechanisms underlying active demethylation are still unclear.1,3 Recently, numerous and intense studies have shown that active demethylation is regulated by many enzymes including cytidine deaminase (activation-induced deaminase, AID; apolipoprotein B mRNA-editing enzymes, Apobec), G/T mismatch DNA glycosylase (thymine-DNA glycosylase, TDG; methyl CpG binding domain protein 4, MBD4), methylcytosine dioxygenase (ten-11 translocation, Tet), and DNA repair-related factors such as poly-(ADP-ribose) polymerase (PARP), uracil DNA glycosylase (UNG), and MutS homolog 2/6 (MSH2/6).1,3,6,7 In one of these proposed demethylation pathways, 5mC is modified to 5-hydroxymethylcytosine (5hmC) by Tet proteins, and 5hmC is thought to be a novel epigenetic mark for gene regulation.8-10
Zebrafish is a common animal model and has been used to study regeneration of various organs.11-13 It is well known that dedifferentiation processes such as the loss of molecular markers for differentiated cells, re-expression of molecular markers for progenitor cells, and restart of cell proliferation occur during regeneration in amphibians and zebrafish.13,14 Although epigenetic modifications are thought to be critical for the dedifferentiation processes in regeneration,15,16 only 2 studies concerning DNA methylation have been reported thus far. These studies showed that the demethylation of a Xenopus elongation factor 1-α (ef1-α):EGFP transgene in transgenic zebrafish (Tg[ef1-α:EGFP]) was observed during fin regeneration17 and that the sonic hedgehog gene expression is correlated with the methylation status of the limb-specific sonic hedgehog enhancer in Xenopus limb regeneration.18 However, despite these findings, the status and changes of DNA methylation during regeneration remain largely unknown.
It has been already reported that during early zebrafish development, the dynamic change of the 5mC or 5hmC distribution was observed in zebrafish embryos and larvae.19-22 However, the distribution of these epigenetic markers during regeneration has not yet been reported. In this study, we analyzed the spatial and temporal changes of 5mC or 5hmC distribution during zebrafish fin regeneration by using dot blot assays and immunohistochemical analyses.
Results
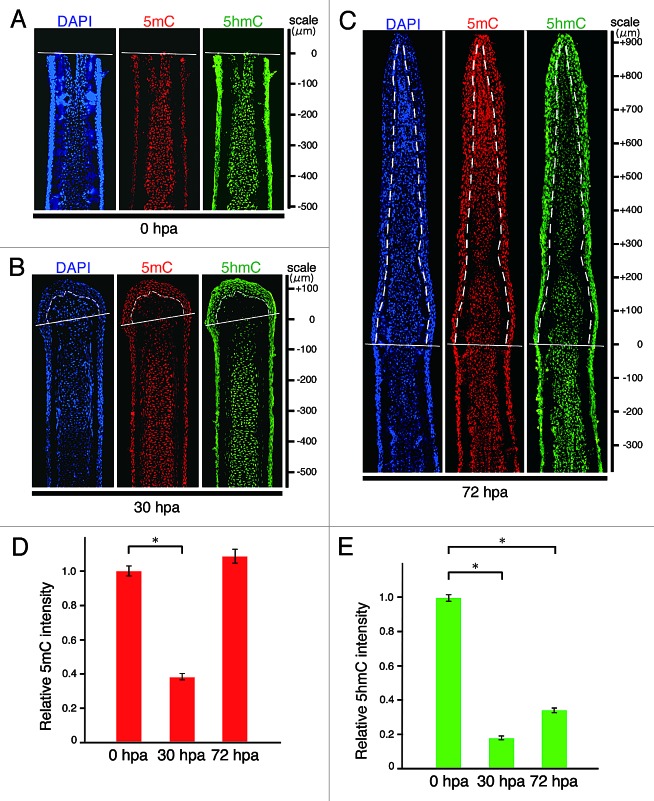
Because knowledge concerning changes in 5mC or 5hmC level during zebrafish fin regeneration is limited, we evaluated the level of the two epigenetic marks by using dot blot assays in genomic DNA from amputated fins at 0, 30, and 72 h post-amputation (hpa). The levels of both 5mC and 5hmC at 30 hpa were significantly lower than those at 0 hpa (Fig. S1). These results suggest that both epigenetic marks are transiently downregulated in the early stages of fin regeneration. To explore the spatial distribution of 5mC or 5hmC in the fin regenerates, we next performed immunohistochemical staining by using longitudinal sections of the regenerates (Fig. S2). The intra-ray cells, which show the same level of 5mC or 5hmC fluorescent signal, were uniformly distributed within the 500 μm proximal region to the amputation plane at 0 hpa (Fig. 1A). At 0 hpa, the fluorescent level of 5mC or 5hmC did not change from that before amputation (Fig. S3). However, we found that both signals start to reduce in the cells adjacent to the amputation plane at around 12 hpa and these reduced signals are evident by 18 hpa (Fig. S4). At 30 hpa, the fluorescent signals of 5mC and 5hmC in the blastema and intra-ray cells within the 150 μm proximal region to the amputation plane were markedly lower than those in the intra-ray cells at 0 hpa (Fig. 1B, D, and E). In the blastema cells, the level of 5mC signal was almost restored at 72 hpa (Fig. 1C and D), whereas the level of 5hmC signal at 72 hpa remained lower than that at 0 hpa (Fig. 1C and E). The 5hmC level was gradually upregulated by the end of regeneration (14 d post amputation [dpa]) (Fig. S5 and data not shown). Interestingly, high levels of 5mC and 5hmC in the epidermal and inter-ray cells are maintained before and after regeneration by immunohistochemical staining (Fig. 1; Figs. S2 and S4, and data not shown). Because these epidermal and inter-ray cells are included in dot blot assays, the relative 5mC or 5hmC level at 30 hpa observed in the dot blot assays could be higher than that observed in the immunohistochemical analyses (Fig. 1; Fig. S1).
