Abstract
The biological function of NF-κB1 (p50) in the regulation of protein expression is far from well understood owing to the lack of a transcriptional domain. Here, we report a novel function of p50 in its regulation of p53 protein translation under stress conditions. We found that the deletion of p50 (p50–/–) impaired arsenite-induced p53 protein expression, which could be restored after reconstitutive expression of HA-p50 in p50–/– cells, p50–/– (Ad-HA-p50). Further studies indicated that the amounts of p53 mRNA, p53 promoter-driven transcription activity and p53 protein degradation were comparable between wild-type and p50–/– cells. Moreover, we found that p50 was crucial for Akt/S6 ribosomal protein activation via inhibition of the translation of the PH domain and leucine-rich repeat protein phosphatases 1 (PHLPP1), a phosphatase of Akt. Further studies showed that p50-mediated upregulation of miR-190 was responsible for the inhibition of PHLPP1 translation by targeting the 3′-untranslated region of its mRNA. Collectively, we have identified a novel function of p50 in modulating p53 protein translation via regulation of the miR-190/PHLPP1/Akt-S6 ribosomal protein pathway.
Keywords: p50, p53 translation, miR-190, PHLPP1, Akt/S6 ribosomal protein
INTRODUCTION
Tumor suppressor p53 is a transcription factor that is responsible for the transcriptional regulation of several important genes implicated in cell cycle control, DNA repair and apoptosis.1–4 The p53 protein expression is regulated at the levels of transcription, translation, protein modification and turnover, and cellular compartmentalization, as well as association with other proteins.5–9 For example, p53 is upregulated in cell response to genotoxic agents, such as ionizing radiation, and certain environmental stress chemicals, through multiple mechanisms, including transcription, protein modification and degradation.1–4 Arsenite is a well-known environmental human carcinogen and an anticancer therapeutic agent.10–12 Previous studies have shown that arsenite exposure elevates p53 protein expression in various cells.13 However, the molecular mechanisms underlying the upregulation of p53 protein expression are far from well understood.
NF-κB consists of five members in mammalian cells, including NF-kB1 (p50), NF-κB2 (p52), Rel A (p65), Rel B and c-Rel.14 Unlike other members, both p50 and p52 are lacking a transactivation domain.15 To function as a transcription factor, p50 forms a heterodimer with p65, Rel B or c-Rel.16 It has been reported that p50 homodimers can translocate into the nucleus and bind with the NF-κB-binding sites of its target genes. Although deficiency of p50 blocks NF-κB activation and attenuates neointimal hyperplasia,17 p50 homodimer alone cannot act as a transcription factor to regulate NF-κB downstream genes’ expression. For this reason, biological functions of p50 are much less studied in comparison with the other members of the NF-κB family. Recently, we demonstrated that p50 can mediate growth arrest and DNA damage (GADD) 45a protein expression by inhibiting GADD45α protein degradation via increasing its de-ubiquitination.18 Here we report that p50 modulates p53 expression at the translational level by regulating miR-190/PH domain and leucine-rich repeat protein phosphatases 1 (PHLPP1)/Akt-S6 ribosomal protein pathway.
RESULTS
P50 protein regulated p53 protein translation
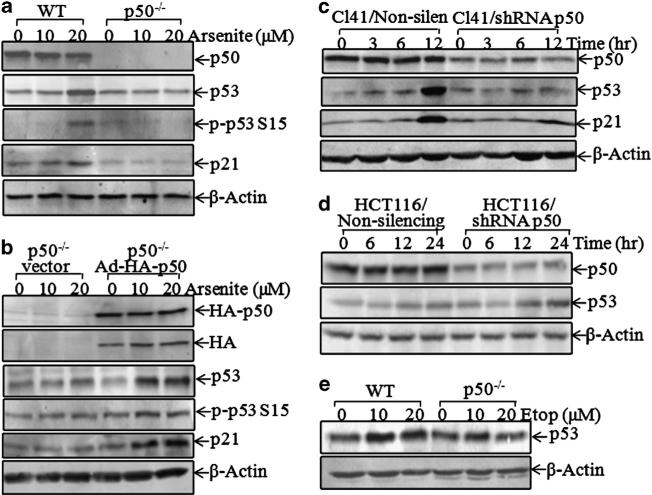
Previous studies have shown that arsenite exposure elevates p53 protein expression in various cells.13 To evaluate whether p50 is involved in arsenite-induced p53 protein expression, p53 expression level in p50–/– cells was compared with that of wild-type (WT) murine embryonic fibroblasts (MEFs) following arsenite exposure. As shown in Figure 1a, a deletion of p50 in MEFs (p50–/–) resulted in the loss of p53 protein induction following arsenite exposure, whereas basal levels of p53 protein expression were comparable between WT and p50–/– cells (Figure 1a). Consistently, phosphorylation of p53 at Ser15 and the induction of p21, a p53-regulated downstream gene, were also impaired in p50–/– cells (Figure 1a). The critical role of p50 for arsenite-induced p53 protein expression and function was further verified by the finding that reconstituted expression of p50 in p50–/– cells using Ad-HA-p50 could restore the induction of p53 protein expression and p21 expression following arsenite exposure (Figure 1b). In addition, p50 short hairpin RNA (shRNA) was used to knockdown endogenous p50 expression and to investigate the p53 induction following arsenite treatment. As shown in Figure 1c, knockdown of p50 in normal mouse epidermal JB6 Cl41 cells led to a marked reduction of p53 protein expression following arsenite exposure. A similar result was reproduced with the introduction of p50 shRNA into normal MEFs (data not shown). However, knockdown of p50 expression by p50 shRNA in human colon cancer HCT116 cells (HCT116/shRNA p50) did not show any observable inhibition of p53 protein induction by arsenite compared with Non-silencing HCT116 cells (HCT116/Non-silencing) (Figure 1d). It is known that p53 expression is regulated at the transcriptional and protein degradation levels owing to oxidative stress,19,20 and that oxidative status in cancer cells is distinct from normal cells.21 We therefore anticipate that the distinctions of oxidative status between normal and cancer cells might be responsible for the differential role of p50 in the regulation of p53 protein expression observed in normal and cancer cell lines. As arsenite is used as an effective chemotherapeutic medication of certain types of leukemia, we aimed to evaluate whether p50 could also regulate p53 induction by other anticancer drugs, such as etoposide, a widely used chemotherapeutic agent with p53 inductive activity at both translational and post-translational levels.22–26 As shown in Figure 1e, etoposide-induced p53 protein expression was partially inhibited in p50–/– cells (Figure 1e), revealing that p50 might mediate p53 upregulation either at the transcriptional level or at the translational level.
