Abstract
Despite recent advances in understanding the molecular mechanisms of autism spectrum disorders (ASD), the current treatments for these disorders are mostly focused on behavioral and educational approaches. The considerable clinical and molecular heterogeneity of ASD present a significant challenge to the development of an effective treatment targeting underlying molecular defects. Deficiency of SHANK family genes causing ASD represent an exciting opportunity for developing molecular therapies because of strong genetic evidence for SHANKs as causative genes in ASD and the availability of a panel of Shank mutant mouse models. In this article we review the literature suggesting the potential for developing therapies based on molecular characteristics and discuss several exciting themes that are emerging from studying Shank mutant mice at the molecular level and in terms of synaptic function.
Keywords: Autism Spectrum Disorders, Mouse model, SHANK family protein, Synapses, Brain stimulation
Introduction
Despite the significant progress in recognizing and understanding the etiology of autism spectrums disorders (ASD) in the last decade, we have made few advances in the area of treatments and interventions (Blenner et al., 2011; State, 2010; Volkmar et al., 2009). The current therapeutic options are mostly restricted to programs of behavioral modification such as applied behavioral analysis (ABA), early start Denver model, and Treatment and Education of Autistic and related Communication handicapped Children (TEACCH) (Dawson et al., 2010; Kasari and Lawton, 2010; McPheeters et al., 2011; Myers and Johnson, 2007; Taylor et al., 2012; Warren et al., 2011). These interventions are primarily based on behavioral and educational approaches linked to autistic behaviors but not targeting the underlying biological causes (Vismara and Rogers, 2010). The outcome of these behavioral therapies is quite variable and a vigorous validation for their efficacy is still warranted (Grindle et al., 2012; Hayward et al., 2009; Taylor et al., 2012). Because of the considerable clinical and molecular heterogeneity of ASD that has become apparent from the last decade of research (Betancur, 2011; Devlin and Scherer, 2012; Georgiades et al., 2012), one of the critical questions is whether there is a common pathophysiology at the molecular and circuitry levels underlying ASD that can be targeted for interventions (Dolen et al., 2010; Geschwind, 2008; Geschwind and Levitt, 2007; Kelleher and Bear, 2008; Smith and Ehlers, 2012). Currently, there is no medication available to specifically treat the core symptoms of ASD despite the fact that the use of drugs targeting behavioral presentations is common in clinical practice (Carrasco et al., 2012; McPheeters et al., 2011; Myers, 2007; Volkmar, 2001). Two medications, risperidone and aripiprazole, have been approved by the Food and Drug Administration (FDA) to treat the comorbidities commonly seen in ASD (Marcus et al., 2011a; Marcus et al., 2011b)(McPheeters et al, 2011)(McCracken et al., 2002). Similar to behavioral interventions, there is little biological evidence to specifically supports these treatments. Also, the safety profile and efficacy of these treatments in children with ASD remain to be further investigated (Huffman et al., 2011; Panagiotopoulos et al., 2010).
Genetic defect of SHANKs in ASD
The discovery of genetic defects in a sub-set of ASD patients offers a unique opportunity to explore therapeutic approaches for the core symptoms that is based on the underlying biological mechanism (Devlin and Scherer, 2012; Malhotra and Sebat, 2012; Smith and Ehlers, 2012; Toro et al., 2010). SHANK family genes (SHANKs) causing ASD (Shankopathies) probably represent one of the best opportunities for this direction of research. SHANK family genes include SHANK1, SHANK2, and SHANK3 and encode proteins with 5 protein-protein interaction domains including ankrin repeats (ANK), SH3, PDZ, proline-rich region with Homer binding, and SAM (Grabrucker et al., 2011; Kreienkamp, 2008; Sheng and Kim, 2000). SHANK proteins are scaffolding proteins enriched at the post synaptic density (PSD) of excitatory synapses (Naisbitt et al., 1999). Since the first report of a SHANK3-specific mutations in ASD in 2007 (Durand et al., 2007), there is now genetic evidence supporting the involvement of all SHANK family genes in ASD (Berkel et al., 2010; Berkel et al., 2012; Leblond et al., 2012; Moessner et al., 2007; Sato et al., 2012). Evidence for SHANK3 causing ASD is particularly strong because it involves different types of genetic defects such as microdeletions and point mutations, and the findings have been independently replicated in different ASD patient cohorts (Durand et al., 2007; Gauthier et al., 2009; Moessner et al., 2007).
Mutation mechanism underlying the Shankopathies
All types of genetic mutations of SHANKs found in ASD are reported in heterozygotes (Moessner et al., 2007). Microdeletion of SHANK3 in 22q13.2 deletion syndrome (i.e. Phelan-McDermid syndrome) is the most common molecular defect that accounts for more than 95% of cases reported in the literature (Phelan, 2007). Point mutations or small intragenic mutations contribute to a small percentage of SHANK causing ASD cases studied (Berkel et al., 2010; Berkel et al., 2012; Bonaglia et al., 2011; Moessner et al., 2007). Chromosome translocation with a breakpoint within the SHANK3 gene has also been reported (Bonaglia et al., 2005). The fact that microdeletions usually disrupt entire SHANK genes generally supports haploinsufficiency as the molecular mechanism underlying the pathogenesis in these patients. For point mutations, particularly missense mutations of SHANK2 and SHANK3 found in ASD, the possibility of a gain of function mechanism may also be considered.
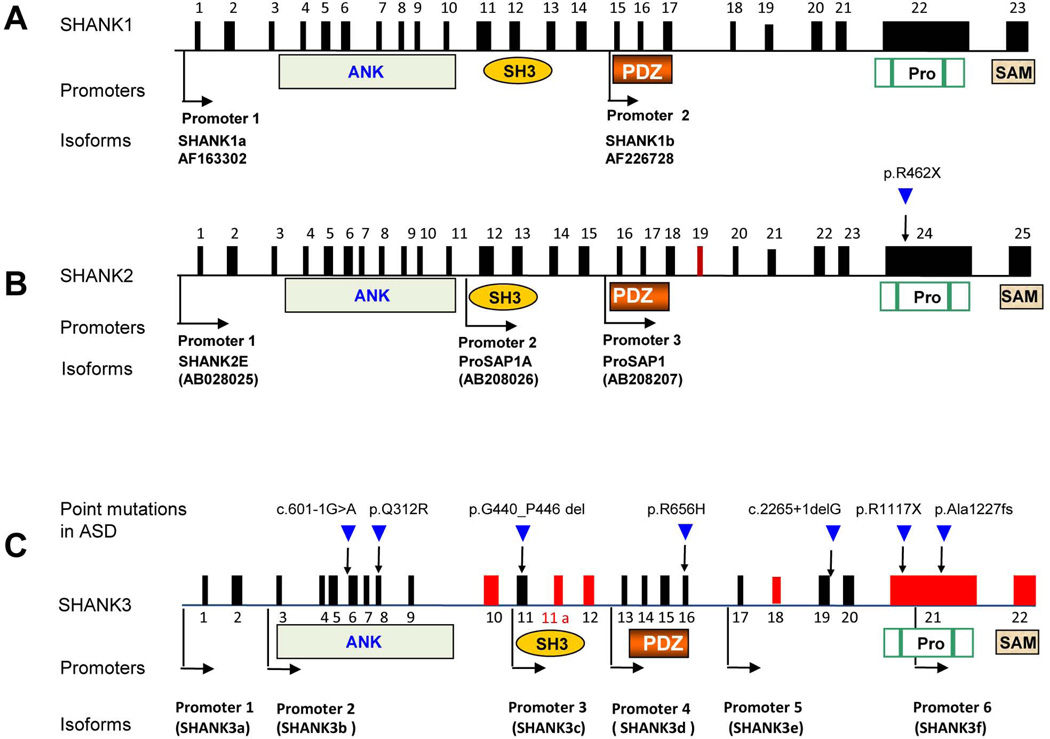
One of the interesting features observed in all SHANK family genes is complex transcriptional structure and extensive isoforms resulting from multiple intragenic promoters and extensive alternative splicing of coding exons in SHANKs (Durand et al., 2011; Leblond et al., 2012; Lim et al., 1999; Wang et al., 2011; Wilson et al., 2003) (Figure 1). For example, 3 promoters and brain-region-specific alternative splicing of several coding exons are reported in SHANK2 (Leblond et al., 2012). SHANK3 has 6 promoters and extensive splicing of several coding exons that result in an array of SHANK3 isoforms with different combinations of the 5 protein domains (Wang et al., 2011). These data indicate that individual point mutations in different exons of SHANK genes may only disrupt selective isoforms of SHANKs in ASD. A similar phenomenon is suggested in SHANK1 but the details remain to be characterized (Lim et al., 1999). Because each isoform has a combination of different protein domains for SHANK proteins, the function of each isoform at synapses is predicted to be different based on the study of domain specific mutations by RNAi in cultured neurons (Roussignol et al., 2005; Sala et al., 2001). The interesting hypothesis to be tested in humans is whether isoform-specific disruption of SHANKs contributes to the clinical heterogeneity observed in patients with different types of mutations.