Figure 1. Spatial and temporal distributions of 5mC and 5hmC during regeneration of zebrafish fin. (A–C) Longitudinal sections of wild type fin regenerates that were immunohistochmically stained with antibodies against 5mC (red) and 5hmC (green) at 0 (A), 30 (B), and 72 hpa (C). The fluorescent signals of DAPI (blue) indicate the presence of nuclei. Uniform distributions of 5mC and 5hmC fluorescent signals were observed in the intra-ray cells at 0 hpa (A). The fluorescent signals of both 5mC and 5hmC in the balstema cells and cells within 150 μm proximal to the amputation plane at 30 hpa were lower than those at 0 hpa (B). At 72 hpa, the 5mC level in the blastema cells was almost restored, whereas the 5hmC level in the blastema cells was still lower than that at 0 hpa (C). White lines indicate the amputation planes. Dashed lines outline the basement membrane, which shows the boundary between the epidermis and blastema. Scale bars represent the distance from the amputation plane. (D andE): Quantification of the relative fluorescent signal of 5mC or 5hmC at 0, 30, and 72 hpa. Relative 5mC intensity at 30 hpa, and relative 5hmC intensities at 30 and 72 hpa, were significantly lower than those at 0 hpa. In contrast, relative 5mC intensity at 72 hpa was almost the same as that at 0 hpa. * p < 0.001 by Student’s t- test. Error bars represent the standard error.
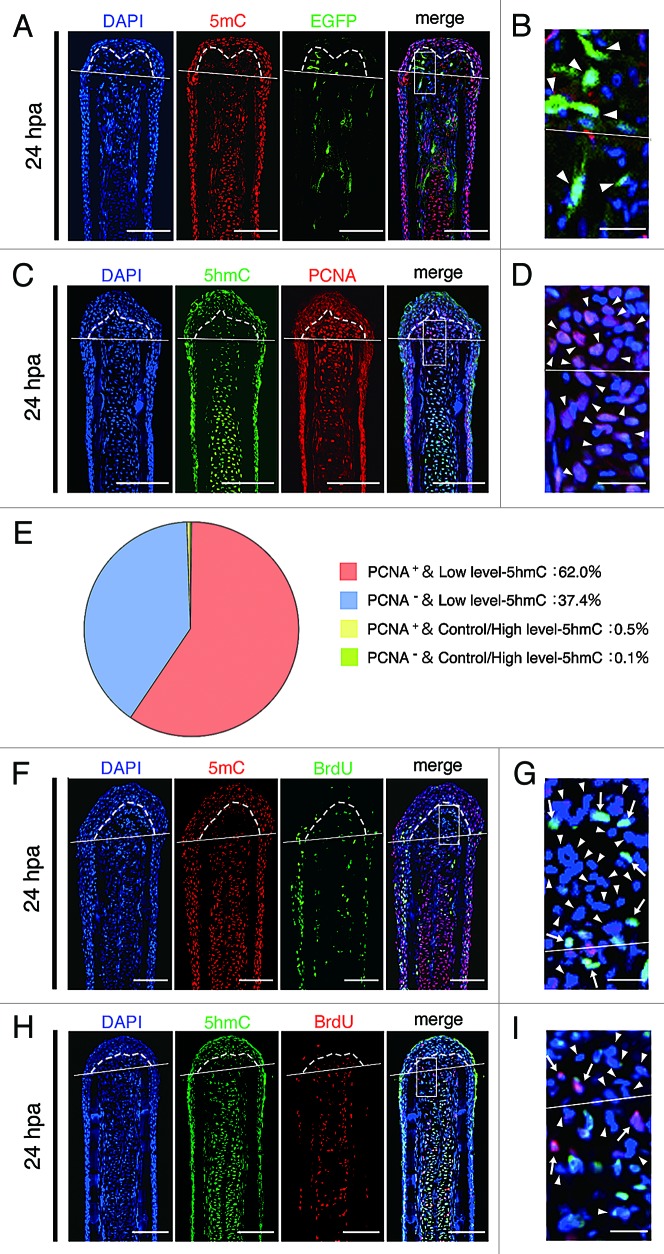
To test whether the intra-ray cells, which show reduced levels of 5mC and 5hmC, are dedifferentiated cells, we examined the expression of Xenopus ef1-α:EGFP transgene by using Tg(ef1-α:EGFP)23 and proliferating cell nuclear antigen (PCNA), which is detected throughout the G1, S, and G2/M phases.24 Although the expression of Xenopus ef1-α:EGFP transgene was detected during development (data not shown), EGFP fluorescence was lost in adult fin (Fig. S6). After amputation, this transgene was re-expressed in some of the blastema cells and cells adjacent to the amputation plane (Fig. 2A; Fig. S6G and K), indicating that theses EGFP+ cells are dedifferentiated cells. Immunohistochemical staining using a 5mC antibody revealed that these EGFP+ cells show reduced level of 5mC at 24 hpa (Fig. 2A and B). Moreover, the majority of the blastema cells showing low-5hmC are PCNA-positive at 24 hpa (Fig. 2C–E). These results suggest that these blastema cells are dedifferentiated cells and are re-entering the cell cycle. In addition, these results suggest that demethylation during fin regeneration may be a replication-dependent process (i.e., passive demethylation). To investigate this possibility, we examined the relationship between the demethylation and cell proliferation in the blastema cells and cells adjacent to the amputation plane by analyzing the bromodeoxyuridine (BrdU) incorporation. Most blastema cells and cells adjacent to the amputation plane, which show reduced level of 5mC or 5hmC, did not incorporate the BrdU until 24 hpa (Fig. 2F–I), indicating that these cells are still in the G1 phase, and not in the S or G2/M phase, and the active demethylation processes are associated with the reduction of both 5mC and 5hmC in the blastema cells and cells adjacent to the amputation plane until 24 hpa.