Figure 1.
P50 regulated p53 expression following arsenite and etoposide exposure. (a, b and e) WT and p50–/– MEFs were exposed to arsenite or etoposide for 12 h, and the cell extracts were subjected to western blotting for the detection of p53, phosphor-p53 at Ser15, p21, HA and b-actin. (c and d) shRNA p50 was stably transfected into human colon cancer HCT116 cells (c) and mouse epidermal JB6 Cl41 cells (d). P53 protein expression was determined by western blotting, and β-actin was used as protein-loading controls.
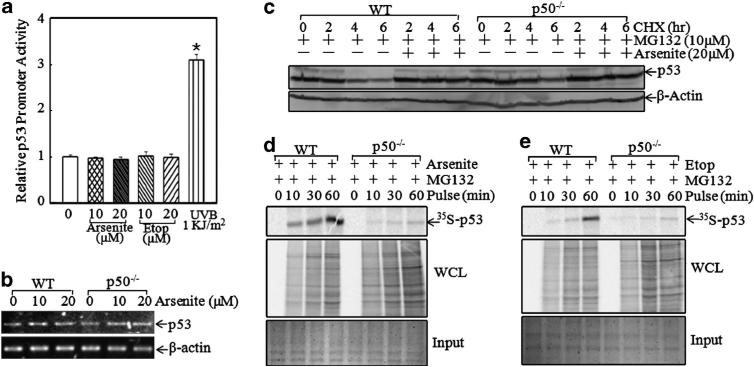
To test whether arsenite induced p53 expression at the transcriptional level, WT MEFs were transfected with p53 promoter luciferase reporter. The result indicated that neither arsenite nor etoposide showed an observable effect on p53 promoter activity, whereas ultraviolet B radiation showed a significant induction (Figure 2a), suggesting that p53 protein induction due to arsenite exposure did not occur at the transcriptional level. p53 mRNA level was further compared between WT and p50–/– cells, and it was almost comparable between WT and p50–/– cells (Figure 2b). Following this, the p53 protein degradation rate was evaluated. As shown in Figure 2c, after MG132 pretreatment for 4 h, the medium containing MG132 was replaced with a medium containing cycloheximide, or cycloheximide plus arsenite, as indicated. The results showed that the p53 protein degradation rate in p50–/– cells was slightly lower than that of WT cells in the absence of arsenite. Moreover, arsenite treatment inhibited the p53 protein degradation in a similar pattern in both WT and p50–/– MEFs, indicating that the defect of p53 protein induction by arsenite in p50–/– cells was not regulated at the protein degradation level. It suggested that p50 might regulate p53 expression at the protein translational level. To this end, short-term pulse-labeling assay was performed to examine p53 protein translational process in both WT and p50–/– cells. As shown in Figure 2d, the incorporation of 35S-methionine/cysteine into newly synthesized p53 protein was gradually increased along with the incubation time in WT cells, whereas the p53 protein synthesis rate was markedly reduced in p50–/– cells, indicating that the synthesis of p53 protein in p50–/– cells was impaired in comparison with WT cells following arsenite exposure. Importantly, etoposide-induced new p53 protein synthesis was also blocked in p50–/– cells (Figure 2e). These results strongly supported our notion that p50 was crucial for p53 protein translation following either arsenite or etoposide exposure.
Figure 2.
P50 regulated p53 protein translation following exposure to arsenite and etoposide (etop). (a) P53 promoter activity was analyzed by measuring p53 promoter-driven luciferase activity following exposure to arsenite or etoposide for 12 h. Ultraviolet B (UVB) was used as a positive control. *Significant increase in comparison with medium control (P<0.05). (b) P53 mRNA level in WT and p50–/– cells was analyzed using RT–PCR. (c) After pretreatment with MG132 (10 μm) for 4 h, cycloheximide (CHX) (10 μm) in combination with or without arsenite (20 μm) was added into the culture medium and incubated for the indicated time, and the cells were then extracted for the determination of p53 protein expression by western blotting. (d and e) WT and p50–/– MEFs were exposed to 20 μm arsenite (d) or 10 μm etoposide (e) for 9 h. Newly synthesized p53 protein was monitored by pulse assay using 35S-labeled methionine/cysteine, and WCL stands for whole cell lysate. Coomassie blue staining was used for protein loading control.
Akt/S6 ribosomal protein pathway was downregulated in p50–/– cells
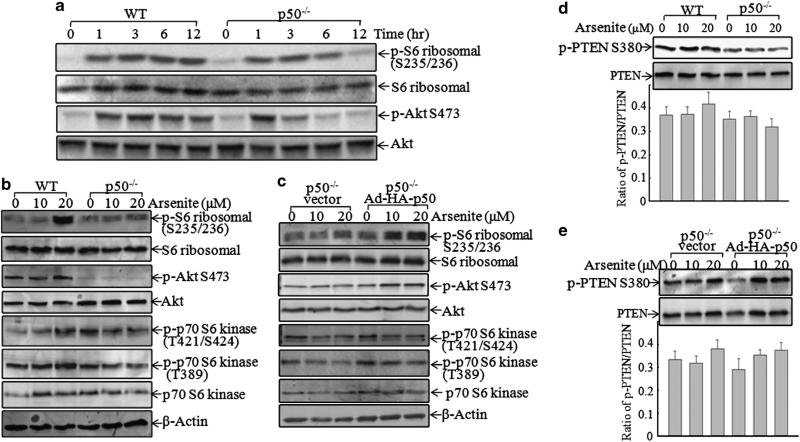
Ribosomal protein S6 has an important role in the regulation of protein translation,27 and its phosphorylations at Ser235/236 in the C terminus were required for enhancing the affinity for m7GpppG cap of mRNA.28 Thus, the Akt/S6 ribosomal protein activation was determined following arsenite exposure. In the early time phase (1–3 h after exposure), arsenite exposure caused an increase in S6 ribosomal protein phosphorylations at Ser235/ 236 in both WT and p50–/– cells. However, the phosphorylations in p50–/– cells were markedly inhibited at 6 and 12 h in comparison with those in WT cells (Figure 3a). Consistently, the phosphorylation of Akt at Ser473 showed similar changes between WT and p50–/– MEFs (Figure 3a). The findings were further verified by the facts that the introduction of HA-p50 back into p50–/– cells was able to restore the phosphorylations of both S6 ribosomal protein at Ser235/236 and Akt at Ser473 (Figure 3c). In contrast to p50 regulation of Akt/S6 ribosomal protein phosphorylation, ectopic expression of HA-p50 in p50–/– cells did not show restoration of phosphorylations of p70 S6 kinase at either T389/421 in p50–/– cells (Figure 3c), although p50 deficiency led to the inhibition of phosphorylations of p70 S6 kinase at T389/421 (Figure 3b). The results suggested that p50-mediated regulation of S6 ribosomal protein phosphorylations at Ser235/236 might be controlled by Akt, but not by p70 S6 kinase. As the phosphorylations of both S6 ribosomal protein at Ser235/236 and Akt at Ser473 in the early phase did not show any difference between WT and p50–/– MEFs, but their activation in the late phase showed a marked difference between these two cells, we anticipated that the kinase responsible for the activation of Akt/S6 ribosomal protein pathway was not affected in p50–/– cells, whereas the major difference in the phosphorylations of Akt/S6 ribosomal protein during the late phase could be due to p50-mediated phosphatase alteration.
