Abstract
Glioblastoma multiforme (GBM) is the most common malignant adult brain tumor. Standard GBM treatment includes maximal safe surgical resection with combination radiotherapy and adjuvant temozolomide (TMZ) chemotherapy. Alarmingly, patient survival at five-years is below 10%. This is in part due to the invasive behavior of the tumor and the resulting inability to resect greater than 98% of some tumors. In fact, recurrence after such treatment may be inevitable, even in cases where gross total resection is achieved. The Cancer Genome Atlas (TCGA) research network performed whole genome sequencing of GBM tumors and found that GBM recurrence is linked to epigenetic mechanisms and pathways. Central to these pathways are epigenetic enzymes, which have recently emerged as possible new drug targets for multiple cancers, including GBM. Here we review GBM treatment, and provide a systems approach to identifying epigenetic drivers of GBM tumor progression based on temporal modeling of putative GBM cells of origin. We also discuss advances in defining epigenetic mechanisms controlling GBM initiation and recurrence and the drug discovery considerations associated with targeting epigenetic enzymes for GBM treatment.
Keywords: epigenetics, glioblastoma, statistical modeling, drug discovery
Surgical and Pharmacological Management of GBM
In 90% of cases, GBMs arise de novo as primary tumors without progression from lower grade tumors while secondary GBMs originate from previously diagnosed low-grade astrocytomas. Maximal safe resection of a primary GBM is the mainstay of treatment and confers improved prognosis. Patients who receive a surgical resection greater than 98% of the tumor volume have a prognosis of 13.1 mo compared with 8.8 mo in patients from whom less of the tumor is resected.1
Because GBMs have infiltrating cells, the entire tumor cannot be removed. For this reason, most GBM patients will follow a standard treatment regimen after the tumor is resected. This consists of 6 weeks of external beam radiation 5 times a week plus oral temozolomide daily. Temozolomide (TMZ) is an alkylating agent whose therapeutic benefit arises from its ability to alkylate/methylate DNA; methylation damages DNA and triggers tumor cell death. However, some tumor cells are able to repair this type of DNA damage by expressing an enzyme called O-6-methylguanine-DNA methyltransferase (MGMT), thereby diminishing the therapeutic efficacy of TMZ.
Unfortunately, most patients will have a recurrence of GBM within 6.9 mo of their original diagnosis. Essentially all GBMs recur after initial therapy. Re-operation and re-radiation are treatment options for only a minority of patients; 80% or more of GBM recurrences occur in the same area as the original tumor, precluding additional radiation therapy because of toxicity concerns.
Greater knowledge of the cellular, genetic, and epigenetic origin of GBM is the key for advancing GBM treatment. Clinical researchers are analyzing freshly resected tumors for genetic and epigenetic modifications in collaboration with genomics and drug discovery groups. These studies are coupled to derivation of cell lines from patients’ tumors in the hope of identifying and developing personalized drug treatment regimens.
Cell of Origin for Glioblastoma
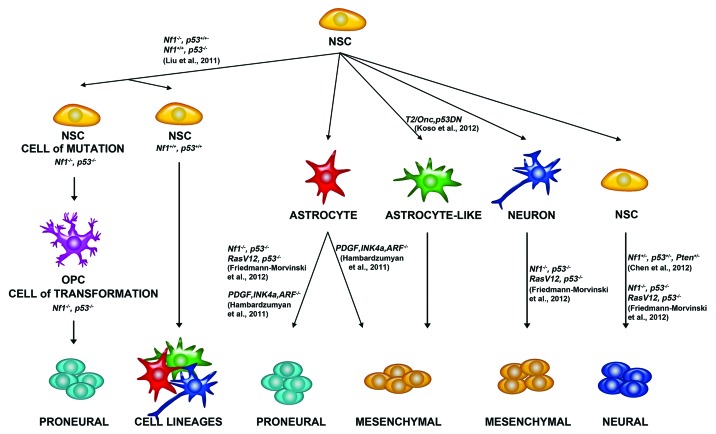
A major drug discovery challenge is defining the cellular origin of GBM since it is difficult to develop a successful GBM treatment without first uncovering the responsible cell type to eliminate. Considering that epigenetic modifications, enzymes, and non-coding RNAs are often cell type specific makes these cellular elements prime targets for identifying the cell-of-origin. However, such determinations are often difficult since cells acquiring a mutation (cell of mutation) may not be the same as cell of origin (ref. 2; Fig. 1). For instance, it is possible that neural stem cells pass on mutations to downstream progeny such as oligodendrocyte precursor cells (OPCs), which are putative glioma cells of origin. This is the model Liu et al., 2012 proposed by labeling different cell populations using mosaic analysis with double markers (MADM) in mice.3 Briefly, glioma was induced by sporadic introduction of specific Cre-mediated deletion of Neurofibromin-1 (Nf1) and p53 in neural stem cells, via nestin or GFAP promoter mediated expression. After Cre mediated-recombination and proliferation of the neural stem cells, the progeny that contained homozygous deletion of Nf1 and p53 was correlated with green fluorescent protein (GFP) expression while wild-type cells were labeled red with red fluorescent protein (RFP). By performing single cell analysis, the authors determined that OPCs expanded or increased upon Nf1 and p53 mutation, while neural stem cells or other lineages were not overrepresented. Consistent with this notion, introducing Nf1 and p53 mutations directly into OPCs in vitro induced gliomagenesis,3 which is seen clinically in several genetic diseases with predisposition to glioma including Li-Fraumeni syndrome (TP53 mutation) and Neurofibromatosis type 1.
Figure 1. Possible cells of origin of glioma. Studies in mouse models have shown that various cell types can give rise to glioma. Neural stem cells (NSCs) give rise to other Neural Stem Cells, Astrocytes, Astrocyte-like cells, and neurons. Liu et al. 2011 demonstrated that NSCs give rise to OPCs, which can give rise to glioma. Friedmann-Morvinski et al. 2012 demonstrated that astrocytes and neurons can give rise to glioma. Hambardzumyan et al. 2011 demonstrated that astrocytes can give rise to glioma after PDGF overexpression and Ink4a, and ARF deletion. Koso et al. 2012 demonstrated that overexpression of a mutagenic Sleeping Beauty (SB) transposon (T2/Onc2) along with a dominant negative p53 in astrocyte like cells can give rise to glioma. Chen et al., 2012 demonstrated that NSCs could give rise to glioma after Nf1, p53, and Pten deletion. Neural Stem Cells can give to proneural, mesenchymal, and neural cell lineages.