Figure 2. Downregulation of 5mC or 5hmC level is associated with active demethylation. (A and B): Longitudinal sections of Tg(ef1-α:EGFP) fin regenerates that immunohistochemimally stained with an antibody against 5mC at 24 hpa. Merged images revealed that some cells, which show low level of 5mC, are EGFP fluorescence positive in the blastema cells and cells adjacent to amputation plane (A, arrowheads in B). The boxed area in A is shown enlarged in B. (C–E) Longitudinal sections of wild type fin regenerates that were co-stained with antibodies against 5hmC and PCNA at 24 hpa (C). The boxed area in C is shown enlarged in D. Quantification of the relative 5hmC intensity and PCNA in the blastema area at 24 hpa (E). Merged images and quantification data revealed that the majority of the blastema nuclei (62.0%) show PCNA-positive and low level-5hmC (E, C, arrowheads in D). Control/High level-5hmC: control or high level of 5hmC, Low level-5hmC: low level of 5hmC. (F–I) Longitudinal sections of wild type fin regenerates that were co-stained with antibodies against 5mC/5hmC and BrdU at 24 hpa, respectively. BrdU is not incorporated in almost all blastema cells and cells adjacent to amputation plane that show lower level of 5mC or 5hmC (FandH, arrowheads in GandI). However, BrdU is incorporated in some blastema cells and cells adjacent to the amputation plane, which show lower level of 5mC or 5hmC (arrows in G and I). The boxed areas in F and H are shown enlarged in G and I, respectively. White lines indicate the amputation planes. Dashed lines outline the basement membrane, which shows the boundary between the epidermis and blastema. Scale bars:100 μm in A, C, F, and H; 20 μm in B, D, G, and I.
Recently, numerous and intense investigations have shown that active demethylation is regulated by specific enzymes, including members of the cytidine deaminase family, G/T mismatch DNA glycosylase, and methylcytosine dioxygenase, as well as DNA repair factors.1,3,6,7 Moreover, a recent paper showed that many demethylation-related genes are expressed during regeneration of zebrafish retina.25 Therefore, we next examined the expressions of 11 demethylation- and DNA repair-related genes (growth arrest and DNA damage 45ba; gadd45ba, gadd45bb, gadd45 g, aid, apobec2a, apobec2b, tdg, mbd4, tet2, tet3, and parp1) during fin regeneration by using quantitative real-time PCR (qPCR). Expressions of aid and apobec2a were not detected before and after fin amputation (data not shown). Although the relative expressions of gadd45ba, gadd45bb, apobec2b, tdg, mbd4, tet2, tet3, and parp1 were not markedly upregulated after fin amputation, the relative expression of gadd45 g was significantly upregulated at 18 and 30 hpa (Fig. 3). These data also show that downregulation of 5mC and 5hmC is associated with active demethylation during fin regeneration.