Figure 3.
The Akt/S6 ribosomal protein pathway was a mediator for p50-regulated p53 translation. Cells were exposed to (a) arsenite (20 μm) for an indicated time or (b) arsenite at indicated doses for 12 h. The cell extracts were subjected to western blotting. (c) P50–/– cells were infected with Ad-HA or Ad-HA-p50 for 12 h, and then exposed to arsenite at the indicated doses for 12 h. The cell extracts were subjected to western blotting. (d and e) The indicated cells were exposed to arsenite for 12 h, and the cell extracts were subjected to western blotting for the detection of p-PTEN at Ser380 and total PTEN. The densitometric analyses of the bands were performed using the ImageQuant 5.2 software (GE Healthcare). The results shown are representative of three independent experiments. HA, hemagglutinin; PTEN, phosphatase and tensin homolog.
PHLPP1 was a downstream phosphatase that was responsible for p50-mediated activation of Akt/S6 ribosomal protein
Phosphatase and tensin homolog (PTEN) is a tumor suppressor that negatively regulates the phosphatidylinositol 3-kinase/Akt pathway by dephosphorylation of the lipid second message, PI(3,4,5)P3, a product of phosphatidylinositol 3-kinase.29 The phosphorylation of PTEN at Ser380 is critical for its protein stability30 and for repressing phosphatase activation.31 Therefore, the total PTEN protein expression and phosphor-PTEN at Ser380 were evaluated between WT and p50–/– MEFs following arsenite exposure. The results showed that there was no significant difference in either PTEN protein expression or phosphorylation among WT, p50–/– and p50–/– (Ad-HA-p50) cells (Figures 2d and e), suggesting that p50-mediated regulation of Akt/S6 ribosomal protein activation is through a PTEN-independent pathway.
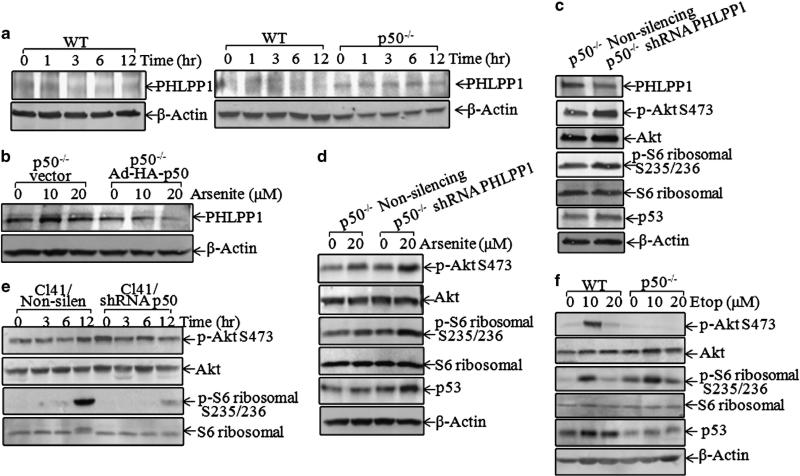
PHLPP1 is an important Akt phosphatase responsible for dephosphorylating Akt at Ser473.32 To test whether PHLPP1 is involved in p50-mediated regulation of activation of Akt/S6 ribosomal protein pathway upon arsenite exposure, the effect of arsenite exposure on PHLPP1 protein expression was first determined. The results indicated that arsenite exposure markedly downregulated PHLPP1 protein expression in WT cells (Figure 4a, left panel). Depletion of p50 (p50–/–) led to a markedly increased basal level of PHLPP1 expression, and arsenite did not cause an obvious downregulation of PHLPP1 in p50–/– cells compared with that observed in WT cells (Figure 4a, right panel). Consistently, ectopic expression of Ad-HA-p50 in p50–/– cells reduced PHLPP1 expression and restored the downregulation of PHLPP1 caused by arsenite treatment (Figure 4b). The data suggested that PHLPP1 might be a mediator for p50 in the activation of the Akt/S6 ribosomal protein pathway. This notion was greatly supported by our further studies showing that the knockdown of PHLPP1 in p50–/– cells resulted in a significant increase of the basal phosphorylations levels of Akt/S6 ribosomal protein (Figure 4c). More importantly, introduction of shRNA PHLPP1 in p50–/– cells restored arsenite-induced p53 expression, as well as phosphorylations of Akt/S6 ribosomal protein (Figure 4d). Moreover, knockdown of p50 in JB6 Cl41 cells also impaired arsenite-induced phosphorylations of Akt at Ser473 and S6 ribosomal protein at Ser235/236, in comparison with Non-silencing control transfectants (Figure 4e). These results indicated that PHLPP1 was a downstream phosphatase of p50 that had a role in the mediation of phosphorylations of Akt at Ser473 and S6 ribosomal protein at Ser235/236 following arsenite treatment.
Figure 4.
PHLPP1 downregulation by p50-mediated Akt/S6 ribosomal protein activation and p53 protein expression. (a and b) PHLPP1 expression levels were compared between WT, p50–/–, p50–/– (vector) and p50–/– (Ad-HA-p50) following exposure to 20 μm arsenite. (c and d) Indicated stable transfectants were used to analyze PHLPP1, phosphor-Akt, phosphor-S6 ribosomal protein and p53 with or without arsenite treatment for 12 h. (e and f) JB6 Cl41 transfectants were exposed to arsenite (20 μm) for the indicated time points (e), or WT and p50–/– cells were exposed to etoposide for 12 h (f). The cells were extracted and protein samples were subjected to western blotting.