By contrast to Liu et al.,3 Koso and colleagues4 suggested that the cell of origin in some GBM is not an OPC but an astroglial-like cell. They further postulated that originating mutations can occur in NSCs (Fig. 1). They introduced a mutagenic transposon in Nestin-Cre mice along with dominant negative p53-(DN) and observed 100% glioma formation. Thus, it may be likely that multiple cells of origin give rise to glioma. Further, studying the genetic and epigenetic landscape of human OPCs and other cells as they are differentiating could uncover epigenetic enzymes and pathways misregulated in gliomagenesis.
Recent studies suggest that epigenetic modification determination during neural differentiation will likely give insight into de-differentiation processes, which may give rise to GBM. Verma and colleagues demonstrated that neurons de-differentiate and become tumor-initiating cells in mouse models of glioma (ref. 5; Fig. 1). They used a modification of the Sleeping Beauty system, which concurrently deleted p53 and Nf1 using shRNAs targeting these transcripts. In this modified system, LoxP sites flank RFP, which is deleted using specific Cre expression. Since the presence of GFP is constitutive and is not deleted, a GFP/RFP ratio could be attained and mosaic analysis could be performed after Cre mediated deletion (Fig. 1). Since they used a Cre construct that is only expressed in neurons (Synapsin I-Cre), they could follow Cre mediated recombination in neurons via loss of RFP. Performing this analysis, they demonstrated that mature neurons (NeuN and Tuj1 positive) not expressing RFP, GFAP (astrocyte marker), and Ki-67 (proliferation marker), induced glioma formation. To provide further evidence that neurons can induce glioma, they isolated cortical neurons from Synapsin I-Cre mice, transduced them with shRNAs targeting p53 and Nf1 in vitro and demonstrated that they give rise to high-grade glioma when injected in immune-compromised mice. Similarly, they used Nestin-Cre and GFAP-Cre mice to demonstrate that mutation of neural stem cells or astrocytes can produce high-grade glioma in mice (Fig. 1). Parallel studies showed that targeting astrocytes promotes glioma formation. Hambardzumyan et al.,6 overexpressed PDGFB in astrocytes derived from Ink4a-ARF−/− mice (Fig. 1), and observed robust glioma induction in different brain regions. Collectively these findings suggest that glioma may arise from either de-differentiating neural stem cells or astrocytes.
Importantly, de-differentiating neurons or astrocytes that give rise to glioma might be the very cells that are resistant to TMZ therapy and induce tumor recurrence.7 Parada and colleagues provided evidence that cancer stem cells may be responsible for GBM recurrence by using a nestin-ΔTK-IRES-GFP mouse model that labels quiescent subventricular adult neural stem cells, and breeding this transgenic mouse to a well-established mouse model of gliomagenesis (hGFAP-Cre; Nflfl/+; p53fl/fl; Ptenfl/+) (ref. 7; Fig. 1). As expected, TMZ treatment eliminated tumors in these mice, but, surprisingly, the cells that returned and proliferated were GFP+, suggesting that they were neural stem cells. Further, treatment with ganciclovir, which targets cells containing a modified version of the herpes simplex thymidine kinase (ΔTK), eliminated these cells and reduced tumor growth in vivo. These exciting studies suggest the presence of cancer stem cells in glioma and are supported by concurrent studies in papilloma and other cancers.8 They further suggest that understanding the epigenetic changes in cancer stem cells after TMZ treatment is key to identifying small molecule inhibitors of GBM recurrence.
Genetic and Epigenetic Characteristics of Glioblastoma
The remarkable advances in defining the GBM cell of origin have been paralleled by insights into the genetic and epigenetic underpinnings of this disease. GBM was the first cancer studied by The Cancer Genome Atlas (TCGA; ref. 9) where sequencing of over 200 different tumors identified the EGFR, PDGFR, PI3K, NF1, TP53, and Rb pathways as misregulated in GBM.9 Other studies have uncovered mutations or fusions of other genes such as IDH1/IDH2 and FGFR, respectively, in subsets of GBM patients.10-12
However, there are only a few GBM mutational “drivers,” suggesting that we may have to expand our search for “drivers” beyond somatic mutations to understand the genomic networks misregulated in GBM.13 In simple terms, a driver event is usually defined as one that occurs early in tumorigenesis and occurs in pathways considered critical to the development of any of the hallmarks of cancer.14-16 A study in collaboration with The Cancer Genome Atlas (TCGA) Research Network proposed four subtypes of GBM based on genomic profiling of hundreds of human samples.17 These four subtypes have been named “proneural,” “mesenchymal,” “classical,” and “neural.” Proneural GBMs show altered expression of PDGFRA, IDH1 and TP53 mutation and loss of heterozygosity (LOH) along with PTEN mutation and CDKN2A loss. Mesenchymal GBMs have deletion of NF1, mutation of TP53 and PTEN, and loss of CDKN2A. Classical GBMs are typified by EGFR amplification and lack of PTEN, and CDKN2A. Finally, the Neural GBMs show a strong expression of neuron markers and genes associated with neuron projection and axon and synaptic transmission. These subgroups might develop from different cells of origin. Moreover, Verhaak et al.17 discovered that aggressive treatment significantly reduces the mortality in Classical and Mesenchymal subgroups but does not significantly improve survival in the Neural and Proneural groups (p > 0.05). Subtype and MGMT methylation status were not significantly correlated, indicating that a patient’s response can be evaluated independent of MGMT status.
Supporting evidence for a potential driving event may include data showing that the event occurs in a substantial fraction of GBM patient samples. For example, Schwatzentruber et al.15 demonstrated that somatic mutations in the H3.3-ATRX-DAXX chromatin-remodeling pathway frequently occur in pediatric GBMs and are associated with alternative lengthening of telomeres and genomic instability.