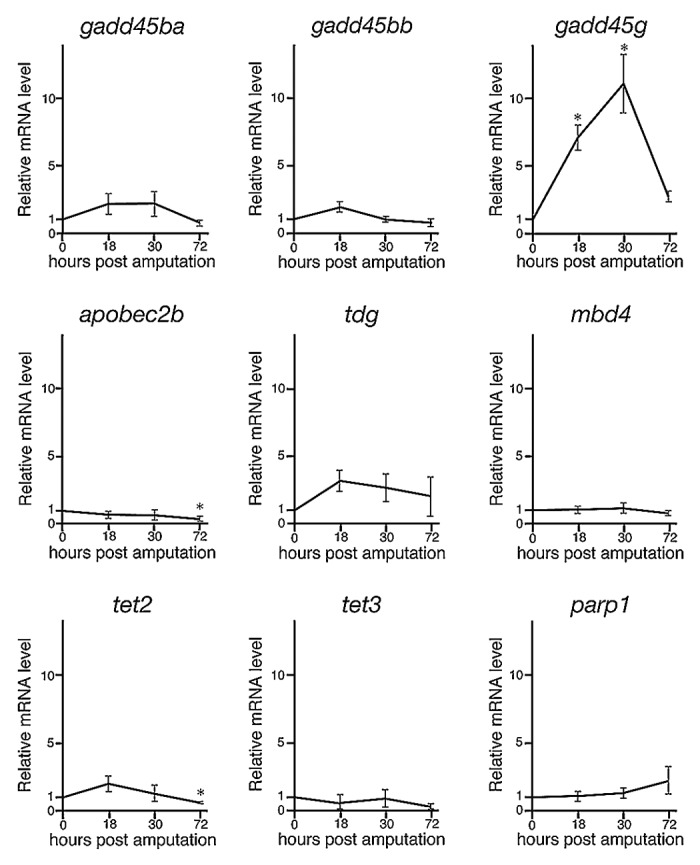
Figure 3. The relative expressions of demethylation- and DNA repair-related genes during regeneration of zebrafish fin by qPCR. The expression of demethylation- and DNA repair-related genes were examined via qPCR at 0, 18, 30, and 72 hpa. The expression level at 0 hpa is set as 1.0, and the relative mRNA levels (y-axis) of each gene were shown at 18, 30, and 72 hpa. The relative expressions of gadd45ba, gadd45bb, apobec2b, tdg, mbd4, tet2, tet3, and parp1 were not markedly upregulated after regeneration. In contrast, the relative expression of gadd45 g was significantly upregulated at 18 and 30 hpa. * p < 0.001 by Student’s t-test. Error bars represent the standard error of four independent experiments.
Discussion
A previous report revealed that, after fin amputation, mesenchymal tissue is disorganized and intra-ray cells, which are proximally located (up to 160 μm away from the amputation plane), migrate to the amputation plane.11,12,26 In this study, we showed that the intra-ray cells, which exhibit reduced level of 5mC or 5hmC, are not observed within 160 μm from the amputation plane at 0 hpa (Fig. 1). Our results, combined with the findings of a previous report, suggest that the levels of 5mC and 5hmC in the intra-ray cells are reduced during or after migration to the amputation plane.
Our results indicated that although the majority of the blastema cells are PCNA positive, almost all of them did not incorporate the BrdU by 24 hpa. There are 2 possible explanations for these results: (1) these blastema cells are still in the G1 phase and (2) the DNA repair process is active in the blastema cells at 24 hpa, because PCNA is a component of the base excision repair process and is thought to be active in demethylation pathways.3 PCNA possibly functions in DNA repair process of active demethylation in the blastema cells at 24 hpa. In either case, our results suggest that the reduction of 5mC or 5hmC is thought to be an epigenetic marker for dedifferentiation.
The main finding of our study is that the levels of both 5mC and 5hmC are transiently reduced in the dedifferentiated blastema cells and the cells adjacent to the amputation plane independent of cell proliferation by 24 hpa. This is the first report to suggest that global and active DNA demethylation in dedifferentiated cells occurs during regeneration. Previous studies have reported that the global and active DNA demethylation is also observed in reprogramming process of the paternal pronuclei in fertilized mouse zygotes and primordial germ cells.1-3 Moreover, numerous studies have shown that DNA demethylation influences gene transcription, DNA replication, etc., and that 5hmC is involved in the regulation of gene expression.1-10 Therefore, the reduction of 5mC and 5hmC during fin regeneration may lead to dedifferentiation, in which genes for fin regeneration are re-expressed and the cell cycle is restarted. Further studies will be necessary to elucidate the relationship between dedifferentiation and DNA demethylation during zebrafish fin regeneration.