Figure 1. SHANK family gene structure, mutations, and protein domains.
The gene structures are deduced from cDNAs of AF163302 for SHANK1, AB208205 for SHANK2, and AB569469 for SHANK3 deposited in GenBank. The promoters are shown in arrows and the alternatively spliced exons are indicated in red. Microdeletions of SHANK1, SHANK2, and SHANK3 and point mutations of SHANK2 and SHANK3 are reported in ASD. Exon 11a of SHANK3 is a newly identified exon. The positions of six identified promoters are indicated as black arrows. The exons in red are alternatively spliced. The positions of point mutations are indicated as blue arrows and the nature of point mutations are as described above the arrow. c.601-1G>A splicing mutation in intron 5 (Hamdan et al., 2011), p.Q312R in exon 8 (Moessner et al., 2007), p.G440_P446del in exon 11 (Waga et al., 2011), p.R656H in exon 16 (Waga et al., 2011), c.2265+1delG splicing mutation in intron 19 (Gauthier et al., 2009), p.R1117X (Gauthier et al., 2010) and p.Ala1227fs in exon 21 (Durand et al., 2007). Protein domains are shown and aligned to corresponding exons (Pro, proline rich region).
The pathophysiology of SHANK causing ASD and Shank mutant mice
The progress to model human SHANK mutations in mutant mice has been impressive. Mutant mice for all Shank family genes have been produced. The Shank1 mutant mouse model was first produced well before the discovery of involvement of SHANK1 in ASD (Hung et al., 2008; Silverman et al., 2011; Wohr et al., 2011). The PDZ domain is disrupted in Shank1 mutant mice. The phenotypes at both the synaptic and behavioral levels in Shank1 mutant mice are unexpectedly mild. Shank1 mutant mice have altered PSD protein composition, reduced size of dendritic spines, smaller and thinner PSDs, weaker basal synaptic transmission, but normal synaptic plasticity. Behaviorally, Shank1 mutant mice have normal social interaction behavior, increased anxiety, reduced ultrasonic vocalizations, and impaired contextual fear memory. Unexpectedly, Shank1 mutant mice display enhanced performance in a spatial learning task. Two mutant models for Shank2 were reported recently (Schmeisser et al., 2012;Won et al., 2012). Schmeisser et al. created Shank2 exon 7 deletion mutant mice (Shank2 Δex7). In these animals long-term potentiation (LTP) in hippocampal CA1 synapses was increased but no change in long-term depression (LTD) was observed (Schmeisser et al., 2012). Impairment in social interaction, increased stereotypical behavior, hyperactivity, and altered ultrasonic vocalization patterns were found in Shank2 Δex7−/− mice. In contrast, Won et al. generated a slightly different Shank2 mutant mouse where exons 6–7 were deleted (Shank2 Δex6–7) (Won et al., 2012). Shank2 Δex6–7−/− mice also display abnormal synaptic function and ASD-like behaviors. Both exon 7 and exon 6–7 deletions resulted in frame shift mutations shortly after exon 7 which suggest that the two mutations should have very similar consequences for the Shank2 protein. There are similarities and differences between Shank2 Δex7 Shank2 Δex6–7 mice. Intriguingly, LTP in the hipppocampal CA1 region was reduced in Shank2 Δex6–7−/− mice, and this is opposite to Shank2 Δex7−/− mice. The behavioral profile of Shank2 Δex6–7−/− mice is similar to Shank2 Δex7−/−. The explanation for the apparent discrepancy in hippocampal LTP in two very similar Shank2 mutations is not immediately clear and further investigation is warranted.
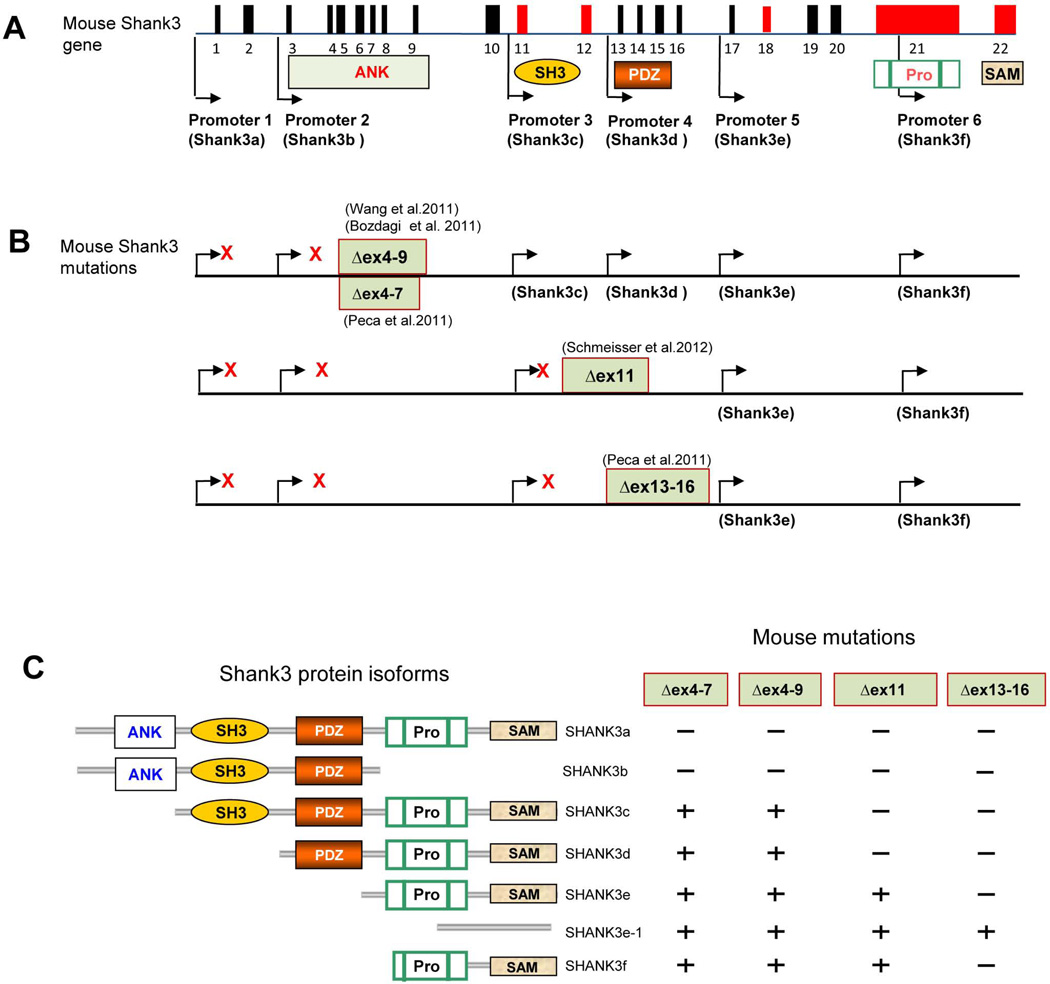
Recently, four different laboratories have developed five lines of mutant mice carrying different mutations in Shank3 (Figure 2) (Bozdagi et al., 2010; Peca et al., 2011; Schmeisser et al., 2012; Wang et al., 2011). The mutations include deletions of exons 4–9 by two groups with slightly different design of [(Δe4–9Buxbaum(B)) (Bozdagi et al., 2010) and Δe4–9Jiang (J) (Wang et al., 2011)], exons 4–7(Δe4–7)(Peca et al., 2011), deletion of exon 11(Schmeisser et al., 2012) and exons 13–16 (Δe13–16) (Peca et al., 2011). Because all of these deletions cause a frame shift for targeted transcripts, they all result in either a truncated Shank3 protein or possible disruption of full length RNA or protein isoforms due to the instability of shortened RNA or protein. Based on our knowledge of alternative Shank3 promoters and alternative splicing of coding exons, each of these mice is expected to disrupt different Shank3 isoforms but not completely disrupt the Shank3 gene (Wang et al., 2011).
