Abstract
DNA methylation is an important molecular-level phenotype that links genotypes and complex disease traits. Previous studies have found local correlation between genetic variants and DNA methylation levels (cis-meQTLs). However, general mechanisms underlying cis-meQTLs are unclear. We conducted a cis-meQTL analysis of the Genetics of Lipid Lowering Drugs and Diet Network data (n = 593). We found that over 80% of genetic variants at CpG sites (meSNPs) are meQTL loci (P-value < 10−9), and meSNPs account for over two thirds of the strongest meQTL signals (P-value < 10−200). Beyond direct effects on the methylation of the meSNP site, the CpG-disrupting allele of meSNPs were associated with lowered methylation of CpG sites located within 45 bp. The effect of meSNPs extends to as far as 10 kb and can contribute to the observed meQTL signals in the surrounding region, likely through correlated methylation patterns and linkage disequilibrium. Therefore, meSNPs are behind a large portion of observed meQTL signals and play a crucial role in the biological process linking genetic variation to epigenetic changes.
Keywords: DNA methylation, meSNP, meQTL, epigenome-wide study, Infinium Human Methylation 450K BeadChip
DNA methylation plays an important role in gene regulation. As such, it is an important intermediate molecular phenotype that links genotypes and other macro-level phenotypes and may contribute to the missing heritability.1 Despite their prominent physiologic role, the genetic determinants of DNA methylation patterns are poorly understood. There is evidence of correlation between genetic variation at specific loci and the quantitative trait of DNA methylation.2-5 Additionally, prior studies6,7 revealed that genetic variants at CpG sites (meSNPs) are likely to disrupt the substrate of methylation reactions and thus drastically change the methylation status at a single CpG site. However, it is unclear if meSNPs represent a major class of methylation-associated loci (meQTLs). Moreover, it is unclear if meSNPs modulate the methylation status of nearby CpG sites. Most existing meQTL studies2-5 are based on relatively small-sized samples and low-resolution methylation microarrays, wherein meSNPs are sparsely represented. Additionally, current practices of meQTL analysis typically exclude probes that contain sequence variants to limit misclassification due to alterations in probe hybridization.2,3 Recently, we generated high-resolution genotyping and epigenetic data from the Genetics of Lipid Lowering Drugs and Diet Network (GOLDN),8 yielding a considerable amount of meSNPs with information on both genotype and methylation status. These data enabled us to investigate the impact of meSNPs on the methylation status of the CpG site harboring the SNP, as well as on surrounding CpG sites.
We used CD4+ T-cells harvested from frozen buffy coat samples isolated from peripheral blood for methylation measurement. We used CD4+ T-cells simply due to their abundance and physiological role in the immune response. While that ensures higher validity of findings, our results may not be generalizable to other cell populations. After genotype imputation and quality control (see Supplemental Methods), we had data for 461,281 CpGs of n = 593 GOLDN participants. All participants had previously been genotyped at 906,600 loci using the Affymetrix Genome-Wide Human SNP Array 6.0, as described in prior publications.9,10 MACH software (Version 1.0.16) was used to impute non-genotyped SNPs using HapMap Phase II (release 22, Human Genome build 36, hg18) as a reference, resulting genotypes of 2,529,001 SNPs.
We first performed a genome-wide cis-meQTL analysis of GOLDN data. We restricted the analysis to SNP/CpG site pairs within 20 kb of each other, and removed the pairs in which the SNP was located on the methylation probe as described below, yielding 12,347,772 pairs for the final analysis. We used linear mixed models, fit using the lmekin function of the Kinship package in R,11 to regress the methylation level (β value) of a CpG site on the genetic variant at a SNP site, adjusting for covariates (age, sex, and recruitment center) as fixed effects and family structure as a random effect. Adjusting top principal components of phenotype matrix is an effective approach to reduce experimental artifacts and enhance power for gene expression quantitative trait analysis.12,13 Therefore, we adjusted for the top four principal components as the proportion of explained variance in methylation drops off sharply after the fourth principal component. We identified 465,649 cis-meQTL signals, defined as any SNP/CpG site pair within 20 kb distance of each other that reached the nominal P-value cutoff of 10−9. We used the significant association of random SNP/CpG site pairs as an upper bound of false discoveries, a strategy used in expression QTL analysis.12 While that approach overestimates the true false discovery rate (FDR) due to ignoring any potential true trans-meQTLs, it is easier to compute and preserve all family structure and covariates. Using 130 million pairs, we estimated that the nominal P-value of 10−9 as equivalent to the FDR of 0.000004, and the nominal P-value of 10−200 as FDR < 7.7 × 10−9 (no random pairs with p < 10−200). As expected, these P-values far exceed the cutoffs used in previous reports2-5 due to the high density of the M450K chip.