The results of qPCR revealed that although gadd45 g expression is significantly upregulated at 18 and 30 hpa, the expression level of 8 demethylation- and DNA repair-related genes (gadd45ba, gadd45bb, apobec2b, tdg, mbd4, tet2, tet3, and parp1) is maintained or downregulated after amputation (Fig. 3). Because of the lack of obvious enzymatic activity, Gadd45 family proteins have been linked to DNA demethylation and the DNA repair process as adaptor proteins.7 Recent studies reported that DNA demethylation or DNA repair is promoted when Gadd45 forms a complex with Aid, Apobec, Mbd4, TDG, or other factors for nucleotide excision repair.6,7 Therefore, it is possible that the upregulation of zebrafish gadd45 g is critical for DNA demethylation and DNA repair processes during fin regeneration. In addition, although Tet has also been implicated in DNA demethylation because of the conversion of 5mC to 5hmC, the 5mC level was reduced simultaneously with the 5hmC level. One possible explanation for this result is that the conversion of 5mC to 5hmC is so fast that the differences between the levels of 5mC and 5hmC cannot be detected by immunohistochemical staining. Functional analysis of demethylation- or DNA repair-related genes through knockdown experiments is required to validate their role in the changes of 5mC or 5hmC levels during fin regeneration.
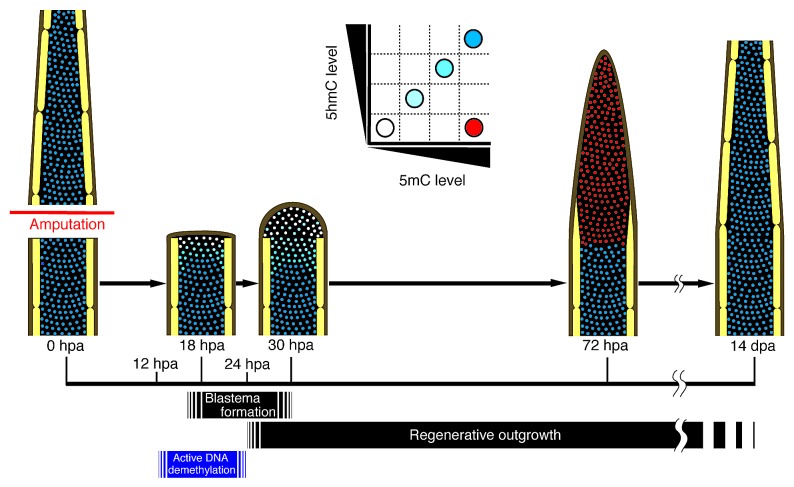
Based on our results, we propose a model for the spatial and temporal DNA methylation profile during zebrafish fin regeneration (Fig. 4). The levels of both 5mC and 5hmC in the dedifferentiated blastema cells and cells adjacent to the amputation plane are transiently reduced from 12 hpa, and the downregulation of 5mC and 5hmC levels is independent of cell proliferation by 24 hpa. It is thought that after 24 hpa, the dedifferentiated blastema cells begin to proliferate, and this proliferation leads to regenerative outgrowth. In addition, we found that DNA demethylation- and repair-related genes are expressed during fin regeneration, and particularly, expression of gadd45 g is increased after fin regeneration. Taken together, these results show that transient reduction of 5mC and 5hmC levels is associated with active DNA demethylation during zebrafish fin regeneration.
Figure 4. Spatial and temporal changes of 5mC and 5hmC levels during regeneration of zebrafish fin. A proposed model for 5mC or 5hmC level during regeneration of zebrafish fin based on our findings. Schematic representations of a longitudinal section of a fin ray. The level of 5mC or 5hmC in the cells adjacent to the amputation plane starts to reduce from approximately at 12 hpa, and the number of demethylated cells increases until 24 hpa. Subsequently, these demethylated cells start to proliferate, so that the number of blastema cells is increased. By 72 hpa, the 5mC level is upregulated, probably because DNA remethylation occurs in the blastema cells. In contrast to that of 5mC, the 5hmC level is still reduced in the blastema cells at 72 hpa and is gradually recovered by 14 dpa.
Materials and Methods
Ethics statement
All animal experiments were conducted according to relevant national and international guidelines ‘Act on Welfare and Management of Animals’ (Ministry of Environment of Japan). Ethics approval from the Hiroshima University Animal Research Committee (HuARC) was not sought since this law does not mandate protection of fish.
Zebrafish husbandry and fin amputation
Adult zebrafish and zebrafish embryos were maintained under a 14 h day/ 10 h night cycle at 28.5 °C. Transgenic zebrafish XIG8A (Tg[ef1-α:EGFP])23 was obtained from National Institute of Genetics.