Another approach to discovery is to consider the epigenetic drivers of gliomagenesis. Several reviews have detailed the histone and DNA modifications specific to GBM that can be used to expand the current search for “drivers.”18-21 For example, Strum et al.22 incorporated the mutational status of H3F3A and IDH1 with differences in global methylation patterns in GBMs to identify 6 distinct epigenetic subgroups, which correlate with distinct clinical characteristics. Here we will concentrate on microRNAs (approximately 22 nucleotide RNAs) and long non-coding RNAs (greater than 200 nucleotide RNAs) that affect gene expression through regulation of mRNA stability and transcription regulation. MicroRNAs are non-coding RNAs, which bind to microRNA response elements (MREs) in target mRNAs. Once the miRNA is loaded into the RISC complex (RNA-induced silencing complex), the miRNA/RISC complex binds the target mRNA, thereby modulating its stability. miRNAs dysregulated in glioma include miR10b, which is expressed in glioma tumors and stem cells, but not neuronal progenitors, mature glia, or neurons.23-26 miR10b controls GBM cell and stem cell cycle traverse and is correlated with poor prognosis (http://tcga-data.nci.nih.gov/tcga). The targets and pathways controlled by miR10b are under investigation. However, network analysis of miR10b targets and follow-up studies will be required to fully understand its role in GBM cell proliferation.
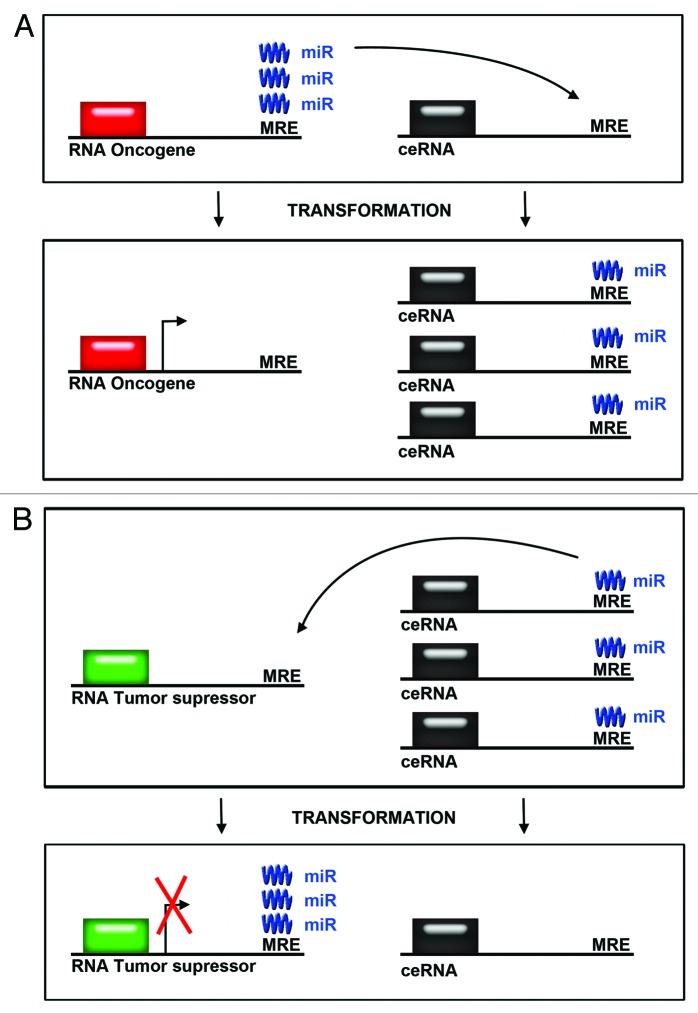
Three elegant studies elucidated a microRNA network, which regulates RNA-RNA interactions in oncogenic pathways controlling GBM.27-29 While one of the studies utilized a melanoma model, all three studies concentrated on an established GBM tumor suppressor, PTEN. PTEN is a phosphatase, which opposes the PI3K pathway, a known GBM driver. PTEN levels are controlled by miR-26A, which is amplified in GBM.27-29 The current thinking is that in addition to well-established PTEN somatic mutations, PTEN levels are controlled by miRNAs, which may be misregulated in tumors. What these papers demonstrate is that miRNA networks are more complex than we had previously anticipated. Specifically, competitive endogenous RNAs (ceRNAs) control the amount of each miRNA species (Fig. 2). In the case of miR-26A, if there is an excess of mRNAs that contain the microRNA recognition elements (MREs) relative to PTEN mRNA, PTEN protein levels should rise. By contrast, removing a PTEN mRNA competitor should lower PTEN levels since there would be more miRNA to bind PTEN mRNA. (Fig. 2A). This is what the authors observed: lowering PTEN competitors reduced PTEN levels and accelerated tumor growth. Similarly, Fang and colleagues30 found that an increase of versican 3′-UTR produced competition for 4 different miRNAs resulting in higher versican, fibronectin, and CD34 mRNA and protein levels, thereby leading to cell proliferation and tumor growth (Fig. 2B). Interestingly, in order to shed light on these mechanisms Sarver and Subramanian31 built an open access database where competing endogenous mRNA rankings are generated using conserved mRNA-miRNA interactions. The user enters an mRNA for which they are interested in finding potential competing mRNAs, and the tool returns potential ceRNA regulators. This useful database allows us to delineate pathways controlling glioma progression based on an understanding of miRNA-ceRNA networks.
Figure 2. Competing endogenous RNA levels modulate expression of oncogenes and tumor suppressors. (A) The levels of oncogenes can be modulated by an increase in levels in competing endogenous RNAs. microRNAs are titrated away from mRNAs encoding oncogenes when the levels of competing endogenous RNAs are increased. This leads to increased expression of oncogenes. (B) The levels of tumor suppressor proteins are modulated by decreases in the levels of competing endogenous RNAs. microRNAs bind to RNAs of tumor suppressors, thereby reducing tumor suppressor protein expression after transformation.
miRNA-ceRNA networks are likely further controlled by long non-coding RNAs (lncRNAs), which control global gene repression.32-35 lncRNAs control multiple tumor suppressor proteins and oncogenes.27-29,32-35lncRNAs modulate transcription, regulate post-transcriptional RNA processing, influence translation,36 and alter DNA methylation and chromatin architecture through local (cis) and long distance (trans) mechanisms.37 Through interactions with transcription factors, co-activators and/or repressors, lncRNAs can affect different aspects of gene transcription to form a fine-tuned complex regulatory network. lncRNAs also modulate gene expression by recruiting chromatin remodeling complexes like histone methyltransferases to specific genomic loci.