Although the M450K chip is not designed for profiling methylation levels at CpG sites harboring SNPs, valid methylation measurements can nevertheless be obtained through careful analysis. The M450K chip is known to have a “probe effect,” i.e., the SNP/CpG site pair with the SNP on the probe may be enriched for meQTLs. While Illumina recommends caution for the probe effect when the SNP is within 10 bp of the interrogated CpG site, we observed that the probe effect is present throughout the entire length of the 50 bp probe (Fig. S2). Therefore, we removed all pairs with SNPs on the probe from this analysis. However, it is important to note that, among the two chemistries in the M450K platform—Infinium I and Infinium II—only Infinium I measurements, whose probe covers the entire CpG dinucleotide, are subject to the probe effects of SNPs at the CpG site itself. For the 1 bp extension of Infinium II chemistry, position 0 on a forward probe and position 1 on a reverse probe do not hybridize to interrogated DNA and thus still provide valid methylation measurements (see Supplementary Methods). After excluding probes that matched to non-unique positions in the hg19 genome as well as the 95 sites located on Infinium I probes, we first investigated these 812 meSNPs for which both methylation level and genotype were directly observed or imputed. See Table S1 for a complete list of these meSNPs.
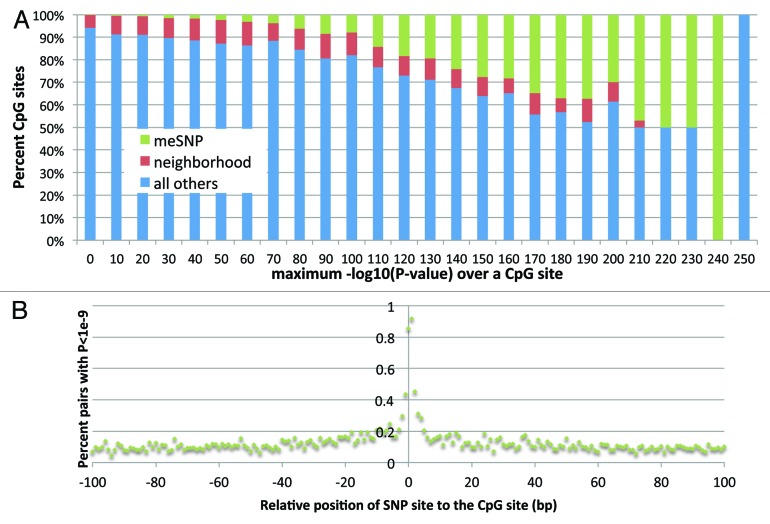
Many of the most statistically significant meQTLs were co-localized to the 812 meSNPs (Fig. 1). 85% of meSNPs interrogated by forward strand probes and 92% by reverse strand probes were cis-meQTLs (Fig. 1). These SNPs were strongly negatively associated with the methylation of the CpG site (Table 1). Cis-meQTL signals remained strong at 1–3 bp away from the CpG site (20–45%), but sharply decreased to < 20% for most sites > 3 bp away from the CpG site. The signal attenuated to approximately 5% of pairs separated by the distance of 1,000 bp and 3% for pairs at 20,000 bp, still well above the 0.0003% for random pairs in the entire genome. Interestingly, we observed that pairs with SNPs on the probe do have inflated meQTL signals (Fig. S2). Absence of a significant meQTL signal at meSNPs was mainly due to the methylation being already depleted for the CpG allele or to very low genetic variation. Therefore, we conclude that meSNPs indeed disrupt the methylation and that the M450K chip provides accurate measurement of that phenomenon.