Adult wild type 3–6-mo-old zebrafish (AB/Tüebingen strain) were used for all experiments. For caudal fin amputation, fish were anesthetized using tricaine, and approximately two-third of fins were cut with a blade. After fin amputation, these fish were allowed to regenerate in the aquarium until defined time points at 28.5 °C.
Dot blot assays
Dot blot assays were performed as described previously.27,28 Immediately after fin amputation (0 hpa), the fins were cut within 1000 μm from the amputation plane, and at 30 and 72 hpa, the blastema regions were cut. For the dot blot assays, 3, 6, and 3 amputated fins at 0, 30, and 72 hpa, respectively were used, and the experiments for 5mC and 5hmC were repeated 6 times, respectively. After lysis of the fins or blastema, genomic DNA was purified using the Gentra Puregene Tissue Kit (Qiagen) according to the manufacturer’s instructions. The concentration of purified genomic DNA in each regeneration stages was measured using a NanoDrop 1000 (Thermo Fisher Scientific). The genomic DNA (6.25, 25, or 100 ng) was loaded onto a nylon membrane (Amersham Hybond N+, GE Healthcare Life Sciences) and was crosslinked using the UV crosslinker CL-1000 (UVP Inc.). The following primary antibodies were used: anti-5mC mouse monoclonal antibody at 1:500 (Calbiochem) and anti-5hmC rabbit polyclonal antibody at 1:1000 (Active motif). The following secondary antibodies were used: anti-rabbit IgG-horseradish peroxidase (HRP) antibody at 1:250000 (Santa Cruz Biotechnology) and anti-mouse IgG-HRP antibody at 1:250000 (GE Healthcare). The signal of 5mC or 5hmC was exposed to X-ray film by using the ECL Prime Western Blotting Detection System (GE Healthcare Life Sciences) according to the manufacturer’s instructions. The intensities of these signals were scanned and the images were analyzed using the ImageJ software (NIH). p-values were calculated using a Student’s t-test.
Immunohistochemical analyses
The amputated fins were fixed in 4% paraformaldehyde in 0.1 M phosphate-buffered saline (PBS) overnight at 4 þC. After fixation, the fins were treated with PBS containing 30% sucrose overnight, and were embedded with Tissue-Tek O.C.T. compound (Sakura Finetek). The embedded fins were frozen and sectioned to 14 μm thickness by using a Leica CM3050S. The sections were dehydrated through a methanol/PBS-0.1% Tween (PBS) series, incubated in 2 N HCl for 30 min at 28.5 °C, and then neutralized with PBS for 5 min. After neutralization, the sections were blocked with PBDT (PBS, 1% DMSO, and 0.5% Tween20) containing 5% sheep serum for 3 h at room temperature and then incubated in PBDT with primary antibody/antibodies overnight at 4 þC. The following primary antibodies were used: anti-5mC mouse monoclonal antibody at 1:1000 (Calbiochem); anti-5hmC rabbit polyclonal antibody at 1:2000 (Active motif); anti-PCNA mouse monoclonal antibody at 1:1000 (Sigma); anti-BrdU rat monoclonal antibody at 1:200 (Abcam); anti-BrdU mouse monoclonal antibody at 1:100 (Amersham). The following secondary antibodies were used: Alexa Fluor® 488 goat anti-rabbit IgG antibody at 1:500 (Invitrogen, Life Technologies Corp.); Alexa Fluor® 488 goat anti-rat IgG antibody at 1:500 (Invitrogen, Life Technologies Corp.); Alexa Fluor® 594 goat anti-mouse IgG antibody at 1:500 (Invitrogen, Life Technologies Corp.). 4',6-diamidino-2-phenylindole (DAPI) was used for nuclei staining at a concentration of 1:500. For the negative control, we confirmed that no fluorescent signals are observed without a primary antibody or a secondary antibody (data not shown) and that the immunostaining pattern of 5mC or 5hmC is not changed when a pre-absorbed primary antibody for 5mC or 5hmC is used (data not shown).