The various regulatory roles of lncRNAs may play a crucial role in GBM development and progression. For example, the lncRNA MEG3 has been implicated in glioma cell proliferation.38 Interestingly, MEG3 expression is associated with differential methylation. Han et al.39 investigated lncRNA expression between GBM and normal samples, and discovered several lncRNAs implicated in glioma signaling. Their findings suggest that two lncRNAs, ASLNC22381 and ASLNC20819, which target IGF-1 may be important in GBM progression and recurrence. Since some lncRNAs interact with miRNAs,40 it will be important to further investigate their relationship in glioma. Katsushima et al.41 investigated miRNAs in a glioma stem cell (GSC) model, identifying miR-1275 as being associated with GSC differentiation, GBM heterogeneity, and tumor cell proliferation. Interestingly, miR-1275 expression was shown to be associated with histone H3 lysine 27 trimethylation (H3K27me3), and subsequent studies have shown a molecular interplay between lncRNAs and H3K27me3 in gene silencing.42 This implicates lncRNAs as potential factors in the complex genomic interactions in glioma. Unpublished data from our laboratory identified several lncRNAs that are differentially expressed in GBM compared with control tissues (Pastori et al., unpublished observations). However, the challenge is defining whether lncRNA up or downregulation in GBM is a driver or passenger event. We argue here that determining this is only possible using systems biology and network modeling approaches.
Epigenetic Enzymes as Therapeutic Targets in Glioblastoma
Determining the miRNAs, ceRNAs, and lincRNAs that control levels of epigenetic enzymes will be critical in elucidating whether these enzymes are GBM drivers. Epigenetic enzymes are gaining considerable attention due to their druggability and overexpression in certain cancers. Our group profiled 150 epigenetic enzymes in 27 GBM human samples using the Nanostring platform (Daniel et al., unpublished observations). Several chromatin writers, readers and erasers appear to be differentially expressed in GBM. Consistent with prior reports,43,44 we found that EZH2 is upregulated in GBM samples (Daniel et al., unpublished observations). Ezh2 is one of the most studied epigenetic enzymes as a lysine methyltransferase that modifies histones, thereby repressing expression of some tumor suppressor proteins including Cdkn2a/b.45 Ezh2 may also promote cancer cell proliferation by modifying non-histone proteins to create methyl-degrons recognized by ubiquitin ligases that induce cell cycle traverse.46 Further, Ezh2 interacts with lncRNAs that are overexpressed in multiple tumors.47-50 Ezh2 levels are high in GBM relative to normal brain tissue and Ezh2 is required for GBM stem cell maintenance.43,44 But, whether Ezh2 is a good therapeutic target in GBM is unclear since somatic mutations have not been described in GBM.11 By contrast, Ezh2 mutations are present in lymphoma where Ezh2 inhibitor sensitivity is correlated with mutational status.51,52 Highly specific small-molecule Ezh2 inhibitors have been recently described that potently reduce lymphoma cell proliferation in vitro and in vivo.51,52 These inhibitors are considerably more potent at reducing proliferation of cells harboring Ezh2 mutations than those bearing wild type Ezh2.51,52 However, GBM cells containing aberrant modulation of Ezh2 and Ezh2 binding RNAs may be equally sensitive to small molecule Ezh2 inhibitors. For instance, Ezh2 mRNA is regulated by miRNA-101, a microRNA downregulated in GBM.53,54 Thus, comparing the Ezh2/miRNA-101 network including ceRNAs in normal and GBM cells may be a means of validating Ezh2 as a therapeutic target in GBM. In addition, whether long non-coding and natural antisense transcripts regulate Ezh2 abundance in GBM should be determined to help validate Ezh2 as a target in GBM (Fig. 2). Similar analyses for other epigenetic enzymes that are likely to be regulated by miRNAs, ceRNAs, and lncRNAs should be performed in order to gain a global view of their regulation in GBM. After determining putative epigenetic enzymes misregulated in glioma using network analysis, prioritization of those enzymes required for GBM stem cell survival will be crucial. Mouse models of GBM, which delete different driver mutations, can then be used to help validate the prioritized enzymes as drug targets in GBM.
Drug Discovery Challenges in Glioblastoma
Once integrative modeling of neuronal differentiation and miRNA and lncRNA networks is performed, the next challenge is to identify epigenetic targets for GBM treatment and subsequently develop therapeutic strategies for drug discovery. Ongoing clinical trials are testing HDAC inhibitors for the treatment of GBM (NCT01378481, NCT00302159). Further, the identification of IDH1/2 mutations in GBM suggests that metabolic pathways may be attractive targets for GBM.55 However, the drug discovery challenges associated with targeting epigenetic enzymes or lncRNA-miRNA-protein interactions in GBM are the same as targeting any cell or target in the brain, which is protected by the blood brain barrier (BBB). The BBB protects the brain by forming a highly selective barrier that blocks the entry of large, hydrophilic molecules. The BBB also makes delivery of drugs affecting GBM and other neurologic disorders challenging. Moreover, drug transporters effectively pump small molecules from the brain. These include the multidrug resistance protein (MDR) and multidrug resistance-associated protein (MRP; ref. 56). The first multidrug resistance protein studied, P-glycoprotein, is considered to be one of the most important transporters at the BBB, since mice lacking this transporter have higher levels of small molecules in their brain.56 Further, drugs such as verapamil and cyclosporine A, which effectively inhibit P-glycoprotein activity in vitro, substantially increase brain retention of anti-cancer agents such as vincristine.56 Thus, it may be therapeutically advantageous to simultaneously inhibit an epigenetic target and P-glycoprotein. This could in theory be done pharmacologically by developing compounds that target both classes of proteins. However, this may be difficult and may be better achieved by performing network analysis to determine that a specific epigenetic enzyme/microRNA/lncRNA pathway is responsible for maintaining P-glycoprotein levels, and subsequently identifying small molecules that disrupt that network.
Epigenetic Systems Biology and Regulatory Networks
Recent attempts to characterize the GBM epigenetic/genetic landscape have utilized integrative, systems biology approaches that include multiple types of high-throughput data.18 The types of data involved can include genetic mutations, RNA expression, as well as methylation and protein expression. This poses challenges for statisticians, mathematicians, and bioinformaticians, as well as computational and medical researchers. Recent approaches have focused on examining the latent or underlying biological pathways in data repositories from glioma studies and the correlations or relationships between them. For example, Fronza et al.57 examined the interactions between groups of mRNAs and miRNAs in four glioma data sets and identified miRNA clusters, which they validated as being associated with survival in GBM. Wuchty et al.58 also examined miRNAs and mRNAs in order to discover significant miRNA-mRNA interactions in GBMs. Kunkle et al.59 integrated data on genetic variants and genes responsive to environmental exposures, along with various networking databases, to identify genes and pathways involved in gene-environment interactions, which may play a role in GBM development. The above studies use associations between different types of genomic data to infer possible regulatory systems and disease pathways.