Figure 2. Targeted Mutations in Shank3 Mice.
(A) Schematic of mouse gene structure deduced from cDNA AB230103 deposited in GenBank. The promoters are shown in arrows and the alternatively spliced exons are indicated in red. (B) Schematic of Shank3 mutant mice. The positions of targeted mutations in five different lines of Shank3 mutant mice are shown. The transcripts that are predicted or confirmed to be disrupted (red X) or intact in each mutant line of mice are indicated. Exon 21 is spliced out in known Shank3b and Shank3e-1 isoforms. Whether the exon 21 is spliced out in transcripts from other promoters has not been determined (Bangash et al., 2011; Bozdagi et al., 2010; Peca et al., 2011; Wang et al., 2011). (C) The predicted isoform-specific expression of Shank3 mRNA and protein in Shank3 mutant mice. The “−” indicates that the isoform is disrupted and “+” indicates the isoform remains intact. The full complement of Shank3 mRNA and protein isoforms that derive from combinations of alternative promoters and mRNA splicing remains unknown. Therefore, the pattern of isoform-specific expression and disruption by specific mutations is likely more complex than indicated.
Shank3 mutant mice have been extensively characterized by biochemical, ultrastructural, electrophysiological, and behavioral approaches. Notably, synapse structure phenotypes vary with specific Shank3 mutations, are different in different brain regions, and display developmental heterogeneity. This could be due to differential spatial and temporal expression of other Shank family members or due to compositional variation across different populations of glutamatergic synapses. Dendritic branching and spine areas were increased in medium spiny neurons (MSNs) in the striatum of Δex13–16−/− mice but PSD thickness and length were decreased at corticostriatal synapses in these mice (Peca et al., 2011). Similar findings, however, were not found in hippocampal CA1 synapses in Δex4–9J−/− (Wang et al., 2011) and Δex11−/− mice (Schmeisser et al., 2012). Spine length was increased in CA1 hippocampus of Δex4–9J−/− mice (Wang et al., 2011), and spine density was decreased in the striatum and CA1 hippocampus of Δex13–16−/− and Δex4–9J−/− mice, respectively. Activity-induced spine growth by theta burst stimulation in cultured brain slices was attenuated at CA1 synapses of Δex4–9B+/− mice (Bozdagi et al., 2010). Similarly, electrophysiological studies of these mutant mice have revealed variable findings. Measurements of miniature excitatory postsynaptic current (mEPSC) frequency and amplitude, paired pulse ratio, input/output (I/O) curves, fiber volley, and population spikes indicated that synaptic transmission was reduced at hippocampal CA1 synapses of Δex4–9B+/− mice (Bozdagi et al., 2010), but not in mice bearing Δe4–9J−/− (Wang et al., 2011), or Δe13–16−/− (Peca et al., 2011). The explanation for the difference between Δe4–9B+/− and Δe4–9J−/− is not immediately clear. One possibility is that these mutations induce different cryptic splicing as described in Δe4–9J−/− mice (Wang et al., 2011). Another possibility is that heterozygous mutations may produce a dominant gain-of-function phenotype which differs from the phenotype of homozygous deletions. In striatum, the frequency of mEPSCs and amplitude of population spikes were significantly decreased in Δex13–16−/− mice (Peca et al., 2011). Presynaptic responses as measured by paired pulse ratio and input/output curves were not altered at corticostriatal synapses in Δex13–16−/− or Δex4–7−/− mice (Peca et al., 2011). Hippocampal LTP was reduced at CA1 synapses of Δex4–9J−/− and Δex4–9B+/−. (Bozdagi et al., 2010; Wang et al., 2011). Alterations in mGluR-dependent LTD was not evident in the Δex4–9B+/− mice as induced by PP-LFS (Bozdagi et al., 2010). However, acute knockdown of Shank3 in cultured neurons decreases mGluR-dependent plasticity (Verpelli et al., 2011), suggesting differences in effects of Shank3 on mGluR1/5 signaling over development and pointing to the need for cautious interpretation regarding the pathogenic versus compensatory roles of synaptic phenotypes observed in Shank3 mutant mice.
Extensive behavioral analyses were performed in all Shank3 mutant mice on different genetic backgrounds using different protocols. The most notable and consistent observations are reduced social interaction and affiliation behaviors in all mutant mouse lines (Bozdagi et al., 2010; Peca et al., 2011; Wang et al., 2011). Repetitive behaviors measured by increased self-grooming in the home cage and behavioral inflexibility in the reverse Morris water maze and hole board test were observed in Δe4–9J−/− mice (Wang et al., 2011). Significantly increased self-grooming leading to skin lesions are also observed in Δex11−/− and Δex13–16−/− mice (Peca et al., 2011). The number, frequency, and duration of ultrasonic vocalizations were altered in a sex-specific manner in Δe4–9J−/− and Δe4–9B+/− mice (Bozdagi et al., 2010; Wang et al., 2011).
Deciphering the relationship between the phenotypic diversity and the molecular diversity of Shank3 mutations remains a significant challenge. It is tempting to speculate that the phenotypic diversity in Shank3 mutant mice reflects the clinical heterogeneity in SHANK3 mutations found in human ASD patients. Since each mutation has a different impact on Shank3 isoform expression, a simple hypothesis is that the diversity of phenotypes in Shank3 mutant mice reflects the molecular diversity of Shank3. However, analysis of heterozygotes and homozygotes, performing different measurements in different brain regions as well as in animals with different genetic backgrounds and ages could all contribute to the observed phenotypic heterogeneity. Further investigations comparing different lines of Shank3 mice side by side is necessary to resolve these discrepancies and provide validated data for future pre-clinical trials.
Therapeutic approaches for Shankopathies
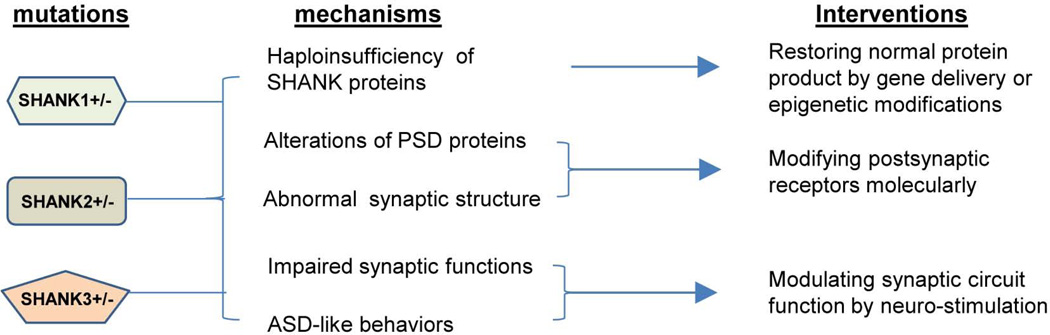
Further investigations in Shank mutant mice or in humans are warranted to fully understand the pathophysiology of Shankopathies. However, several exciting themes related to the development of treatments of SHANK causing ASD are emerging from the knowledge learned from human genetics and mutant mouse models (Figure 3). Two general questions are worth discussing prior to focusing on SHANK specific treatment strategies. The first question is the critical window of development underlying the pathophysiology of ASD in general. Although ASD are classified as neurodevelopmental disorders clinically and presumably have a developmental origin, the evidence in the human literature as to whether the pathophysiology of ASD is the result of mainly a developmental versus functional defect, or a combination of both is very limited (Bale et al., 2010; Rubenstein, 2010; Zoghbi, 2003). Little is known about the developmental window that is critical for the expression of core behavioral features of ASD (LeBlanc and Fagiolini, 2011). Clinically, in the typical course of ASD, particularly for the mild and moderate cases, there may be normal development during the first 12–16 months before significant signs and symptoms of ASD emerge (Lord and Bishop, 2009). One of the best characterized examples is Rett syndrome (Zoghbi, 2003, 2005). However, this traditional view is challenged by recent reports from neuroimaging studies using more sensitive and higher resolution techniques which suggest an early developmental defect may be present before clinically apparent symptoms emerge (Dawson et al., 2004; Wolff et al., 2012; Zwaigenbaum et al., 2009). In the case of SHANK3 causing ASD, most PMS patients display signs of neurological impairments such as hypotonia at birth or developmental delay during infancy (Phelan, 2007). No reports are in the literature on the neurological deficits or neuroimaging studies for the cases with SHANK3 only mutations. Only one neuroimaging study of 8 cases of 22q13.3 deletion including SHANK3 is reported (Philippe et al., 2008). The major findings are thin or morphologically atypical corpus callosum, localized dysfunction of the left temporal polar lobe, and amygdala hypoperfusion. Because other genes in addition to SHANK3 are also deleted in 22q13.3 deletion syndrome, it could not be determined whether the deficiency of SHANK3 or other genes, or both are responsible for these imaging finding as well as early neurological impairments. In all Shank mutant mice, no apparent structural or histological defects are reported (Bozdagi et al., 2010; Peca et al., 2011; Schmeisser et al., 2012; Wang et al., 2011; Won et al., 2012). These observations do not indicate the presence of an earlier developmental defect associated with the deficiency of Shanks but the possibility of a subtle or ultra-structural defect in specific cell lineages cannot be completely ruled out.