It is known that etoposide causes DNA double-strand breaks,33 a genotoxic stress that usually leads to halt of cap-dependent protein translation, whereas internal ribosome entry site-dependent translation initiation machinery maintains the expression of certain critical proteins involved in cell growth, survival and death during cellular stress, for example, p53.34 Thus, etoposide has been known to increase p53 protein expression by regulating cap-independent, internal ribosome entry site-dependent p53 translation.26 In contrast, arsenite is rarely found to function as a mutagen, rather it majorly regulates gene expression via modulation of either transcription, protein degradation or epigenetic mechanisms.35 Consistently, our results showed that although p53 protein expression and Akt phosphorylation at Ser473 were reduced in p50–/– cells following etoposide treatment, S6 ribosomal protein phosphorylation at Ser235/236 was not inhibited in p50–/– cells following etoposide treatment (Figure 4f). Collectively, our results suggest that although p50 was crucial for cap-dependent p53 protein translation via activation of the Akt/S6 pathway in arsenite response, p50-mediated p53 protein translation is through S6 ribosomal protein-independent mechanisms upon etoposide treatment.
Akt/S6 ribosomal protein activation was critical for p53 protein translation following arsenite treatment
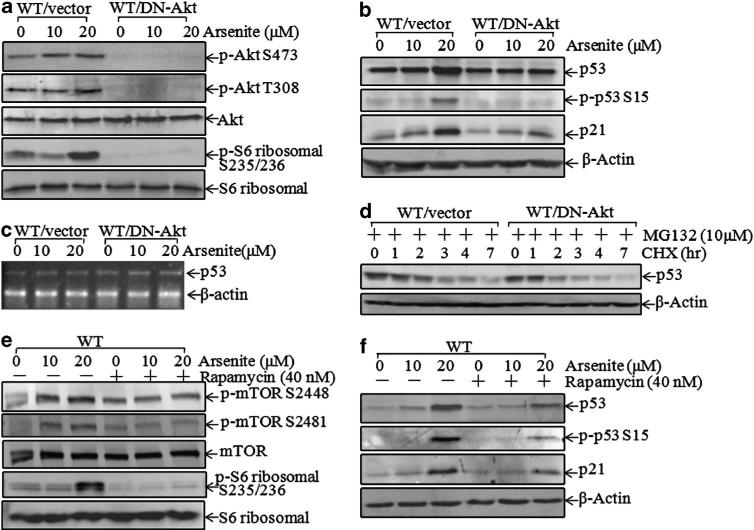
To clarify whether the Akt/S6 ribosomal protein pathway has a role in p53 protein translation upon arsenite exposure, the construct of dominant-negative (DN)-Akt29,36 was stably introduced into WT cells and a stable transfectant, WT/DN-Akt, was established. As shown in Figure 5a, the introduction of DN-Akt into WT cells showed a marked inhibition of Akt phosphorylation and subsequently blocked arsenite-induced S6 ribosomal protein phosphorylations at Ser235/236 (Figure 5a), suggesting that S6 ribosomal protein phosphorylations at Ser235/236 were mediated by activated Akt in arsenite response. Moreover, p53 induction and its regulated p21 expression by arsenite were also impaired in WT/DN-Akt transfectant in comparison with WT/vector cells (Figure 5b), indicating that Akt/S6 ribosomal protein activation was important for p53 induction by arsenite. However, mRNA expression levels of p53 were comparable between WT/vector and WT/DN-Akt cells (Figure 5c). In addition, the p53 protein degradation rate in WT/DN-Akt cells showed only a marginal increase in WT/vector cells (Figure 5d), revealing that the regulation of p53 induction by the Akt/S6 ribosomal protein pathway in arsenite response was not primarily through protein degradation. To test the role of S6 ribosomal protein in p53 production, rapamycin, a specific mammalian target rapamycin (mTOR) inhibitor, was used. As shown in Figure 5e, pretreatment of cells with rapamycin inhibited phosphorylations of both mTOR at Ser2448/2481 and S6 ribosomal protein at Ser235/236. Consistently, p53 protein induction and its regulated p21 expression were also impaired by rapamycin (Figure 5f).
Figure 5.
Arsenite induced p53 translation through Akt/S6 ribosomal protein activation. (a and b) Cells were exposed to arsenite for 12 h, and the cell extracts were subjected to western blotting for the detection of phosphor-Akt, phosphor-S6 ribosomal protein, p53, phosphor-p53 at Ser15 and p21. (c) The mRNA level of p53 in WT (vector) and WT (DN-Akt) cells was analyzed using RT–PCR. (d) After pretreatment with MG132 (10 μm) for 4 h, cycloheximide (CHX) (10 μm) was added to cells and incubated for the indicated additional time. Cells were extracted and subjected to western blotting for the determination of p53 protein degradation. (e and f) After pretreatment of cells with rapamycin (40 nm) for 30 min, cells were exposed to arsenite at the indicated doses for 12 h. Cells were extracted and cell extracts were subjected to western blotting for the determination of phosphor-mTOR, phosphor-S6 ribosomal protein, p53, phosphor-p53 and p21.