A particularly interesting way to gain therapeutic insights for glioma is to examine the epigenetic landscape of neural differentiation. As stated previously, determining the epigenetic modifications of neuronal precursors as they are differentiating is important if OPCs, neurons and differentiated astrocytes indeed give rise to GBM in humans. Increasing evidence suggests that epigenetic pathways play a critical role in the regulation of neurogenesis.60 In order to examine the epigenetics of differentiation, we must consider biological processes over time; the modeling of such processes may involve methods from time series analysis, Markov chain modeling, and dynamic Bayesian networks.61
Traditional time series methods allow for the modeling of temporal processes where measurements at different time points are correlated or dependent.62 They can be used with real-valued continuous data, as well as discrete data, and can be particularly useful in cyclical contexts. This is because time series methods allow the use of periodic functions to model changes in a variable over several, repeating cycles. Of course most time series methods were developed for series of some length—from tens to thousands of time points—and for series involving a single response variable. Cell cycle studies, particularly in epigenetics, may involve hundreds or thousands of short series, one for each epigenetic or genomic variable and each cell cycle. This leads us to methods that find clusters of transcripts and describe these clusters.63,64 These clusters may correspond to groups of transcripts being targeted by a specific miRNA, a pool of miRNAs, or a specific ncRNA. Different clusters may also correspond to transcripts involved in specific biological processes associated with differentiation as well as with the GBM transcriptome (Fig. 3).
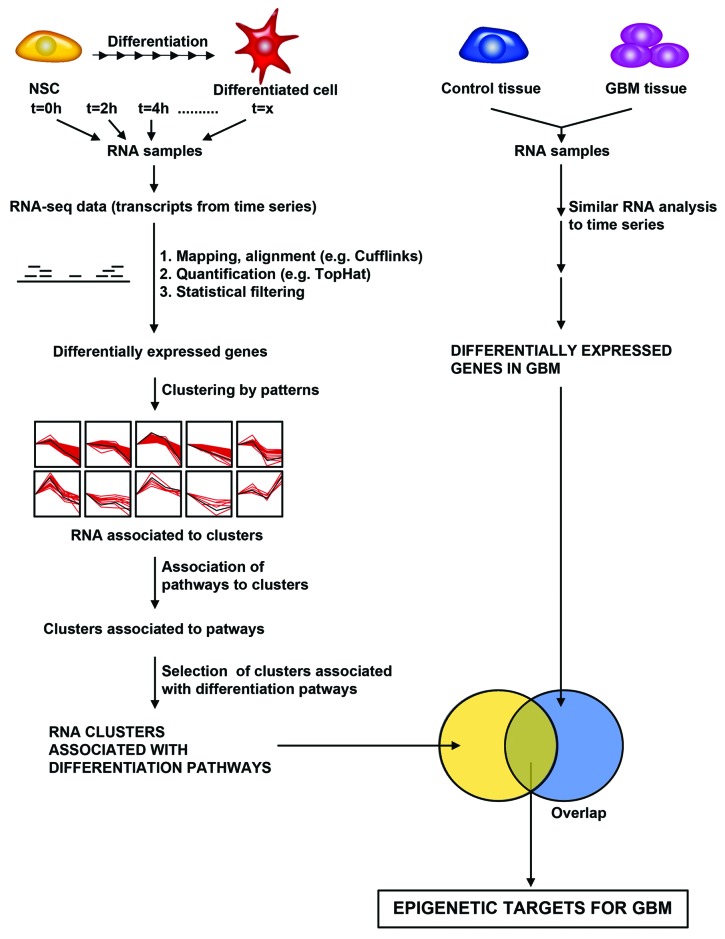
Figure 3. A bioinformatics and statistical pipeline for identifying epigenetic targets for GBM from transcriptome data. Hypothetical pipeline for identifying epigenetic targets in GBM based on differentially expressed pathways in both differentiating neural stem cell and GBM. Left Panel: Differentiating neural stem cells are analyzed for changes in RNA transcript levels by performing RNA-sequencing analysis of differentiating cells. RNA sequencing yields transcripts expressed over time. Mapping/alignment of transcripts using human genome is performed using Tophat and quantification of aligned transcripts is then performed using Cufflinks, or similar bioinformatics pipeline. Statistical filtering by t-tests or analysis of variance after quantification yields differentially expressed genes. Clustering of genes by patterns is then performed to identifying RNAs that are associated with differentiation pathways. Right Panel: RNA-sequencing of GBM and control tissue is performed to identify differentially expressed genes using the same bioinformatics pipeline utilized in analyzing differentiating neural stem cells. The degree of overlap of those transcripts, which are differentially expressed during differentiation, and in GBM is then calculated to identify epigenetic targets in GBM.