Figure 3. Therapeutic approach for Shankopathies.
The proposed therapeutic approaches for SHANK family gene causing ASD based on the molecular and circuit mechanism.
The second important question directly relevant to treatment is the reversibility of neurological impairment in postnatal human brains. There are now several examples in mouse models where neurological impairments can be reversed in postnatal brains (Daily et al., 2011; Guy et al., 2007; Han et al., 2012; Krueger and Bear, 2011). In mutant mice lacking Mecp2, genetic restoration of Mecp2 during late development reverses the neurological impairments including defective synaptic plasticity and abnormal behaviors in adult mice (Giacometti et al., 2007; Guy et al., 2007; Kerr et al., 2012). Similarly, in the Angelman syndrome (AS) mouse model, virus mediated delivery of the AS causing gene Ube3a resulted in rescued LTP and amelioration of abnormal behaviors in adult mice (Daily et al., 2011). In the case of fragile X syndrome, pharmacological treatments given postnatally ameliorated major neurological impairments by both mGluR5 and GABA receptor modulators both in humans and the mouse model (Berry-Kravis et al., 2012; Dolen et al., 2007; Henderson et al., 2012; Michalon et al., 2012). Similar examples are also reported in other ASD mouse models (Bhattacharya et al., 2012; Silverman et al., 2012). These examples, although still limited, are exciting because they support the concept of reversibility in postnatal brains in animal models. It remains untested and an ongoing subject of debate whether the same may be accomplished in humans for these or other CNS related disorders.
Drugs to enhance glutamatergic synaptic activity
Shank family proteins interact with both ionotropic (NMDA and AMPAR receptors) and metabotropic glutamate receptors (mGluRs) at the PSD through different protein interaction domains (Grabrucker et al., 2011; Gundelfinger et al., 2006; Kreienkamp, 2008). However, how the interactions between Shanks and different types of receptors are coordinated is poorly understood. Biochemical analysis of Shank mutant mice has led to a general conclusion that there is reduced synaptic function mediated by glutamate receptors (Bozdagi et al., 2010; Hung et al., 2010; Schmeisser et al., 2012; Wang et al., 2011; Won et al., 2012). This result then raises the interesting question as whether pharmacological approaches which enhance glutamate receptor activity may have therapeutic benefit to Shankopathies. This hypothesis has been tested in Shank2 mutant mice (Won et al., 2012). Treatment with a positive allosteric modulator of the metabotropic glutamate receptor 5 (mGluR5), which enhances NMDA receptor function via mGluR5 activation, normalizes NMDA receptor function and markedly enhances social interaction in Shank2 mutant mice (Won et al., 2012). These data support a basic premise to test different glutamate receptor agonists such as the NMDA agonist D-cyclosine or other mGluR positive allosteric modulators for mGluR1/5 in Shank mouse models (Smith and Ehlers, 2012). The anticipated challenges for using these receptor modulators are their specificity and selectivity. For instance, Shank proteins have different expression patterns in different brain regions or synapses (Bockers et al., 2004; Leblond et al., 2012). Enhancing glutamate receptor activity in neurons that do not express SHANKs may have detrimental effect due to disrupting the balance of circuits mediated by SHANK proteins.
Molecular restoration of SHANK proteins
Because haploinsufficiency of SHANKs is predicted to be the major molecular mechanism for SHANK causing ASD in the majority of cases caused by the deletions of entire SHANK genes, an interesting possibility for treatment is whether the transcription of SHANKs can be up-regulated from the non-mutated allele by a molecular approach. The isoform specific expression of SHANK3 is epigenetically regulated (Beri et al., 2007; Ching et al., 2005; Maunakea et al., 2010). SHANK3 has multiple CpG islands in the gene body which harbor multiple intragenic promoters (Wang et al., 2011). These CpG islands display tissue specific DNA methylation and other epigenetic marks both in humans and mice (Beri et al., 2007; Ching et al., 2005). The isoform-specific expression of SHANK3 could be modified by DNA methylation inhibitors and histone deacetylase (HDAC) inhibitors, as well as methylation promoters in cultured neurons (Beri et al., 2007; Maunakea et al., 2010). Whether these epigenetic modifications may have similar impact in vivo has not been investigated. A drug screen to discover compounds that can up regulate SHANK3 from the non-mutated allele would be an attractive direction for future investigation. It should be noted that the successful use of this approach has been reported in Angelman syndrome using a Ube3a reporter fusion protein approach in mice (Huang et al., 2012). Similarly, multiple CpG islands and brain tissue specific methylation are also found in SHANK1 and SHANK2 and an epigenetic mechanism may also be involved in regulating the expression of these genes.
Conceptually, genetic restoration of SHANK proteins by gene delivery is an ideal approach in individuals with SHANK mutations. The rationale for this approach is straight forward as in gene therapies proposed in other genetic disorders. The challenge is that SHANK proteins are structural proteins in synapses. The delivery of exogenous SHANK3 protein inside of cells to reach the proper sub-cellular targets in brains is the major obstacle for this experimental design. A decade ago, gene delivery to the brain was limited to stereotaxic injection of viral vectors into the brains of laboratory animals (Mueller et al., 2012). More recently, advancements in vector design and the exploration of alternative routes of administration have made efficient global central nervous system (CNS) gene delivery a possibility despite other remaining significant challenges (Guggenhuber et al., 2010). The most popular CNS gene delivery vector is adeno-associated virus (AAV) (Mueller et al., 2012). Lentivirus-based vectors also play an increasingly significant role in CNS-directed gene therapy because they have the advantage of a larger packaging capacity than AAV (Manfredsson and Mandel, 2011). Multiple groups have now reported in detail the ability of AAV9 vectors to cross the blood brain barrier (BBB) and transduce neurons and astrocytes following intravenous injection in neonatal and adult mice, cats, and nonhuman primates (Duque et al., 2009; Foust et al., 2009). Strategies employing intravenous delivery of AAV9 vectors have successfully treated spinal muscular atrophy (Foust et al., 2010) and mucopolysaccharidosis (MPS IIIB) in mice (Fu et al., 2011). Using a similar design, several studies have treated lysosomal storage diseases in animal models (Li et al., 2012; Sun et al., 2008). Proof of principle studies have also been reported for Rett syndrome and Angelman syndrome, two typical neurological disorders that share a number of similarity with SHANK causing ASD (Daily et al., 2011; Gadalla et al., 2012). The major variables influencing the feasibility of any gene therapy approach include 1) whether a secreted factor can be utilized or if the therapeutic gene product is limited to cell autonomous effects, 2) what the range of effective and tolerated gene expression is, and 3) whether the expressed products can pass BBB and what type of delivery efficiency is required for a meaningful therapeutic effect.