P50 downregulated PHLPP1 translation by increasing miR-190 expression
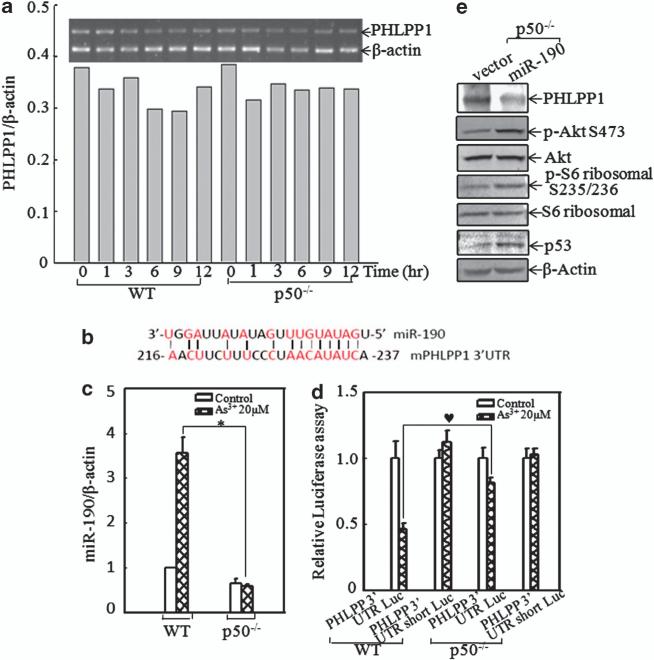
To evaluate the molecular mechanisms underlying p50-regulated PHLPP1 expression, we first determined the mRNA levels of PHLPP1 in both WT and p50–/– cells exposed to arsenite. The results showed that arsenite did not change PHLPP1 mRNA expression in either cells (Figure 6a), suggesting that the downregulation of PHLPP1 expression by arsenite did not occur by affecting transcription or mRNA stability. It had been reported that microRNA, a class of ~22-nucleotide noncoding small RNAs, was able to bind to the 3′-untranslated region (UTR) of target genes and inhibit their translation,37 and miR-190 could regulate human PHLPP1 protein translation.38 As shown in Figure 6b, miR-190 also had potential binding sites in 3′-UTR sequence of mouse PHLPP1 mRNA. We compared arsenite-induced miR-190 expression between WT and p50–/– cells. The results from the real-time polymerase chain reaction (PCR) assay indicated that arsenite treatment resulted in a significant induction of miR-190 in WT cells, whereas this induction was totally impaired in p50–/– MEFs under the same experimental conditions (Figure 6c). To test whether miR-190 was involved in the translational regulation of PHLPP1, PHLPP1 3′-UTR luciferase reporter or PHLPP1 3′-UTR luciferase reporter with the deletion of miR-190 binding sites (named as 3′-UTR short luciferase reporter) was transiently co-transfected with pRL-TK into WT and p50–/– cells, respectively. The transfectants were exposed to 20 μm arsenite for 12 h, and the luciferase activity was determined and normalized to pRL-TK activity. The results showed that PHLPP1 3′-UTR luciferase activity upon arsenite exposure was significantly increased in p50–/– cells compared with that in the WT cells, whereas this increased luciferase activity was not observed in p50–/– cells transfected with PHLPP1 3′-UTR short luciferase reporter (Figure 6d). These results clearly indicated that p50 expression was crucial for arsenite-induced miR-190 expression and that miR-190 could suppress PHLPP1 translation by interacting with binding sites of PHLPP1 mRNA 3′-UTR. To verify whether miR-190 could regulate PHLPP1 expression in p50–/– cells, a construct containing miR-190 was transfected into p50–/– cells, and the stable transfectants were named p50–/– miR-190. In comparison with p50–/– vector cells, the PHLPP1 protein level was downregulated in p50–/– miR-190 cells, and subsequently phosphorylations of Akt at Ser473 and S6 ribosomal protein at Ser235/236 were markedly increased (Figure 6e). Furthermore, p53 protein expression was elevated in p50–/– miR-190 cells (Figure 6e). These results demonstrated that p50 downregulated the translation of PHLPP1 by increasing miR-190 expression.
Figure 6.
P50 attenuated PHLPP1 translation via upregulation of miR-190 expression. (a) Total RNA was extracted from WT and p50–/– cells upon 20 μm arsenite treatment for the indicated time periods. PHLPP1 mRNA level was determined by RT–PCR and quantified using the ImageQuant 5.2 software (GE Healthcare). Data are representative of one of three independent experiments. (b) The binding site of miR-190 in 3′-UTR of mouse PHLPP1 was analyzed. The sites shown were relative to the starting site of mouse PHLPP1 3′-UTR. (c) WT and p50–/– cells were exposed to 20 μm arsenite for 9 h. Total RNA was extracted using an miRNeasy Mini kit (Qiagen) according to the manufacturer's instruction. After reverse transcription using the miScript PCR system (Qiagen), real-time PCR was conducted using an miR-190-specific primer. β-Actin was used as a control. *Significant inhibition of miR-190 expression in p50–/– cell in comparison with that in WT cells following arsenite treatment (P<0.05). The value was shown as mean±s.d. from three independent experiments. (d) For testing the role of miR-190 on PHLPP1 translation, PHLPP1 3′-UTR luciferase reporter or PHLPP1 3′-UTR short luciferase reporter was transiently transfected into WT or p50–/– cells, respectively. Luciferase activity was detected, as described in ‘Materials and methods’, following treatment of cells with arsenite for 12 h. The symbol (♥) indicates a significant increase of arsenite-induced PHLPP1 3′-UTR luciferase activity in p50–/– cells compared with that in WT cells (P<0.05). The value was showed as mean±s.d. from three independent experiments.
P50-mediated miR-190/PHLPP1 expression was critical for cell apoptotic response due to arsenite exposure
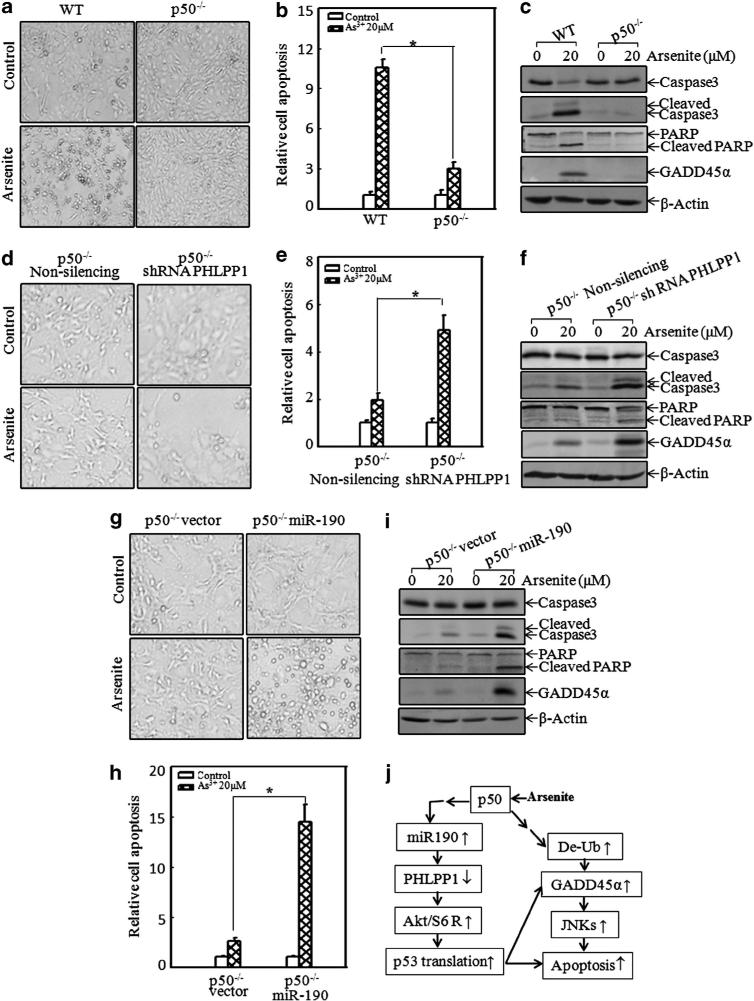
To test whether p50-mediated miR-190/PHLPP1/p53 had a role in arsenite-induced cell apoptosis, we first compared cell apoptotic response between WT and p50–/– MEFs. The results showed that p50 was crucial for arsenite-induced cell apoptosis (Figures 7a–c), which was consistent with our previous studies.18 We next evaluated the role of PHLPP1 in arsenite-induced cell apoptosis using p50–/– shRNA PHLPP1 cells and the control Non-silencing cells. As shown in Figures 7d–f, specific knockdown of PHLPP1 in p50–/– cells rendered p50–/y1 cells more sensitive to apoptotic response as compared with Non-silencing transfectants, suggesting that PHLPP1 is a p50 downstream target responsible for the resistance of p50–/– cells to apoptotic response. Moreover, we observed the effect of overexpression of miR-190 on apoptotic induction by arsenite in p50–/– cells. Our results showed that the introduction of miR-190 into p50–/– cells increased the apoptosis due to arsenite exposure (Figures 7g–i). Our results demonstrated that the regulation of miR-190/PHLPP1 was at least one of the major mechanisms responsible for p50-mediated cell apoptosis following arsenite exposure (Figure 7j).