Image quantification
Immunohistochemical staining images were captured using an Olympus FV1000-D confocal microscope with the same exposure times. The fluorescent signal intensities were measured in intra-ray nuclei within 500 μm of the amputation plane at 0 hpa, or at 24, 30, and 72 hpa in blastema nuclei, by using the FluoView software. The 5mC or 5hmC signal intensity of individual nucleus in an amputation site at 0 hpa, or at 24, 30, and 72 hpa in a blastema was normalized by its own DAPI signal intensity as an internal control. For non-amputated fins, the signal intensity of 5mC or 5hmC was measured in intra-ray nuclei at the approximate two-thirds mark of the fins, a common site of amputation in this study, and was normalized by the cells’ own DAPI signal intensity. The mean 5mC or 5hmC intensity of 600 randomly chosen intra-ray nuclei within 500 μm from the amputation plane in 6 different fish (approximately 100 nuclei per fish fin) is set as 1.0 at 0 hpa. Relative 5mC or 5hmC intensity of amputated fins at 24, 30, or 72 hpa is the normalized value divided by the mean 5mC or 5hmC intensity at 0 hpa, respectively. 600 randomly chosen blastema nuclei from 6 different fish (approximately 100 nuclei per fish fin) were used to calculate the relative 5mC or 5hmC intensity at 24, 30, or 72 hpa. The relative 5mC or 5hmC intensity of non-amputated fins is the normalized value divided by the mean 5mC or 5hmC intensity at 0 hpa, respectively. 600 randomly chosen nuclei at a position of approximately two-thirds of the non-amputated fins from 6 different fish (approximately 100 nuclei per fish fin) were used to calculate the relative 5mC or 5hmC intensity. At 24 hpa, the relative 5hmC intensity of an individual blastema nucleus was classified as the control, or as a high level of 5hmC (Control/High level-5hmC) if the intensity was greater than or equal to 0.98. A low level of 5hmC (Low level-5hmC) classification was given if the intensity was less than 0.98 (5hmC intensity at 0 hpa: mean ± standard error, 1.0 ± 0.02). P-values were calculated using a Student’s t-test.
BrdU incorporation assays
BrdU incorporation assays were performed as described previously.29 Fin-amputated fish were allowed to regenerate in the fish water containing with 50 μg/ml BrdU until 24 hpa. After incubation, the regenerating fins were cut and BrdU-labeled cells were detected as described above.
Quantitative real-time PCR (qPCR)
For qPCR analyses, total RNA was extracted from the regenerating fins within 1000 μm from the amputation plane at 0 hpa (3 regenerates per extraction), 18 hpa (5 regenerates per extraction), and 30 hpa (5 regenerates per extraction), and from the blastema at 72 hpa (3 regenerates per extraction) using TRIzol (Invitrogen, Life Technologies Corp.). Three hundred nanograms of DNase-treated RNA was reverse transcribed using oligo-(dT) primers and Reverse transcriptase XL (Takara). Reverse transcription (RT)-PCR was performed as a means of RT-negative control to test for genomic DNA contamination; no amplification was observed in any of the samples confirming that there was no contamination (data not shown). qPCR for 9 genes (gadd45ba, gadd45bb, gadd45 g, apobec2b, tdg, mbd4, tet2, tet3, and parp1) was performed in tetraplicates by using the Thermal Cycler Dice® Real Time System, SYBR® Premix Ex Taq™ (TaKaRa Bio Inc.) according to the manufacturer’s instructions. Because both zebrafish actb1 and ribosomal protein L13a (rpl13a) were stably expressed in fins or fin regenerates (data not shown), actb1 was used as reference gene for qPCR. All primer pairs were designed to span an intron-exon boundary to prevent amplification of genomic DNA and the amplified signals were confirmed to be a single band by gel electrophoresis. No-template control was performed for 9 genes to confirm the specificity of qPCR (data not shown). Detailed qPCR conditions used to amplify each of nine genes, actb1, and rpl13a are listed in Table S1. P-values were calculated using a Student’s t-test.
Supplementary Material
Acknowledgments
We thank Drs. Koichi Kawakami for providing the transgenic fish XIG8A; Hiroshi Ide and Toshiaki Nakano for NanoDrop 1000; Institute for Amphibian Biology for cryostat sectioning. We also thank the members of Kikuchi and Atsushi Suzuki laboratories in Hiroshima University for helpful discussion and critical comments. This study was supported by grants from Grant-in-Aid for Scientific Research from the MEXT (KAKENHI 23616002) to Y.K., and Hiroshima University Alumni Association Research Grant and Hiroshima University Support Foundation Research Grant to K.H.