Figure 1. meSNPs are enriched for the most significant meQTL signals. (A) The percent of CpG sites that are meSNP, within 20 kb neighborhood of a meSNP (“neighborhood”), or “all others.” For each CpG site, the measure of its meQTL signals is the maximum of –log10(P-value) of its SNP/CpG pairs. For each bin of –log10(P-value), we plotted the percent of each type of CpG sites. (B) Proportion of SNP/CpG site pairs with meQTL signal, defined as P-value < 10−9, stratified by distance. 96 meSNPs at the position 0 and 716 meSNPs at position 1 are represented. On average, 138 non meSNP pairs per position are within position < 50 bp, and 283 pairs per position between 50 and 100 bp, reflecting the change in effect due to filtering out the “probe effect.”
Table 1. Summary of 812 cis-meQTLs at meSNPs probed by Infinium II chemistry.
| Strand | Position within CpG | Ref | Alt | Count | Mean -log10Pval |
Mean effect size β | %meQTL* | |
|---|---|---|---|---|---|---|---|---|
| F |
0 |
C |
A |
6 |
102.0 |
-0.207 |
83.33% |
|
| F |
0 |
C |
G |
11 |
95.2 |
0.151 |
90.91% |
|
| F |
0 |
C |
T |
79 |
108.3 |
-0.229 |
84.81% |
|
| R |
1 |
G |
A |
606 |
115.4 |
-0.216 |
92.41% |
|
| R |
1 |
G |
C |
48 |
74.5 |
0.111 |
87.50% |
|
| R | 1 | G | T | 62 | 103.1 | -0.217 | 88.71% | |
% of pairs with p < 10−9. See Supplementary Methods for an interpretation of the effect sizes
We subsequently investigated the potential impact of meSNPs on the methylation status of nearby CpG sites, located within 100bp. It is notable that the M450K chip only directly measures the methylation status at CpG sites of the reference genome, and thus only SNPs that disrupting CpG sites are directly observable. To study the potential effect of all CpG-gain SNPs to nearby CpG sites, we conducted a bioinformatics annotation of all SNPs in the GOLDN and the 1000 Genomes Project phase 1 release. All SNPs were classified into four categories in terms of changing CpG sites on the reference genome: CpG-loss, which eliminates a CpG site; CpG-gain, which creates a CpG site; CpG-shift, which represents a variant between CCG and CGG that shifts the position of a CpG site by 1 bp; and CpG-irrelevant. CpG-loss and CpG-gain SNPs are only defined relative to the reference genome; both CpG-loss and CpG-gain have an allele that disrupts the substrate of methylation and thus are likely to be meQTLs. All 812 meSNPs represented on the M450K chip and were successfully genotyped in GOLDN are either CpG-loss (803, 99%) or CpG-shift (9, 1%). We scanned all 39 million SNPs from the 1000 Genomes Project phase 1 release and all 2.5 million SNPs in the GOLDN imputed variant set, resulting in the classification of SNPs as shown in Figure S3. Overall, we found that 14% of GOLDN SNPs are CpG-loss and 14% are CpG-gain. Similar proportions were observed in the 1000 Genomes Project Phase 1 SNPs (Fig. S3).