Figure 7.
Introduction of shRNA PHLPP1 and miR-190 into p50–/– cells restored its apoptotic response following arsenite exposure. (a, d and g) WT and p50–/–, p50–/– (non-silencing) and p50–/– (shRNA PHLPP1), p50–/– (vector) and p50–/– (miR-190) cells were exposed to 20 μm arsenite for 24 h. The photos were taken under the microscope. (b, e and h) Cells were exposed to 20 μm arsenite for 24 h, and then collected for flow cytometry analysis. *Significant alterations in apoptosis in comparison with the corresponding control transfectants (P<0.05). The value was shown as mean±s.d. from three independent experiments. (c, f and i) Cell extracts with the same treatment as above were subjected to western blot assay for the detection of caspase 3, poly (ADP-ribose) polymerase (PARP) and their cleaved fragments. (j) The potential schematic of p50 regulation of p53 protein translation and apoptosis following arsenite exposure.
DISCUSSION
P50 exists in mammalian cells either as a homodimer or a heterodimer with p65, RelB or c-Rel.15 As p50 lacks the transcriptional domain, it exerts its biological function as a transcription factor mainly by forming a heterodimer with p65, RelB or c-Rel.15 However, the function of the p50 homodimer has been the subject of few studies. Our previous studies demonstrated that p50 mediated arsenite-induced cell apoptosis by regulating GADD45α protein degradation.18 Here we identified a novel function of p50 in the regulation of p53 protein expression at the translational level. Our results further indicated that p50-mediated p53 translation was regulated by the activation of the Akt/S6 ribosomal protein pathway, which resulted from the downregulation of the PHLPP1 expression via enhancement of the miR-190 level. Combined with the positive role of p50 in the manipulation of GADD45α/JNK pathway activation,18 it was concluded that p50 was involved in arsenite-induced cell apoptosis through increment of either p53 translation or upregulation of the GADD45α/JNK pathway in a transcriptional- or post-translational-dependent manner, as elucidated in Figure 7j.
P53 is a tumor suppressor that has a crucial role in circumventing cancer development.2 Regulation of p53 expression at the transcriptional or protein degradation levels has been studied extensively. For example, oxidant stress, such as arsenite or ultraviolet, can induce p53 gene transcription. MDM2, the E3 ubiquitin ligase of p53, promotes p53 nuclear export and degradation through binding with p53 directly.39–41 Recent reports have also shown that arsenite enhances mutant p53 degradation.42 However, translational regulation of p53, particularly under oxidant stress such as arsenite exposure, is largely unexplored. Our current studies demonstrated a crucial role of p50 in arsenite-induced p53 translation. We showed that the p53 protein induction by arsenite was impaired in p50–/– cells, which could be restored by the introduction of p50 into p50–/– cells. In contrast to the protein level, p53 mRNA level and protein degradation rate were comparable between WT and p50–/– cells, suggesting that p50 might regulate p53 translation. Ribosomal protein S6 is a well-known mediator of protein translation.28 Our results indicated that phosphorylations of Akt/S6 ribosomal protein induced by arsenite were markedly inhibited in p50–/– cells in comparison with that in WT cells, indicating that Akt/S6 ribosomal protein may be a p50 downstream target that is responsible for regulating p53 protein translation. This notion was greatly supported by our results indicating that inhibition of Akt/S6 ribosomal protein by overexpression of DN-Akt in WT cells attenuated p53 protein induction, but not p53 mRNA and protein degradation, whereas activation of Akt/S6 ribosomal protein by the introduction of miR-190 into p50–/– cells markedly increased in p53 protein expression. Taken together, our results demonstrated that p50 has a critical role in p53 protein translation due to arsenite exposure.
One of p53's biological functions is to trigger cell apoptosis.43–45 P53 also has a vital role in maintaining chemosensitivity and radiosensitivity for cancer therapy.46,47 Our previous studies showed that p50 has an important role in arsenite-induced cell apoptotic response.18 To determine the role of p50-dependent p53 translation mediated by miR-190/PHLPP1 in apoptotic response upon arsenite exposure, we determined the effect of knockdown of PHLPP1 or introduction of miR-190 on arsenite-induced apoptosis in p50–/– cells. Our results showed that upregulation of p53 expression through knockdown of PHLPP1 or overexpression of miR-190 rendered the p50–/– cells more sensitive to cell apoptosis due to arsenite exposure. These data indicated that miR-190/PHLPP1-regulated p53 translation was crucial for p50-mediated cell apoptosis upon arsenite exposure.