Neuromodulation by transcranial magnetic stimulation as a treatment modality
Repetitive transcranial magnetic stimulation (rTMS) is a technique for non-invasive stimulation of brain via generation of a pulse of high intensity magnetic field by passing a brief electric current through an inductive coil (Pell et al., 2011; Peterchev et al., 2011). Recently, substantial interest in the use of this technique for the treatment of neuropsychiatric disorders such schizophrenia and depression has emerged (Husain and Lisanby, 2011; Rossi et al., 2009; Rossini and Rossi, 2007; Stanford et al., 2011; Wassermann and Lisanby, 2001). One notable development in this regard is that rTMS recently joined electroconvulsive therapy (ECT) as one of FDA approved neuromodulation techniques for treating major depression (Husain and Lisanby, 2011). In terms of therapeutic potential, rTMS paradigms have been shown to reactivate hypoactive structures, inhibit overactive structures, and induce long-term potentiation (LTP)-like effects in human brain and in a few animal studies (Houdayer et al., 2008; Huang et al., 2005). Low frequency (1Hz) stimulation has been shown to suppress excitatory synaptic transmission while high frequency (5–50Hz) or the intermittent form of theta burst stimulation (TBS) may potentiate it (Aydin-Abidin et al., 2008; Pell et al., 2011). The cellular mechanisms underlying these rTMS effects are poorly understood (Funke and Benali, 2011). A factor contributing to this lack of understanding is the considerable variability in the experimental designs employed in prior research (Pell et al., 2011). Nonetheless, several interesting observations in the literature suggest that rTMS may have therapeutic potential by modulating synaptic plasticity in Shankopathies. Chronic treatment with high frequency rTMS in awake animals significantly increases the expression of the AMPAR Glu1A subunit, a key component for synaptic plasticity in hippocampus (Gersner et al., 2011; Newpher and Ehlers, 2009), and also enhances LTP (Ahmed and Wieraszko, 2006; Esser et al., 2006; Hoogendam et al., 2010; Kim et al., 2006). This suggests that the cellular mechanism underlying rTMS may be mediated by modulating the expression synaptic genes. These observations and the consistent finding of impaired synaptic plasticity in ASD mouse models including impaired hippocampus LTP in Shank mouse models (Bozdagi et al., 2010; Shepherd and Katz, 2011; Wang et al., 2011) support that SHANK3 causing ASD would be a good target for investigation into rTMS treatment. In this regard, an advantage of rTMS is that it is non-invasive and relatively limited in risks so that it may be applied to humans immediately without pre-clinical trials. Mouse models of SHANK related ASD then offer an opportunity to dissect the mechanism underlying rTMS treatment.
Future directions
Studies of Shankopathies in humans and mouse models have provided a framework for future investigations of treatment and intervention of ASD. Numerous questions have also emerged from the analysis of SHANK defects in human ASD patients and Shank mutant mice. In humans, natural history studies of genotype and phenotype in patients with various SHANK mutations are critical. A detailed description and comparison of clinical features in patients with different SHANK mutations will provide guidance for modeling human disease in animal models. There is a critical need to directly compare the different Shank mutant mice head to head for cellular, synaptic, circuit, and behavioral phenotypes. Such direct comparisons will allow for more definitive identification of common synaptic defects, circuit endophenotypes, and behaviors. Can mutations in Shanks open the door to a molecular pathway that provides novel therapeutic targets? Much remains to be learned, but it is tempting to consider Shank3 “restoration” in a loose sense as a therapeutic strategy for Phelan-McDermid syndrome, and perhaps more broadly in ASD. Ultimately, the value of Shank mutant mice will depend critically on the ability to use human patients to validate their predictive utility.
Acknowledgement
XMW is supported by Phelan-McDermid syndrome foundation postdoctoral fellowship. The research in YHJ laboratory is supported by Autism Speaks grant and National Institutes of Health grant (MH098114). YHJ is also a scholar of National Institutes of Health grant (5K12-HD0043494-08).
References
- Ahmed Z, Wieraszko A. Modulation of learning and hippocampal, neuronal plasticity by repetitive transcranial magnetic stimulation (rTMS) Bioelectromagnetics. 2006;27:288–294. doi: 10.1002/bem.20211. [DOI] [PubMed] [Google Scholar]
- Aydin-Abidin S, Trippe J, Funke K, Eysel UT, Benali A. High- and low-frequency repetitive transcranial magnetic stimulation differentially activates c-Fos and zif268 protein expression in the rat brain. Exp Brain Res. 2008;188:249–261. doi: 10.1007/s00221-008-1356-2. [DOI] [PubMed] [Google Scholar]
- Bale TL, Baram TZ, Brown AS, Goldstein JM, Insel TR, McCarthy MM, Nemeroff CB, Reyes TM, Simerly RB, Susser ES, et al. Early life programming and neurodevelopmental disorders. Biol Psychiatry. 2010;68:314–319. doi: 10.1016/j.biopsych.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangash MA, Park JM, Melnikova T, Wang D, Jeon SK, Lee D, Syeda S, Kim J, Kouser M, Schwartz J, et al. Enhanced Polyubiquitination of Shank3 and NMDA Receptor in a Mouse Model of Autism. Cell. 2011 doi: 10.1016/j.cell.2011.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Beri S, Tonna N, Menozzi G, Bonaglia MC, Sala C, Giorda R. DNA methylation regulates tissue-specific expression of Shank3. J Neurochem. 2007;101:1380–1391. doi: 10.1111/j.1471-4159.2007.04539.x. [DOI] [PubMed] [Google Scholar]
- Berkel S, Marshall CR, Weiss B, Howe J, Roeth R, Moog U, Endris V, Roberts W, Szatmari P, Pinto D, et al. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat Genet. 2010;42:489–491. doi: 10.1038/ng.589. [DOI] [PubMed] [Google Scholar]
- Berkel S, Tang W, Trevino M, Vogt M, Obenhaus HA, Gass P, Scherer SW, Sprengel R, Schratt G, Rappold GA. Inherited and de novo SHANK2 variants associated with autism spectrum disorder impair neuronal morphogenesis and physiology. Hum Mol Genet. 2012;21:344–357. doi: 10.1093/hmg/ddr470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry-Kravis EM, Hessl D, Rathmell B, Zarevics P, Cherubini M, Walton-Bowen K, Mu Y, Nguyen DV, Gonzalez-Heydrich J, Wang PP, et al. Effects of STX209 (Arbaclofen) on Neurobehavioral Function in Children and Adults with Fragile X Syndrome: A Randomized, Controlled, Phase 2 Trial. Sci Transl Med. 2012;4:152ra127. doi: 10.1126/scitranslmed.3004214. [DOI] [PubMed] [Google Scholar]
- Betancur C. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res. 2011;1380:42–77. doi: 10.1016/j.brainres.2010.11.078. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Kaphzan H, Alvarez-Dieppa AC, Murphy JP, Pierre P, Klann E. Genetic Removal of p70 S6 Kinase 1 Corrects Molecular, Synaptic, and Behavioral Phenotypes in Fragile X Syndrome Mice. Neuron. 2012;76:325–337. doi: 10.1016/j.neuron.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blenner S, Reddy A, Augustyn M. Diagnosis and management of autism in childhood. BMJ. 2011;343:d6238. doi: 10.1136/bmj.d6238. [DOI] [PubMed] [Google Scholar]
- Bockers TM, Segger-Junius M, Iglauer P, Bockmann J, Gundelfinger ED, Kreutz MR, Richter D, Kindler S, Kreienkamp HJ. Differential expression and dendritic transcript localization of Shank family members: identification of a dendritic targeting element in the 3' untranslated region of Shank1 mRNA. Mol Cell Neurosci. 2004;26:182–190. doi: 10.1016/j.mcn.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, Beri S, De Agostini C, Novara F, Fichera M, Grillo L, Galesi O, Vetro A, Ciccone R, et al. Molecular Mechanisms Generating and Stabilizing Terminal 22q13 Deletions in 44 Subjects with Phelan/McDermid Syndrome. PLoS Genet. 2011;7:e1002173. doi: 10.1371/journal.pgen.1002173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, Mani E, Aceti G, Anderlid BM, Baroncini A, Pramparo T, Zuffardi O. Identification of a recurrent breakpoint within the SHANK3 gene in the 22q13.3 deletion syndrome. J Med Genet. 2005 doi: 10.1136/jmg.2005.038604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdagi O, Sakurai T, Papapetrou D, Wang X, Dickstein DL, Takahashi N, Kajiwara Y, Yang M, Katz AM, Scattoni ML, et al. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism. 2010;1:15. doi: 10.1186/2040-2392-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco M, Volkmar FR, Bloch MH. Pharmacologic treatment of repetitive behaviors in autism spectrum disorders: evidence of publication bias. Pediatrics. 2012;129:e1301–e1310. doi: 10.1542/peds.2011-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching TT, Maunakea AK, Jun P, Hong C, Zardo G, Pinkel D, Albertson DG, Fridlyand J, Mao JH, Shchors K, et al. Epigenome analyses using BAC microarrays identify evolutionary conservation of tissue-specific methylation of SHANK3. Nat Genet. 2005;37:645–651. doi: 10.1038/ng1563. [DOI] [PubMed] [Google Scholar]