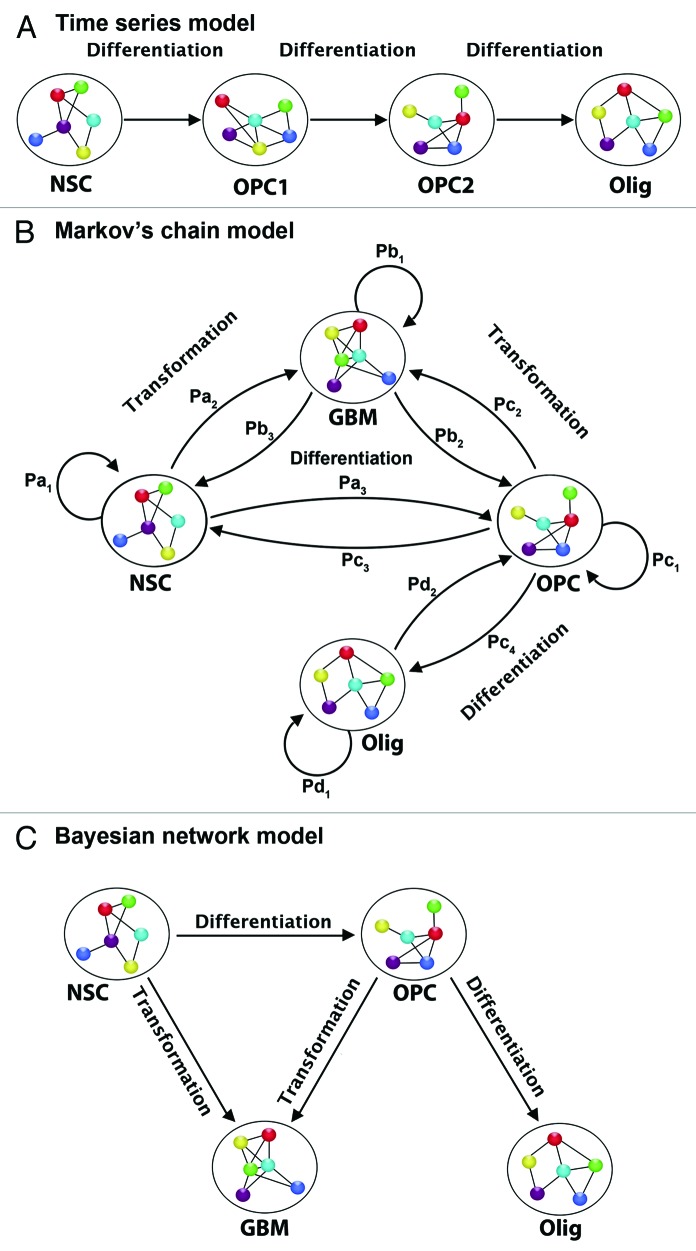
Models based on Markov chain methods62 can view temporal processes as discrete systems, where at each time point the system is in one of a number of possible 'states'. A Markov chain considers the movement of a biological system as a set of transitions among different “states” over time, e.g., the movement of a cellular population through different phases of development or responses to stimuli. Each possible movement among states is assigned a probability expressing the likelihood of that particular move. A hidden Markov model assumes that these states are “hidden” or not directly observed, and must be inferred from the observed data. In order to process epigenetic data of potentially high dimensions, these methods may be coupled with dimension reduction techniques such as clustering, variable selection and shrinkage.63-65 Dimension reduction assumes that the observed high dimensional data in glioma is a complex representation of several lower dimensional genomic processes. For example, we may measure a large group of transcripts from an oncologic pathway such as mTOR or MAPK, but it is plausible that this will not provide completely independent signals from each transcript. Instead, we are likely to see dependent or highly correlated signals. As such the effective dimension of a pathway could be much smaller than the number of transcripts, and dimension reduction/variable selection techniques can help us find a representation of the pathway in this smaller, lower dimensional space. This enables us to model high dimensional epigenomic data using methods that are more effective in lower dimensional contexts (Fig. 4).
Figure 4. Examples of statistical models for neural temporal data. (A) Time series. Each transcript is modeled separately (univariate) or as part of a group (multivariate). The model uses information from previous time points in modeling future time points, and can capture contemporaneous and lagged dependencies among transcripts. (B) Discrete Markov chain model. Each cellular stage is considered a “state” and the chain models the probabilities of moving from one “state” to another in a given time step. Depending on the type of Markov model it may or may not be possible to move both backward and forward in time, and hence for cells to differentiate as well as dedifferentiate. (C) Bayesian network model. If we consider a directed acyclic graph (DAG), then we define a joint probability distribution over cellular states. For each node or state we define a probability distribution for transcription in each state, conditional on transcription in previous states. If we consider a dynamic graphical model (DGM), then we can model each state with a graphical model, and separately model the movement from state to state across time. In this way transcripts can have contemporaneous as well as time-dependent relationships. NSC, neural stem cell; OPC1, oligodendrocyte precursor cell 1; OPC2, oligodendrocyte precursor cell 2; Olig, oligodendrocyte; GBM, glioblastoma cell. Pa1 is the probability that a neural stem cell remains a stem cell from one time point to the next. Pa2 is the probability that a neural stem cell transforms from the current time point to the next time point. Pb3 is the probability that a GBM cell de-differentiates from the current time point to the prior time point. Pb1 is the probability that a GBM cell remains a GBM cell from the current time point to the next. Pc2 is the probability that an oligodendrocyte precursor cell (OPC) transforms from the current time point to the next. Pc1 is the probability that an OPC remains an OPC from the current time point to the next. Pb2 is the probability that a GBM cell de-differentiates into an OPC from the current time point to the next time point. Pc4 is the probability that an OPC differentiates into an oligodendrocyte from the current time point to the next time point. Pc3 is the probability that an OPC dedifferentiates into a neural stem cell from the current time point to the next. Pa3 is the probability that a NSC differentiates into an OPC from the current time point to the next. Pd1 is the probability that an oligodendrocyte will remain an oligodendrocyte from the current time point to the next. Pd2 is the probability that an oligodendrocyte dedifferentiates into an OPC form the current time point to the next. Pa1+Pa2+Pa3 = 1; Pb1+Pb2+Pb3 = 1; Pc1+Pc2+Pc3 = 1.
Another powerful tool for modeling temporal processes is a dynamic graphical model (DGM).66,67 In fact, a hidden Markov model can be thought of as the simplest of DGMs. In brief, DGMs are graphical models—probabilistic models of covariance/dependence among groups of variables—that provide fully Bayesian inference for time series data (for those unfamiliar with Bayesian statistics we recommend Bernardo et al., 2009).68 A graphical model is defined by a graph (V,E), where V represents the vertices and E represents the edges, and a set of properties, which determine a family of probability distributions on the variables represented in the graph. For example, we may have a set of transcripts, which we represent by X1, X2,…, XN, N = |V|. These transcripts can be represented as vertices in the graph with edges between them, and the set of properties for the graph define the probability distributions on the transcripts. These distributions specify how the transcripts co-vary or depend on one another (or not). There are many different types of graphical models; each type is defined by what graphs and what graphical properties are allowed. Many existing cancer cell cycle models neglect the autocorrelation between successive measurements, which has shown to lead to an overestimation of the number of cycling variables.69 Since the model search for a DGM can involve dependencies among transcripts, this type of model can avoid this potential pitfall.
Apart from statistical techniques for epigenomics there are techniques for mathematical modeling of biological networks that represent a key to modern systems biology.70 Ideally, we would like to develop such models to understand the roles of ncRNAs in the regulatory networks that underlie GBM initiation and progression. A recent review by Lim et al.71 describes how complex regulatory functions of cells may be conceptualized as a system of dynamic functions, and, as such, may be modeled by combinations of core network motifs. They posit that in terms of molecular networks or algorithms, it may be possible to limit the space of models for a given cellular regulatory function and hence identify and use such models to generate insights into the regulatory process.