PHLPP1 is a protein phosphatase that was recently discovered to be an Akt phosphatase.32 The loss or decrease of PHLPP family expression has been found in colon cancer tissues, and re-expression of either PHLPP1 or PHLPP2 into HCT116 cells suppresses tumor growth in vivo.48 Our results showed that PHLPP1 protein expression was much higher in p50–/– cells compared with that in WT cells, whereas the increased PHLPP1 expression in p50–/– cells could be inhibited by either introduction of p50 expression or miR-190. Importantly, we found that p50 was critical for miR-190 induction caused by arsenite exposure and that arsenite downregulation of PHLPP1 translation was dependent on binding of miR-190 to PHLPP1 mRNA 3′-UTR. These data demonstrated that p50 was able to specifically downregulate PHLPP1 translation by upregulating miR-190 upon arsenite exposure.
Micro-RNA (miRNA) belongs to the small noncoding RNA family and has an important role in post-transcriptional regulation.37 Most miRNAs are located at the intron of other genes and do not have their own promoter sequence.49 In mammalian cells, primiRNA is gained after transcription with another gene. It is spliced by Drosha in the nucleus into pre-miRNA, and then transferred to the cytoplasm and digested by Dicer into mature miRNAs.37,49 As an important mechanism of post-transcriptional regulation, miRNA can bind to 3′-UTR of the target gene and regulate its translation.37 A current study showed that miR-190 was upregulated in WT cells following arsenite exposure, but this miR-190 induction was impaired in p50–/– experimental cells under the same conditions. This indicated that p50 was required for miR-190 induction by arsenite. The transcription factor YY1 is reported to increase miR-190 expression,50 and p65/p50 complexes can enhance YY1 expression through binding with its promoter.51 Thus, we anticipated that p50 deficiency might affect YY1 expression, subsequently resulting in the reduction of miR-190 expression. Our next study will focus on the mechanisms underlying the p50 regulation of miR-190 expression.
In summary, our results demonstrated a novel function of p50 in regulating p53 translation through targeting the miR-190/PHLPP1/Akt-S6 ribosomal protein pathways. This finding provided new insight into the understanding of p50 regulation of cell apoptotic response, as well as a cross talk among p50, Akt and p53 upon arsenite exposure. A complete understanding of the key molecules involved in carcinogenesis and anticarcinogenesis will help us utilize them as targets for both prevention and therapy of cancers.
MATERIALS AND METHODS
Plasmids, adenovirus, antibodies and other reagents
Akt with T308A and K473A mutants was cloned in the retroviral vector SRα and named DN-Akt.36,52,53 V5-DEST-miR190 and the control constructs were kind gifts from Dr Ping-Yee Law (Department of Pharmacology, University of Minnesota, Minneapolis, MN, USA). HA-p50 and control vector were kindly provided by Dr Jianping Ye (Pennington Biomedical Research Center, Louisiana State University, Baton Rouge, LA, USA), and Ad-HA-p50 and its control Ad-HA were constructed according to the manufacturer's instructions. The shRNA sets for human and mouse p50 were purchased from Open Biosystems (Thermo Fisher Scientific, Huntsville, AL, USA). Human PHLPP1 3′-UTR luciferase reporter and PHLPP1 3′-UTR short luciferase reporter (the binding site of miR-190 was deleted) were cloned into the pGL3 luciferase assay vector, and were kindly provided by Dr Fei Chen (Department of Pharmaceutical Sciences, Wayne State University, Detroit, MI, USA). The antibodies specific for p53, phosphor (P)-p53 Ser15, Akt, P-Akt Thr308, P-Akt Ser473, mTOR, P-mTOR Ser2448, P-mTOR Ser2481, p70 S6 kinase, P-p70 S6 kinase Thr421/Ser424, P-p70 S6 kinase Thr389, PTEN, P-PTEN Ser380, S6 ribosomal protein, P-S6 ribosomal protein Ser235/236, poly (ADP-ribose) polymerase and caspase 3 were purchased from Cell Signaling (Beverly, MA, USA). Antibodies specific for mouse p50 and p21 were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The antibody specific for PHLPP1 was purchased from Bethyl Laboratories Inc. (Montgomery, TX, USA). HA antibody was purchased from Covance Inc. (Princeton, NJ, USA). β-Actin antibody and etoposide were purchased from Sigma (St Louis, MO, USA). Chemicals of MG132, cycloheximide and rapamycin were purchased from Calbiochem (San Diego, CA, USA). The dual luciferase assay substrate was purchased from Promega (Madison, WI, USA).
Cell culture and transfectants
P50–/– and its corresponding WT MEFs were described in our previous publication.18 All MEFs and their transfectants were cultured at 37 1C with 5% CO2 with 10% fetal bovine serum Dulbecco's modified Eagle's medium (DMEM) supplied with 1% penicillin/streptomycin and 2 μm l-glutamine (Life Technologies, Grand Island, NY, USA). WT MEFs were stably transfected with DN-Akt29 or p53 promoter luciferase reporter using FuGENE HD (Roche Applied Science, Indianapolis, IN, USA), following the manufacturer's instructions. The specific shRNAs together with PolyJet DNA In Vitro Transfection Reagent (SignaGen Laboratories, Rockville, MD, USA) were used to knock down PHLPP1 expression or overexpression of miR-190 in p50–/– cells, and stable transfectants, p50–/– (shRNA PHLPP1) and p50–/– (miR-190), were selected by puromycin (Alexis, Plymouth, PA, USA).
Reverse transcription-polymerase chain reaction (RT–PCR)
Cells were treated with 10 and/or 20 μm arsenite for the indicated time points, and were then used for total RNA extraction using TRIzol reagent (Invitrogen, Grand Island, NY, USA), according to the manufacturer's instructions. Total RNA (5 μg) was used for first-strand cDNA synthesis with oligdT(20) primer by SuperScript III First-Strand Synthesis system (Invitrogen). Specific primers (Invitrogen) were used for PCR amplification. The primers used in this study were as follows: p53 (forward: 5′-CACGTACTCTCCTCCCCTCA-3′; reverse: 5′-CTTCTGTACGGCGGTCTCTC-3′), PHLPP1 (forward: 5′-CAAATGGGCTGAGCGCCTCGT-3′; reverse: 5′-GCTGCGACACCACCTTAGACGC-3′) and β-actin (forward: 5′-CCTGTGGCATCCATGAAACT-3′; reverse: 5′-GTGCTAGGAGCCAGAGCAGT-3′). The PCR product was analyzed by agarose gel. The densitometric analyses of the product bands were performed using the ImageQuant 5.2 software (GE Healthcare, Pittsburgh, PA, USA).