- Daily JL, Nash K, Jinwal U, Golde T, Rogers J, Peters MM, Burdine RD, Dickey C, Banko JL, Weeber EJ. Adeno-associated virus-mediated rescue of the cognitive defects in a mouse model for Angelman syndrome. PLoS One. 2011;6:e27221. doi: 10.1371/journal.pone.0027221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson G, Rogers S, Munson J, Smith M, Winter J, Greenson J, Donaldson A, Varley J. Randomized, controlled trial of an intervention for toddlers with autism: the Early Start Denver Model. Pediatrics. 2010;125:e17–e23. doi: 10.1542/peds.2009-0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson G, Webb SJ, Carver L, Panagiotides H, McPartland J. Young children with autism show atypical brain responses to fearful versus neutral facial expressions of emotion. Dev Sci. 2004;7:340–359. doi: 10.1111/j.1467-7687.2004.00352.x. [DOI] [PubMed] [Google Scholar]
- Devlin B, Scherer SW. Genetic architecture in autism spectrum disorder. Curr Opin Genet Dev. 2012;22:229–237. doi: 10.1016/j.gde.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Dolen G, Carpenter RL, Ocain TD, Bear MF. Mechanism-based approaches to treating fragile X. Pharmacol Ther. 2010;127:78–93. doi: 10.1016/j.pharmthera.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Dolen G, Osterweil E, Rao BS, Smith GB, Auerbach BD, Chattarji S, Bear MF. Correction of fragile X syndrome in mice. Neuron. 2007;56:955–962. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsater H, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Perroy J, Loll F, Perrais D, Fagni L, Bourgeron T, Montcouquiol M, Sans N. SHANK3 mutations identified in autism lead to modification of dendritic spine morphology via an actin-dependent mechanism. Mol Psychiatry. 2011 doi: 10.1038/mp.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser SK, Huber R, Massimini M, Peterson MJ, Ferrarelli F, Tononi G. A direct demonstration of cortical LTP in humans: a combined TMS/EEG study. Brain Res Bull. 2006;69:86–94. doi: 10.1016/j.brainresbull.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Funke K, Benali A. Modulation of cortical inhibition by rTMS - findings obtained from animal models. J Physiol. 2011;589:4423–4435. doi: 10.1113/jphysiol.2011.206573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadalla KK, Bailey ME, Spike RC, Ross PD, Woodard KT, Kalburgi SN, Bachaboina L, Deng JV, West AE, Samulski RJ, et al. Improved Survival and Reduced Phenotypic Severity Following AAV9/MECP2 Gene Transfer to Neonatal and Juvenile Male Mecp2 Knockout Mice. Mol Ther. 2012 doi: 10.1038/mt.2012.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier J, Champagne N, Lafreniere RG, Xiong L, Spiegelman D, Brustein E, Lapointe M, Peng H, Cote M, Noreau A, et al. De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proc Natl Acad Sci U S A. 2010;107:7863–7868. doi: 10.1073/pnas.0906232107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier J, Spiegelman D, Piton A, Lafreniere RG, Laurent S, St-Onge J, Lapointe L, Hamdan FF, Cossette P, Mottron L, et al. Novel de novo SHANK3 mutation in autistic patients. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:421–424. doi: 10.1002/ajmg.b.30822. [DOI] [PubMed] [Google Scholar]
- Georgiades S, Szatmari P, Boyle M, Hanna S, Duku E, Zwaigenbaum L, Bryson S, Fombonne E, Volden J, Mirenda P, et al. Investigating phenotypic heterogeneity in children with autism spectrum disorder: a factor mixture modeling approach. J Child Psychol Psychiatry. 2012 doi: 10.1111/j.1469-7610.2012.02588.x. [DOI] [PubMed] [Google Scholar]
- Gersner R, Kravetz E, Feil J, Pell G, Zangen A. Long-term effects of repetitive transcranial magnetic stimulation on markers for neuroplasticity: differential outcomes in anesthetized and awake animals. J Neurosci. 2011;31:7521–7526. doi: 10.1523/JNEUROSCI.6751-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH. Autism: many genes, common pathways? Cell. 2008;135:391–395. doi: 10.1016/j.cell.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Levitt P. Autism spectrum disorders: developmental disconnection syndromes. Curr Opin Neurobiol. 2007;17:103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Giacometti E, Luikenhuis S, Beard C, Jaenisch R. Partial rescue of MeCP2 deficiency by postnatal activation of MeCP2. Proc Natl Acad Sci U S A. 2007;104:1931–1936. doi: 10.1073/pnas.0610593104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabrucker AM, Schmeisser MJ, Schoen M, Boeckers TM. Postsynaptic ProSAP/Shank scaffolds in the cross-hair of synaptopathies. Trends Cell Biol. 2011;21:594–603. doi: 10.1016/j.tcb.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Grindle CF, Hastings RP, Saville M, Hughes JC, Huxley K, Kovshoff H, Griffith GM, Walker-Jones E, Devonshire K, Remington B. Outcomes of a behavioral education model for children with autism in a mainstream school setting. Behav Modif. 2012;36:298–319. doi: 10.1177/0145445512441199. [DOI] [PubMed] [Google Scholar]
- Guggenhuber S, Monory K, Lutz B, Klugmann M. AAV vector-mediated overexpression of CB1 cannabinoid receptor in pyramidal neurons of the hippocampus protects against seizure-induced excitoxicity. PLoS One. 2010;5:e15707. doi: 10.1371/journal.pone.0015707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundelfinger ED, Boeckers TM, Baron MK, Bowie JU. A role for zinc in postsynaptic density asSAMbly and plasticity? Trends Biochem Sci. 2006;31:366–373. doi: 10.1016/j.tibs.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FF, Gauthier J, Araki Y, Lin DT, Yoshizawa Y, Higashi K, Park AR, Spiegelman D, Dobrzeniecka S, Piton A, et al. Excess of de novo deleterious mutations in genes associated with glutamatergic systems in nonsyndromic intellectual disability. Am J Hum Genet. 2011;88:306–316. doi: 10.1016/j.ajhg.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Tai C, Westenbroek RE, Yu FH, Cheah CS, Potter GB, Rubenstein JL, Scheuer T, de la Iglesia HO, Catterall WA. Autistic-like behaviour in Scn1a+/− mice and rescue by enhanced GABA-mediated neurotransmission. Nature. 2012;489:385–390. doi: 10.1038/nature11356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward D, Eikeseth S, Gale C, Morgan S. Assessing progress during treatment for young children with autism receiving intensive behavioural interventions. Autism. 2009;13:613–633. doi: 10.1177/1362361309340029. [DOI] [PubMed] [Google Scholar]
- Henderson C, Wijetunge L, Kinoshita MN, Shumway M, Hammond RS, Postma FR, Brynczka C, Rush R, Thomas A, Paylor R, et al. Reversal of Disease-Related Pathologies in the Fragile X Mouse Model by Selective Activation of GABAB Receptors with Arbaclofen. Sci Transl Med. 2012;4:152ra128. doi: 10.1126/scitranslmed.3004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogendam JM, Ramakers GM, Di Lazzaro V. Physiology of repetitive transcranial magnetic stimulation of the human brain. Brain Stimul. 2010;3:95–118. doi: 10.1016/j.brs.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Houdayer E, Degardin A, Cassim F, Bocquillon P, Derambure P, Devanne H. The effects of low- and high-frequency repetitive TMS on the input/output properties of the human corticospinal pathway. Exp Brain Res. 2008;187:207–217. doi: 10.1007/s00221-008-1294-z. [DOI] [PubMed] [Google Scholar]