What is interesting is that multiple modeling approaches can simulate the same biological phenomenon. Researchers have focused on identifying the most useful and accurate model. From a statistician’s perspective this problem of model selection is one of prediction and robustness. A useful model will be (1) robust to changes in model assumptions and (2) generate accurate predictions in a specific context. Many models involve tuning parameters for which values must be chosen. In some cases methods exist, which provide reasonable estimates for these values, but in other cases the estimates are arbitrary. Ideally we would like any modeling results to be robust to these arbitrary choices, i.e., small changes in the tuning parameters should lead to small changes in model outputs. This gives us confidence that an accurate model reflects a biological truth, not the luck of the analyst.
Historically scientists have emphasized good model fit by selecting a model based on how well it explained the observed data. However, in contexts where the number of variables, p, is much larger than the number of observations, n, this can lead to overfitting. The model fits the observed data so well that it is specific to that data only, and fails to explain the broader biological phenomenon at work. In this scenario, the model cannot predict future observations from the same context as the observed data, nor can it predict observations from similar contexts. We have seen overfitting at work in research involving microarrays as well as other genome-wide technologies.72,73 One way to avoid overfitting is to focus on how well a given model can predict data from similar studies, or data from the same study that is not used in building the model.
The complexity of modeling for systems biology is increasing as investigators collect more complex data types. Issues such as prediction and robustness will be critical in sifting through this data, using modeling approaches to identify and explain potential pharmacologic targets in GBM.
Conclusions
The medical and scientific community has made remarkable advances in defining the cellular, genetic, and epigenetic origin of GBM. Many of these advances have come through studying neurogenesis and epigenetic regulation. The hope is that these advances can be quickly translated into therapeutic benefits for patients who are suffering from this incurable and aggressive disease. Novel therapies are likely to come from epigenetic modifiers, which are becoming attractive drug targets in other cancers such as lymphoma. Epigenetic targets offer an opportunity to modulate aberrant cellular behavior as they exhibit dynamic behavior, as opposed to permanent genetic mutations. However, for these targets to be validated in GBM, detailed network analyses of microRNA/mRNAs and lncRNAs are required. These analyses will also need to be coupled to understanding the role that epigenetic enzymes play in GBM cell of origin survival. Once an epigenetic target is selected, the next challenge will be to generate small molecule inhibitors that can pass the blood brain barrier. Given the likelihood that drug combinations will be required to effectively treat GBM, drug interactions will also have to be considered to identify the effect of each drug in influencing blood brain barrier permeability.
Acknowledgments
We thank all members of the Brain Tumor Initiative at the University of Miami and the Center for Therapeutic Innovation for helpful suggestions. We thank Mohammad Faghihi, Zane Zeier, and Pantelis Tsoulfas for reading the manuscript and for helpful suggestion. This work was partly supported by R01NS067289 to NGA and an IRDI grant from the University of Miami to NGA, JC, and CW. The authors declare no financial conflict of interest.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/25440
References
- 1.Lacroix M, Abi-Said D, Fourney DR, Gokaslan ZL, Shi W, DeMonte F, et al. A multivariate analysis of 416 patients with glioblastoma multiforme: prognosis, extent of resection, and survival. J Neurosurg. 2001;95:190–8. doi: 10.3171/jns.2001.95.2.0190. [DOI] [PubMed] [Google Scholar]
- 2.Chen J, McKay RM, Parada LF. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell. 2012;149:36–47. doi: 10.1016/j.cell.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu C, Sage JC, Miller MR, Verhaak RG, Hippenmeyer S, Vogel H, et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146:209–21. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koso H, Takeda H, Yew CC, Ward JM, Nariai N, Ueno K, et al. Transposon mutagenesis identifies genes that transform neural stem cells into glioma-initiating cells. Proc Natl Acad Sci U S A. 2012;109:E2998–3007. doi: 10.1073/pnas.1215899109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, Marumoto T, Singer O, et al. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 2012;338:1080–4. doi: 10.1126/science.1226929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hambardzumyan D, Cheng YK, Haeno H, Holland EC, Michor F. The probable cell of origin of NF1- and PDGF-driven glioblastomas. PLoS One. 2011;6:e24454. doi: 10.1371/journal.pone.0024454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–6. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. Defining the mode of tumour growth by clonal analysis. Nature. 2012;488:527–30. doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337:1231–5. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–73. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pe’er D, Hacohen N. Principles and strategies for developing network models in cancer. Cell. 2011;144:864–73. doi: 10.1016/j.cell.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalari S, Pfeifer GP. Identification of driver and passenger DNA methylation in cancer by epigenomic analysis. Adv Genet. 2010;70:277–308. doi: 10.1016/B978-0-12-380866-0.60010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–31. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 17.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riddick G, Fine HA. Integration and analysis of genome-scale data from gliomas. Nat Rev Neurol. 2011;7:439–50. doi: 10.1038/nrneurol.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Natsume A, Kondo Y, Ito M, Motomura K, Wakabayashi T, Yoshida J. Epigenetic aberrations and therapeutic implications in gliomas. Cancer Sci. 2010;101:1331–6. doi: 10.1111/j.1349-7006.2010.01545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kreth S, Thon N, Kreth FW. Epigenetics in human gliomas. Cancer Lett. 2012 doi: 10.1016/j.canlet.2012.04.008. In press; [DOI] [PubMed] [Google Scholar]
- 21.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 22.Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22:425–37. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 23.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 24.Ciafrè SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–8. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 25.Gabriely G, Yi M, Narayan RS, Niers JM, Wurdinger T, Imitola J, et al. Human glioma growth is controlled by microRNA-10b. Cancer Res. 2011;71:3563–72. doi: 10.1158/0008-5472.CAN-10-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guessous F, Alvarado-Velez M, Marcinkiewicz L, Zhang Y, Kim J, Heister S, et al. Oncogenic effects of miR-10b in glioblastoma stem cells. J Neurooncol. 2013;112:153–63. doi: 10.1007/s11060-013-1047-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–57. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–81. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–95. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang L, Du WW, Yang X, Chen K, Ghanekar A, Levy G, et al. Versican 3′-untranslated region (3′-UTR) functions as a ceRNA in inducing the development of hepatocellular carcinoma by regulating miRNA activity. FASEB J. 2013;27:907–19. doi: 10.1096/fj.12-220905. [DOI] [PubMed] [Google Scholar]