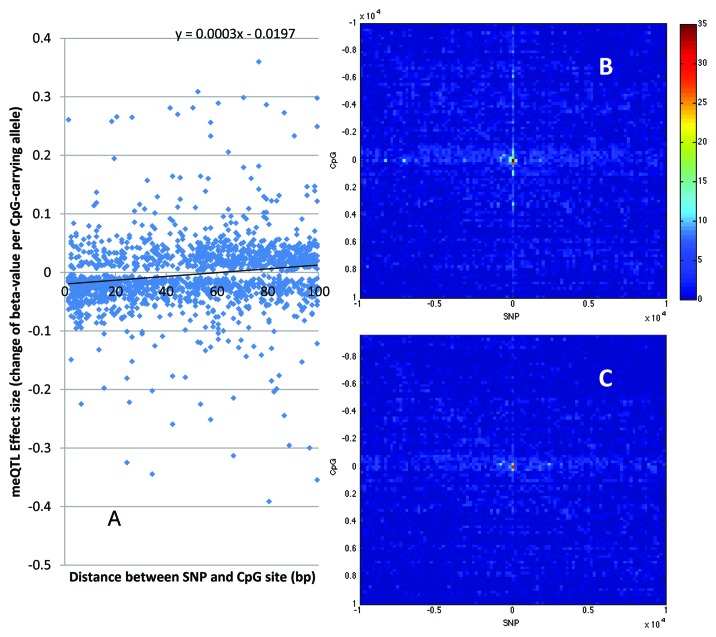
Although the methylation measurements at CpG-gain SNPs and many of the CpG-loss SNPs are not directly observed, we reasoned that they likely follow the same pattern as the 812 CpG-loss SNPs discussed above. To investigate the effect of meSNPs to the methylation at immediate nearby sites, we looked for meQTLs (p < 10−9) between 701 CpG-gain or 861 CpG-loss SNPs with allele frequency between 5% and 95% represented in the GOLDN genotype set and CpG sites within 100 bp (but not at the SNP directly) with M450K measurements in Infinium II chemistry, and found 1,562 such pairs. Through a linear regression of the effect sizes of these meQTL and the distance between the SNP-CpG pairs, we found that, for each CpG-disrupting allele, the methylation of CpG sites at very close distance is also lowered for about 2% (P-value = 3 × 10−11 for the intersect), but the effect is diminishes to flat and even a slight elevation beyond 60 bp (P-value = 4.0 × 10−10 for the slope) (Fig. 2). This result suggests that a CpG-changing SNP can affect not only the methylation status of its own CpG site, but also the methylation patterns surrounding it at very close distance. However, while this may point to an interesting methylation regulation mechanism, additional investigations are warranted to rule out potential technical artifacts such as local hybridization competition.
Figure 2. Effect of meSNPs on methylation at nearby CpG sites. (A) Near-range effect of meSNPs. For 1,562 CpG-changing (including CpG-loss and CpG-gain) SNPs that have significant meQTL (P-value < 10−9) to a CpG site within 100 bp, we plot their SNP-CpG distance (X-coordinate) vs. their meQTL effect sizes (Y-coordinate). Both CpG-loss and CpG-gain SNP genotypes are oriented such that the CpG-carrying allele is coded as 0 and the non-CpG-carrying allele is coded as 1. (B and C) Effect of meSNP beyond immediate neighborhood. We plotted the distribution of cis-meQTL signals surrounding 812 meSNPs in the GOLDN data, before (B) and after (C) adjusting for meSNPs. The center is the location of the meSNPs. The X-axis represents the relative position of the SNP to the central meSNPs, organized into 101 equal-sized bins, each covering 198 bp. The Y-axis represents similar bins for the relative position of the CpG to the meSNPs. Color of the heatmap represents the number of meQTLs found at the distance bin.
Finally, we investigated whether meSNPs may explain meQTL signals at surrounding sites located within 10 kb. Not all meQTL signals are due to independent association signals. In particular, we observe that meQTL signals show a prominent correlation pattern (Fig. 2B). We therefore hypothesize that, in the neighborhood of a meSNP with genotype X and CpG methylation Y, the meQTL signal between genotype X' and CpG methylation Y' is due to their respective correlations to the meSNP pair X and Y: X' to X by linkage disequilibrium, Y' to Y by methylation correlation. Unlike standard mediation analyses, we are not claiming a chain of causative links, but rather to test if the associations X' to Y' and X to Y are independent. Instead of running a full mediation analysis,14 we conducted a partial mediation analysis, i.e., the conditional regression of Y' to X' given Y, in addition to the pairwise meQTL analysis. If they are truly independent associations, regressing Y' to X' conditional on Y should not eliminate the significant meQTL association between Y' and X'.