- Huang HS, Allen JA, Mabb AM, King IF, Miriyala J, Taylor-Blake B, Sciaky N, Dutton JW, Jr, Lee HM, Chen X, et al. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature. 2012;481:185–189. doi: 10.1038/nature10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YZ, Edwards MJ, Rounis E, Bhatia KP, Rothwell JC. Theta burst stimulation of the human motor cortex. Neuron. 2005;45:201–206. doi: 10.1016/j.neuron.2004.12.033. [DOI] [PubMed] [Google Scholar]
- Huffman LC, Sutcliffe TL, Tanner IS, Feldman HM. Management of symptoms in children with autism spectrum disorders: a comprehensive review of pharmacologic and complementary-alternative medicine treatments. J Dev Behav Pediatr. 2011;32:56–68. doi: 10.1097/DBP.0b013e3182040acf. [DOI] [PubMed] [Google Scholar]
- Hung AY, Futai K, Sala C, Valtschanoff JG, Ryu J, Woodworth MA, Kidd FL, Sung CC, Miyakawa T, Bear MF, et al. Smaller dendritic spines, weaker synaptic transmission, but enhanced spatial learning in mice lacking Shank1. J Neurosci. 2008;28:1697–1708. doi: 10.1523/JNEUROSCI.3032-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung AY, Sung CC, Brito IL, Sheng M. Degradation of postsynaptic scaffold GKAP and regulation of dendritic spine morphology by the TRIM3 ubiquitin ligase in rat hippocampal neurons. PLoS One. 2010;5:e9842. doi: 10.1371/journal.pone.0009842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain MM, Lisanby SH. Repetitive transcranial magnetic stimulation (rTMS): a noninvasive neuromodulation probe and intervention. J ECT. 2011;27:2. doi: 10.1097/YCT.0b013e31820c6298. [DOI] [PubMed] [Google Scholar]
- Kasari C, Lawton K. New directions in behavioral treatment of autism spectrum disorders. Curr Opin Neurol. 2010;23:137–143. doi: 10.1097/WCO.0b013e32833775cd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher RJ, 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Kerr B, Soto CJ, Saez M, Abrams A, Walz K, Young JI. Transgenic complementation of MeCP2 deficiency: phenotypic rescue of Mecp2-null mice by isoform-specific transgenes. Eur J Hum Genet. 2012;20:69–76. doi: 10.1038/ejhg.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Kim WR, Chi SE, Lee KH, Park EH, Chae JH, Park SK, Kim HT, Choi JS. Repetitive transcranial magnetic stimulation protects hippocampal plasticity in an animal model of depression. Neurosci Lett. 2006;405:79–83. doi: 10.1016/j.neulet.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Kreienkamp HJ. Scaffolding proteins at the postsynaptic density: shank as the architectural framework. Handb Exp Pharmacol. 2008:365–380. doi: 10.1007/978-3-540-72843-6_15. [DOI] [PubMed] [Google Scholar]
- Krueger DD, Bear MF. Toward fulfilling the promise of molecular medicine in fragile X syndrome. Annu Rev Med. 2011;62:411–429. doi: 10.1146/annurev-med-061109-134644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc JJ, Fagiolini M. Autism: a "critical period" disorder? Neural Plast. 2011;2011:921680. doi: 10.1155/2011/921680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblond CS, Heinrich J, Delorme R, Proepper C, Betancur C, Huguet G, Konyukh M, Chaste P, Ey E, Rastam M, et al. Genetic and Functional Analyses of SHANK2 Mutations Suggest a Multiple Hit Model of Autism Spectrum Disorders. PLoS Genet. 2012;8:e1002521. doi: 10.1371/journal.pgen.1002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Sun B, Nilsson MI, Bird A, Tarnopolsky MA, Thurberg BL, Bali D, Koeberl DD. Adjunctive beta2-agonists reverse neuromuscular involvement in murine Pompe disease. FASEB J. 2012 doi: 10.1096/fj.12-207472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Naisbitt S, Yoon J, Hwang JI, Suh PG, Sheng M, Kim E. Characterization of the Shank family of synaptic proteins. Multiple genes, alternative splicing, and differential expression in brain and development. J Biol Chem. 1999;274:29510–29518. doi: 10.1074/jbc.274.41.29510. [DOI] [PubMed] [Google Scholar]
- Lord C, Bishop SL. The autism spectrum: definitions, assessment and diagnoses. Br J Hosp Med (Lond) 2009;70:132–135. doi: 10.12968/hmed.2009.70.3.40552. [DOI] [PubMed] [Google Scholar]
- Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredsson FP, Mandel RJ. The development of flexible lentiviral vectors for gene transfer in the CNS. Exp Neurol. 2011;229:201–206. doi: 10.1016/j.expneurol.2011.03.013. [DOI] [PubMed] [Google Scholar]
- Marcus RN, Owen R, Manos G, Mankoski R, Kamen L, McQuade RD, Carson WH, Corey-Lisle PK, Aman MG. Aripiprazole in the treatment of irritability in pediatric patients (aged 6–17 years) with autistic disorder: results from a 52-week, open-label study. J Child Adolesc Psychopharmacol. 2011a;21:229–236. doi: 10.1089/cap.2009.0121. [DOI] [PubMed] [Google Scholar]
- Marcus RN, Owen R, Manos G, Mankoski R, Kamen L, McQuade RD, Carson WH, Findling RL. Safety and tolerability of aripiprazole for irritability in pediatric patients with autistic disorder: a 52-week, open-label, multicenter study. J Clin Psychiatry. 2011b;72:1270–1276. doi: 10.4088/JCP.09m05933. [DOI] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken JT, McGough J, Shah B, Cronin P, Hong D, Aman MG, Arnold LE, Lindsay R, Nash P, Hollway J, et al. Risperidone in children with autism and serious behavioral problems. N Engl J Med. 2002;347:314–321. doi: 10.1056/NEJMoa013171. [DOI] [PubMed] [Google Scholar]
- McPheeters ML, Warren Z, Sathe N, Bruzek JL, Krishnaswami S, Jerome RN, Veenstra-Vanderweele J. A systematic review of medical treatments for children with autism spectrum disorders. Pediatrics. 2011;127:e1312–e1321. doi: 10.1542/peds.2011-0427. [DOI] [PubMed] [Google Scholar]
- Michalon A, Sidorov M, Ballard TM, Ozmen L, Spooren W, Wettstein JG, Jaeschke G, Bear MF, Lindemann L. Chronic pharmacological mGlu5 inhibition corrects fragile X in adult mice. Neuron. 2012;74:49–56. doi: 10.1016/j.neuron.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moessner R, Marshall CR, Sutcliffe JS, Skaug J, Pinto D, Vincent J, Zwaigenbaum L, Fernandez B, Roberts W, Szatmari P, et al. Contribution of SHANK3 Mutations to Autism Spectrum Disorder. Am J Hum Genet. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller C, Ratner D, Zhong L, Esteves-Sena M, Gao G. Production and discovery of novel recombinant adeno-associated viral vectors. Curr Protoc Microbiol. 2012;Chapter 14(Unit14D):11. doi: 10.1002/9780471729259.mc14d01s26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers SM. The status of pharmacotherapy for autism spectrum disorders. Expert Opin Pharmacother. 2007;8:1579–1603. doi: 10.1517/14656566.8.11.1579. [DOI] [PubMed] [Google Scholar]
- Myers SM, Johnson CP. Management of children with autism spectrum disorders. Pediatrics. 2007;120:1162–1182. doi: 10.1542/peds.2007-2362. [DOI] [PubMed] [Google Scholar]
- Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, Weinberg RJ, Worley PF, Sheng M. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–582. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- Newpher TM, Ehlers MD. Spine microdomains for postsynaptic signaling and plasticity. Trends Cell Biol. 2009;19:218–227. doi: 10.1016/j.tcb.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Panagiotopoulos C, Ronsley R, Elbe D, Davidson J, Smith DH. First do no harm: promoting an evidence-based approach to atypical antipsychotic use in children and adolescents. J Can Acad Child Adolesc Psychiatry. 2010;19:124–137. [PMC free article] [PubMed] [Google Scholar]
- Peca J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, Lascola CD, Fu Z, Feng G. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011 doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pell GS, Roth Y, Zangen A. Modulation of cortical excitability induced by repetitive transcranial magnetic stimulation: influence of timing and geometrical parameters and underlying mechanisms. Prog Neurobiol. 2011;93:59–98. doi: 10.1016/j.pneurobio.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Peterchev AV, Murphy DL, Lisanby SH. Repetitive transcranial magnetic stimulator with controllable pulse parameters. J Neural Eng. 2011;8 doi: 10.1088/1741-2560/8/3/036016. 036016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan K. In: 22q13.3 Deletion Syndrome. Genetest E-i-c, Pagon Roberta A, Bird Thomas C, Dolan Cynthia R, Stephens Karen., editors. Seattle: Internet; 2007. [Google Scholar]