Glossary
Abbreviations:
- 5mC
5-methylcytosine
- 5hmC
5-hydroxymethylcytosine
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: http://www.landesbioscience.com/journals/epigenetics/article/25653/
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/25653
References
- 1.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–20. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cedar H, Bergman Y. Programming of DNA methylation patterns. Annu Rev Biochem. 2012;81:97–117. doi: 10.1146/annurev-biochem-052610-091920. [DOI] [PubMed] [Google Scholar]
- 3.Franchini DM, Schmitz KM, Petersen-Mahrt SK. 5-Methylcytosine DNA demethylation: more than losing a methyl group. Annu Rev Genet. 2012;46:419–41. doi: 10.1146/annurev-genet-110711-155451. [DOI] [PubMed] [Google Scholar]
- 4.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 5.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–92. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 6.Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–66. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niehrs C, Schäfer A. Active DNA demethylation by Gadd45 and DNA repair. Trends Cell Biol. 2012;22:220–7. doi: 10.1016/j.tcb.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2012;13:7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 9.Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2011;13:28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139:1895–902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akimenko MA, Marí-Beffa M, Becerra J, Géraudie J. Old questions, new tools, and some answers to the mystery of fin regeneration. Dev Dyn. 2003;226:190–201. doi: 10.1002/dvdy.10248. [DOI] [PubMed] [Google Scholar]
- 12.Poss KD, Keating MT, Nechiporuk A. Tales of regeneration in zebrafish. Dev Dyn. 2003;226:202–10. doi: 10.1002/dvdy.10220. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka EM, Reddien PW. The cellular basis for animal regeneration. Dev Cell. 2011;21:172–85. doi: 10.1016/j.devcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slack JM. Amphibian muscle regeneration--dedifferentiation or satellite cells? Trends Cell Biol. 2006;16:273–5. doi: 10.1016/j.tcb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Barrero MJ, Izpisua Belmonte JC. Regenerating the epigenome. EMBO Rep. 2011;12:208–15. doi: 10.1038/embor.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katsuyama T, Paro R. Epigenetic reprogramming during tissue regeneration. FEBS Lett. 2011;585:1617–24. doi: 10.1016/j.febslet.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 17.Thummel R, Burket CT, Hyde DR. Two different transgenes to study gene silencing and re-expression during zebrafish caudal fin and retinal regeneration. ScientificWorldJournal. 2006;6(Suppl 1):65–81. doi: 10.1100/tsw.2006.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yakushiji N, Suzuki M, Satoh A, Sagai T, Shiroishi T, Kobayashi H, et al. Correlation between Shh expression and DNA methylation status of the limb-specific Shh enhancer region during limb regeneration in amphibians. Dev Biol. 2007;312:171–82. doi: 10.1016/j.ydbio.2007.09.022. [DOI] [PubMed] [Google Scholar]
- 19.Martin CC, Laforest L, Akimenko MA, Ekker M. A role for DNA methylation in gastrulation and somite patterning. Dev Biol. 1999;206:189–205. doi: 10.1006/dbio.1998.9105. [DOI] [PubMed] [Google Scholar]
- 20.Mhanni AA, McGowan RA. Global changes in genomic methylation levels during early development of the zebrafish embryo. Dev Genes Evol. 2004;214:412–7. doi: 10.1007/s00427-004-0418-0. [DOI] [PubMed] [Google Scholar]
- 21.MacKay AB, Mhanni AA, McGowan RA, Krone PH. Immunological detection of changes in genomic DNA methylation during early zebrafish development. Genome. 2007;50:778–85. doi: 10.1139/G07-055. [DOI] [PubMed] [Google Scholar]
- 22.Almeida RD, Loose M, Sottile V, Matsa E, Denning C, Young L, et al. 5-hydroxymethyl-cytosine enrichment of non-committed cells is not a universal feature of vertebrate development. Epigenetics. 2012;7:383–9. doi: 10.4161/epi.19375. [DOI] [PubMed] [Google Scholar]
- 23.Urasaki A, Morvan G, Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006;174:639–49. doi: 10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woods AL, Hall PA, Shepherd NA, Hanby AM, Waseem NH, Lane DP, et al. The assessment of proliferating cell nuclear antigen (PCNA) immunostaining in primary gastrointestinal lymphomas and its relationship to histological grade, S+G2+M phase fraction (flow cytometric analysis) and prognosis. Histopathology. 1991;19:21–7. doi: 10.1111/j.1365-2559.1991.tb00890.x. [DOI] [PubMed] [Google Scholar]
- 25.Powell C, Elsaeidi F, Goldman D. Injury-dependent Müller glia and ganglion cell reprogramming during tissue regeneration requires Apobec2a and Apobec2b. J Neurosci. 2012;32:1096–109. doi: 10.1523/JNEUROSCI.5603-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poleo G, Brown CW, Laforest L, Akimenko MA. Cell proliferation and movement during early fin regeneration in zebrafish. Dev Dyn. 2001;221:380–90. doi: 10.1002/dvdy.1152. [DOI] [PubMed] [Google Scholar]
- 27.Tittle RK, Sze R, Ng A, Nuckels RJ, Swartz ME, Anderson RM, et al. Uhrf1 and Dnmt1 are required for development and maintenance of the zebrafish lens. Dev Biol. 2011;350:50–63. doi: 10.1016/j.ydbio.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacKay AB, Mhanni AA, McGowan RA, Krone PH. Immunological detection of changes in genomic DNA methylation during early zebrafish development. Genome. 2007;50:778–85. doi: 10.1139/G07-055. [DOI] [PubMed] [Google Scholar]
- 29.Nechiporuk A, Keating MT. A proliferation gradient between proximal and msxb-expressing distal blastema directs zebrafish fin regeneration. Development. 2002;129:2607–17. doi: 10.1242/dev.129.11.2607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.