We found a considerable attenuation of meQTL signals surrounding meSNPs (Fig. 2C vs. B). In the 10 kb neighborhood of 812 observed meSNPs, the total numbers of meQTLs decreased by 32.8%, from 8,017 to 5,384 after the adjustment. Given the genome-wide distribution of meSNPs, most of which are not directly observed on the M450K chip, we estimate that meQTLs at meSNPs may explain a large portion of observed cis-meQTL signals.
Overall, our analysis provides the first genome-wide profiling of methylation changes at and surrounding SNPs. These meSNPs contribute to the strongest methylation QTL signals as they disrupt the methylation status at CpG sites. Moreover, meSNPs also modulate methylation status at nearby CpG sites and a large proportion of variability in DNA methylation can be explained by genetic variation at the CpG site itself. Our data suggest a potential biological mechanism underlying associations between common genetic variants, epigenetic processes, and disease phenotypes. We conclude that meSNPs may be a crucial class of variants to be considered in interpretation of genomic and epigenomic association results.
Supplementary Material
Submitted
02/28/13
Revised
06/19/13
Accepted
06/21/13
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/25501
References
- 1.Maher B. Personal genomes: The case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 2.Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai SL, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12:R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shoemaker R, Deng J, Wang W, Zhang K. Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res. 2010;20:883–9. doi: 10.1101/gr.104695.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang D, Cheng L, Badner JA, Chen C, Chen Q, Luo W, et al. Genetic control of individual differences in gene-specific methylation in human brain. Am J Hum Genet. 2010;86:411–9. doi: 10.1016/j.ajhg.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gertz J, Varley KE, Reddy TE, Bowling KM, Pauli F, Parker SL, et al. Analysis of DNA methylation in a three-generation family reveals widespread genetic influence on epigenetic regulation. PLoS Genet. 2011;7:e1002228. doi: 10.1371/journal.pgen.1002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hellman A, Chess A. Extensive sequence-influenced DNA methylation polymorphism in the human genome. Epigenetics Chromatin. 2010;3:11. doi: 10.1186/1756-8935-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai CQ, Arnett DK, Corella D, Straka RJ, Tsai MY, Peacock JM, et al. Fenofibrate effect on triglyceride and postprandial response of apolipoprotein A5 variants: the GOLDN study. Arterioscler Thromb Vasc Biol. 2007;27:1417–25. doi: 10.1161/ATVBAHA.107.140103. [DOI] [PubMed] [Google Scholar]
- 9.Irvin MR, Kabagambe EK, Tiwari HK, Parnell LD, Straka RJ, Tsai MY, et al. Apolipoprotein E polymorphisms and postprandial triglyceridemia before and after fenofibrate treatment in the Genetics of Lipid Lowering and Diet Network (GOLDN) Study. Circ Cardiovasc Genet. 2010;3:462–7. doi: 10.1161/CIRCGENETICS.110.950667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aslibekyan S, Kabagambe EK, Irvin MR, Straka RJ, Borecki IB, Tiwari HK, et al. A genome-wide association study of inflammatory biomarker changes in response to fenofibrate treatment in the Genetics of Lipid Lowering Drug and Diet Network. Pharmacogenet Genomics. 2012;22:191–7. doi: 10.1097/FPC.0b013e32834fdd41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atkinson B, Therneau T. kinship: mixed-effects Cox models, sparse matrices, and modeling data from large pedigrees. 2007; R package version 1.1.0–17.
- 12.Liang L, Morar N, Dixon AL, Lathrop GM, Abecasis GR, Moffatt MF, et al. A cross-platform analysis of 14,177 expression quantitative trait loci derived from lymphoblastoid cell lines. Genome Res. 2013;23:716–26. doi: 10.1101/gr.142521.112. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–72. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Aryee MJ, Padyukov L, Fallin MD, Hesselberg E, Runarsson A, et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol. 2013;31:142–7. doi: 10.1038/nbt.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.