- Philippe A, Boddaert N, Vaivre-Douret L, Robel L, Danon-Boileau L, Malan V, de Blois MC, Heron D, Colleaux L, Golse B, et al. Neurobehavioral profile and brain imaging study of the 22q13.3 deletion syndrome in childhood. Pediatrics. 2008;122:e376–e382. doi: 10.1542/peds.2007-2584. [DOI] [PubMed] [Google Scholar]
- Rossi S, Hallett M, Rossini PM, Pascual-Leone A. Safety, ethical considerations, and application guidelines for the use of transcranial magnetic stimulation in clinical practice and research. Clin Neurophysiol. 2009;120:2008–2039. doi: 10.1016/j.clinph.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossini PM, Rossi S. Transcranial magnetic stimulation: diagnostic, therapeutic, and research potential. Neurology. 2007;68:484–488. doi: 10.1212/01.wnl.0000250268.13789.b2. [DOI] [PubMed] [Google Scholar]
- Roussignol G, Ango F, Romorini S, Tu JC, Sala C, Worley PF, Bockaert J, Fagni L. Shank expression is sufficient to induce functional dendritic spine synapses in aspiny neurons. J Neurosci. 2005;25:3560–3570. doi: 10.1523/JNEUROSCI.4354-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JL. Three hypotheses for developmental defects that may underlie some forms of autism spectrum disorder. Curr Opin Neurol. 2010;23:118–123. doi: 10.1097/WCO.0b013e328336eb13. [DOI] [PubMed] [Google Scholar]
- Sala C, Piech V, Wilson NR, Passafaro M, Liu G, Sheng M. Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron. 2001;31:115–130. doi: 10.1016/s0896-6273(01)00339-7. [DOI] [PubMed] [Google Scholar]
- Sato D, Lionel AC, Leblond CS, Prasad A, Pinto D, Walker S, O'Connor I, Russell C, Drmic IE, Hamdan FF, et al. SHANK1 Deletions in Males with Autism Spectrum Disorder. Am J Hum Genet. 2012;90:879–887. doi: 10.1016/j.ajhg.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser MJ, Ey E, Wegener S, Bockmann J, Stempel AV, Kuebler A, Janssen AL, Udvardi PT, Shiban E, Spilker C, et al. Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature. 2012;486:256–260. doi: 10.1038/nature11015. [DOI] [PubMed] [Google Scholar]
- Sheng M, Kim E. The Shank family of scaffold proteins. J Cell Sci. 2000;113(Pt 11):1851–1856. doi: 10.1242/jcs.113.11.1851. [DOI] [PubMed] [Google Scholar]
- Shepherd GM, Katz DM. Synaptic microcircuit dysfunction in genetic models of neurodevelopmental disorders: focus on Mecp2 and Met. Curr Opin Neurobiol. 2011 doi: 10.1016/j.conb.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Smith DG, Rizzo SJ, Karras MN, Turner SM, Tolu SS, Bryce DK, Smith DL, Fonseca K, Ring RH, et al. Negative allosteric modulation of the mGluR5 receptor reduces repetitive behaviors and rescues social deficits in mouse models of autism. Sci Transl Med. 2012;4:131ra151. doi: 10.1126/scitranslmed.3003501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Turner SM, Barkan CL, Tolu SS, Saxena R, Hung AY, Sheng M, Crawley JN. Sociability and motor functions in Shank1 mutant mice. Brain Res. 2011;1380:120–137. doi: 10.1016/j.brainres.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DG, Ehlers MD. Mining and modeling human genetics for autism therapeutics. Curr Opin Neurobiol. 2012 doi: 10.1016/j.conb.2012.03.004. [DOI] [PubMed] [Google Scholar]
- Stanford AD, Corcoran C, Bulow P, Bellovin-Weiss S, Malaspina D, Lisanby SH. High-frequency prefrontal repetitive transcranial magnetic stimulation for the negative symptoms of schizophrenia: a case series. J ECT. 2011;27:11–17. doi: 10.1097/YCT.0b013e3181f41ea3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- State MW. The genetics of child psychiatric disorders: focus on autism and Tourette syndrome. Neuron. 2010;68:254–269. doi: 10.1016/j.neuron.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Young SP, Li P, Di C, Brown T, Salva MZ, Li S, Bird A, Yan Z, Auten R, et al. Correction of multiple striated muscles in murine Pompe disease through adeno-associated virus-mediated gene therapy. Mol Ther. 2008;16:1366–1371. doi: 10.1038/mt.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JL, McPheeters ML, Sathe NA, Dove D, Veenstra-Vanderweele J, Warren Z. A systematic review of vocational interventions for young adults with autism spectrum disorders. Pediatrics. 2012;130:531–538. doi: 10.1542/peds.2012-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro R, Konyukh M, Delorme R, Leblond C, Chaste P, Fauchereau F, Coleman M, Leboyer M, Gillberg C, Bourgeron T. Key role for gene dosage and synaptic homeostasis in autism spectrum disorders. Trends Genet. 2010;26:363–372. doi: 10.1016/j.tig.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Verpelli C, Dvoretskova E, Vicidomini C, Rossi F, Chiappalone M, Schoen M, Di Stefano B, Mantegazza R, Broccoli V, Boeckers TM, et al. Importance of shank3 in regulating metabotropic glutamate receptor 5 (mGluR5) expression and signaling at synapses. J Biol Chem. 2011 doi: 10.1074/jbc.M111.258384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vismara LA, Rogers SJ. Behavioral treatments in autism spectrum disorder: what do we know? Annu Rev Clin Psychol. 2010;6:447–468. doi: 10.1146/annurev.clinpsy.121208.131151. [DOI] [PubMed] [Google Scholar]
- Volkmar FR. Pharmacological interventions in autism: theoretical and practical issues. J Clin Child Psychol. 2001;30:80–87. doi: 10.1207/S15374424JCCP3001_9. [DOI] [PubMed] [Google Scholar]
- Volkmar FR, State M, Klin A. Autism and autism spectrum disorders: diagnostic issues for the coming decade. J Child Psychol Psychiatry. 2009;50:108–115. doi: 10.1111/j.1469-7610.2008.02010.x. [DOI] [PubMed] [Google Scholar]
- Waga C, Okamoto N, Ondo Y, Fukumura-Kato R, Goto Y, Kohsaka S, Uchino S. Novel variants of the SHANK3 gene in Japanese autistic patients with severe delayed speech development. Psychiatr Genet. 2011;21:208–211. doi: 10.1097/YPG.0b013e328341e069. [DOI] [PubMed] [Google Scholar]
- Wang X, McCoy P, Rodriguiz RM, Pan Y, Je HS, Roberts A, Kim C, Berrios J, Colvin JS, Bousquet-Moore D, et al. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren Z, McPheeters ML, Sathe N, Foss-Feig JH, Glasser A, Veenstra-Vanderweele J. A systematic review of early intensive intervention for autism spectrum disorders. Pediatrics. 2011;127:e1303–e1311. doi: 10.1542/peds.2011-0426. [DOI] [PubMed] [Google Scholar]
- Wassermann EM, Lisanby SH. Therapeutic application of repetitive transcranial magnetic stimulation: a review. Clin Neurophysiol. 2001;112:1367–1377. doi: 10.1016/s1388-2457(01)00585-5. [DOI] [PubMed] [Google Scholar]
- Wilson HL, Wong AC, Shaw SR, Tse WY, Stapleton GA, Phelan MC, Hu S, Marshall J, McDermid HE. Molecular characterisation of the 22q13 deletion syndrome supports the role of haploinsufficiency of SHANK3/PROSAP2 in the major neurological symptoms. J Med Genet. 2003;40:575–584. doi: 10.1136/jmg.40.8.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohr M, Roullet FI, Hung AY, Sheng M, Crawley JN. Communication impairments in mice lacking Shank1: reduced levels of ultrasonic vocalizations and scent marking behavior. PLoS One. 2011;6:e20631. doi: 10.1371/journal.pone.0020631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff JJ, Gu H, Gerig G, Elison JT, Styner M, Gouttard S, Botteron KN, Dager SR, Dawson G, Estes AM, et al. Differences in White Matter Fiber Tract Development Present From 6 to 24 Months in Infants With Autism. Am J Psychiatry. 2012 doi: 10.1176/appi.ajp.2011.11091447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H, Lee HR, Gee HY, Mah W, Kim JI, Lee J, Ha S, Chung C, Jung ES, Cho YS, et al. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature. 2012;486:261–265. doi: 10.1038/nature11208. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY. Postnatal neurodevelopmental disorders: meeting at the synapse? Science. 2003;302:826–830. doi: 10.1126/science.1089071. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY. MeCP2 dysfunction in humans and mice. J Child Neurol. 2005;20:736–740. doi: 10.1177/08830738050200090701. [DOI] [PubMed] [Google Scholar]
- Zwaigenbaum L, Bryson S, Lord C, Rogers S, Carter A, Carver L, Chawarska K, Constantino J, Dawson G, Dobkins K, et al. Clinical assessment and management of toddlers with suspected autism spectrum disorder: insights from studies of high-risk infants. Pediatrics. 2009;123:1383–1391. doi: 10.1542/peds.2008-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]