- 31.Sarver AL, Subramanian S. Competing endogenous RNA database. Bioinformation. 2012;8:731–3. doi: 10.6026/97320630008731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–34. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–9. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–9. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–30. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 38.Wang P, Ren Z, Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J Cell Biochem. 2012;113:1868–74. doi: 10.1002/jcb.24055. [DOI] [PubMed] [Google Scholar]
- 39.Han L, Zhang K, Shi Z, Zhang J, Zhu J, Zhu S, et al. LncRNA profile of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis. Int J Oncol. 2012;40:2004–12. doi: 10.3892/ijo.2012.1413. [DOI] [PubMed] [Google Scholar]
- 40.Jalali S, Bhartiya D, Lalwani MK, Sivasubbu S, Scaria V. Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PLoS One. 2013;8:e53823. doi: 10.1371/journal.pone.0053823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katsushima K, Shinjo K, Natsume A, Ohka F, Fujii M, Osada H, et al. Contribution of microRNA-1275 to Claudin11 protein suppression via a polycomb-mediated silencing mechanism in human glioma stem-like cells. J Biol Chem. 2012;287:27396–406. doi: 10.1074/jbc.M112.359109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yap KL, Li S, Muñoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–74. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suvà ML, Riggi N, Janiszewska M, Radovanovic I, Provero P, Stehle JC, et al. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res. 2009;69:9211–8. doi: 10.1158/0008-5472.CAN-09-1622. [DOI] [PubMed] [Google Scholar]
- 44.Orzan F, Pellegatta S, Poliani PL, Pisati F, Caldera V, Menghi F, et al. Enhancer of Zeste 2 (EZH2) is up-regulated in malignant gliomas and in glioma stem-like cells. Neuropathol Appl Neurobiol. 2011;37:381–94. doi: 10.1111/j.1365-2990.2010.01132.x. [DOI] [PubMed] [Google Scholar]
- 45.Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 46.Lee JM, Lee JS, Kim H, Kim K, Park H, Kim JY, et al. EZH2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase complex. Mol Cell. 2012;48:572–86. doi: 10.1016/j.molcel.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 47.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, et al. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24:2615–20. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 50.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–89. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 51.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–12. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 52.Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8:890–6. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 53.Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–9. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smits M, Nilsson J, Mir SE, van der Stoop PM, Hulleman E, Niers JM, et al. miR-101 is down-regulated in glioblastoma resulting in EZH2-induced proliferation, migration, and angiogenesis. Oncotarget. 2010;1:710–20. doi: 10.18632/oncotarget.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carén H, Pollard SM, Beck S. The good, the bad and the ugly: Epigenetic mechanisms in glioblastoma. Mol Aspects Med. 2013;34:849–62. doi: 10.1016/j.mam.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Linnet K, Ejsing TB. A review on the impact of P-glycoprotein on the penetration of drugs into the brain. Focus on psychotropic drugs. Eur Neuropsychopharmacol. 2008;18:157–69. doi: 10.1016/j.euroneuro.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 57.Fronza R, Tramonti M, Atchley WR, Nardini C. Brain cancer prognosis: independent validation of a clinical bioinformatics approach. J Clin Bioinforma. 2012;2:2. doi: 10.1186/2043-9113-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wuchty S, Arjona D, Li A, Kotliarov Y, Walling J, Ahn S, et al. Prediction of Associations between microRNAs and Gene Expression in Glioma Biology. PLoS One. 2011;6:e14681. doi: 10.1371/journal.pone.0014681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kunkle B, Yoo C, Roy D. Discovering gene-environment interactions in Glioblastoma through a comprehensive data integration bioinformatics method. Neurotoxicology. 2013;35:1–14. doi: 10.1016/j.neuro.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 60.Jobe EM, McQuate AL, Zhao X. Crosstalk among Epigenetic Pathways Regulates Neurogenesis. Front Neurosci. 2012;6:59. doi: 10.3389/fnins.2012.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sima C, Hua J, Jung S. Inference of gene regulatory networks using time-series data: a survey. Curr Genomics. 2009;10:416–29. doi: 10.2174/138920209789177610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Allen L. An Introduction to Stochastic Processes with Applications to Biology. 2nd Ed CRC Press/Chapman & Hall, Boca Raton, Fl 2010. [Google Scholar]
- 63.Ernst J, Nau GJ, Bar-Joseph Z. Clustering short time series gene expression data. Bioinformatics. 2005;21(Suppl 1):i159–68. doi: 10.1093/bioinformatics/bti1022. [DOI] [PubMed] [Google Scholar]
- 64.Genolini C, Falissard B. KmL: a package to cluster longitudinal data. Comput Methods Programs Biomed. 2011;104:e112–21. doi: 10.1016/j.cmpb.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 65.Friedman N, Linial M, Nachman I, Pe’er D. Using Bayesian networks to analyze expression data. J Comput Biol. 2000;7:601–20. doi: 10.1089/106652700750050961. [DOI] [PubMed] [Google Scholar]
- 66.Bilmes J. Dynamic Graphic Models. IEEE Signal Process Mag. 2010;34:29–42. [Google Scholar]
- 67.Dobra A, Hans C, Jones B, Nevins JR, Yao G, West M. Sparse graphical models for exploring gene expression data. J Multivariate Anal. 2004;90:196–212. doi: 10.1016/j.jmva.2004.02.009. [DOI] [Google Scholar]
- 68.J. M. Bernardo JOB. A. P. Dawid and A. F. M. Smith (eds). Bayesian Statistics Oxford: Oxford University Press 1999; 6. [Google Scholar]
- 69.Futschik ME, Herzel H. Are we overestimating the number of cell-cycling genes? The impact of background models on time-series analysis. Bioinformatics. 2008;24:1063–9. doi: 10.1093/bioinformatics/btn072. [DOI] [PubMed] [Google Scholar]
- 70.West PA. Time Series: Modeling, Computation, and Inference Chapman & Hall/CRC Texts in Statistical Science) 2010. [Google Scholar]
- 71.Lim WA, Lee CM, Tang C. Design principles of regulatory networks: searching for the molecular algorithms of the cell. Mol Cell. 2013;49:202–12. doi: 10.1016/j.molcel.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ioannidis JP. Why most published research findings are false. PLoS Med. 2005;2:e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ioannidis JP, Allison DB, Ball CA, Coulibaly I, Cui X, Culhane AC, et al. Repeatability of published microarray gene expression analyses. Nat Genet. 2009;41:149–55. doi: 10.1038/ng.295. [DOI] [PubMed] [Google Scholar]