Quantitative RT–PCR for miRNA assay
Cells were treated with arsenite as indicated, and then used for total RNA extraction using the miRNeasy Mini Kit (Qiagen, Valencia, CA, USA). Total RNA (1 μg) was used for reverse transcription. The analysis of miR-190 expression was carried out using the miScript PCR system (Qiagen) and the 7900HT Fast Real-time PCR system (Applied Biosystems, Carlsbad, CA, USA). The primer for miR-190 assay was purchased from Qiagen, and β-actin (Invitrogen) was used as a control (forward: 5′-CCTGTGGCATCCATGAAACT-3′; reverse: 5′-GTGCTAGGAGCCAGAGCAGT-3′). The initial activation was performed at 95 °C for 15 min, followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 55 °C for 30 s and extension at 70 °C for 30 s. Data were analyzed as described in the previous publication.29
Luciferase assay
For the determination of p53 promoter-driven luciferase transcription, WT cells stably transfected with the p53 promoter luciferase reporter were seeded into a 96-well plate. The cells were exposed to arsenite, etoposide or ultraviolet B as indicated, and then lysed for luciferase assay using luciferase substrate (Promega) as described.29 For the determination of PHLPP1 3′-UTR luciferase reporter activity, WT and p50–/– cells were transiently transfected with PHLPP1 3′-UTR luciferase reporter/TK or PHLPP1 3′-UTR short luciferase reporter/TK, respectively. At 12 h after transfection, cells were seeded into a 96-well plate and cultured until they were 70–80% confluent. The cell culture medium was replaced with 0.1% fetal bovine serum DMEM and cultured for another 12 h, and the cells were then exposed to 20 μm arsenite for 12 h. Luciferase activity was determined using the Dual-Luciferase Reporter Assay System (Promega) and analyzed relative to internal TK activity according to the manufacturer's instructions.
Flow cytometry
Cells were seeded into 6-well plates and cultured until they were 70–80% confluent. The cell culture medium was replaced with 0.1% fetal bovine serum DMEM and cultured for another 12 h, and the cells were then exposed to 20 μm arsenite for the indicated duration. All cells were collected by centrifugation at 1500 r.p.m. for 5 min. The cell pellets were washed with ice-cold phosphate-buffered saline one to two times, and then fixed in ice-cold 70% ethanol at 20 1C overnight. The cells were washed with phosphate-buffered saline one or three times, and cell apoptosis was analyzed using flow cytometry (Beckman, Indianapolis IN, USA) after staining for 15 min with propidium iodide (PI) buffer (0.1% Triton X-100, 0.2 mg/ml RNase A, 0.05 mg/ml propidium iodide).
[35S]methionine pulse assays
Cells were exposed to 20 mm of arsenite or etoposide for 9 h, and then incubated with methionine–cysteine-free DMEM (Gibco-BRL, Grand Island, NY, USA) containing 2% dialyzed fetal calf serum (Gibco-BRL) and 50 μm MG132 for 1 h. The cells were then incubated with 2% fetal bovine serum methionine–cysteine-free DMEM containing 35S-labeled methionine/cysteine (250 μCi per dish, Trans 35S-label; ICN) for the indicated time periods. The cells were extracted with lysis buffer (Cell Signaling) containing complete protein inhibitor mixture (Roche) on ice. Total lysate of 500 mg was incubated with anti-p53 antibody-conjugated agarose beads (R&D Systems, Minneapolis, MN, USA) overnight at 4 °C. The immunoprecipitated samples were washed with the cell lysis buffer five times, heated at 100 °C for 5 min and subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis analysis. 35S-labeled p53 protein was detected with the PhosphorImager (Molecular Dynamics, Kent City, MI, USA).
Western blotting
Cells were seeded into 6-well plates and cultured until they were 70–80% confluent. The cells were extracted with cell lysis buffer (10 mm Tris-HCl, pH 7.4, 1% sodium dodecyl sulfate, 1 mm Na3VO4 and proteasome inhibitor) and protein concentration was determined using Nano Drop 2000 (Thermo Scientific, Holtsville, NY, USA). A measure of 30–60 μg of proteins per sample was subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and western blotting was carried as described in our previous report.29 The densitometric analyses of the bands were performed using the ImageQuant 5.2 software (GE Healthcare). The results shown were representative of three independent experiments.
Statistical methods
The Student's t-test was used to determine significant differences. The differences were considered to be significant at .
ACKNOWLEDGEMENTS
We thank Ms Nedda Tichi and Mr Andy Hu for their critical reading of this manuscript. We thank Dr Ping-Yee Law (Department of Pharmacology, University of Minnesota) for providing us with constructs of miR-190 and its control. We also thank Dr Fei Chen (Department of Pharmaceutical Sciences, Wayne State University) for providing us with the PHLPP1 3′-UTR luciferase reporter. This work was partially supported by grants from NSFC81229002, NIH/NCI CA112557, NBRPC2012CB525004 and NIH/NIEHS ES000260.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Fridman JS, Lowe SW. Control of apoptosis by p53. Oncogene. 2003;22:9030–9040. doi: 10.1038/sj.onc.1207116. [DOI] [PubMed] [Google Scholar]
- 2.Muller PA, Vousden KH, Norman JC. P53 and its mutants in tumor cell migration and invasion. J Cell Biol. 2011;192:209–218. doi: 10.1083/jcb.201009059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teodoro JG, Parker AE, Zhu X, Green MR. P53-mediated inhibition of angiogenesis through up-regulation of a collagen prolyl hydroxylase. Science. 2006;313:968–971. doi: 10.1126/science.1126391. [DOI] [PubMed] [Google Scholar]
- 4.Yin Y, Tainsky MA, Bischoff FZ, Strong LC, Wahl GM. Wild-type p53 restores cell cycle control and inhibits gene amplification in cells with mutant p53 alleles. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]
- 5.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 6.Bosari S, Viale G, Roncalli M, Graziani D, Borsani G, Lee AK, et al. P53 gene mutations, p53 protein accumulation and compartmentalization in colorectal adenocarcinoma. Am J Pathol. 1995;147:790–798. [PMC free article] [PubMed] [Google Scholar]
- 7.Heinrichs S, Deppert W. Apoptosis or growth arrest: modulation of the cellular response to p53 by proliferative signals. Oncogene. 2003;22:555–571. doi: 10.1038/sj.onc.1206138. [DOI] [PubMed] [Google Scholar]
- 8.Ryan KM, Phillips AC, Vousden KH. Regulation and function of the p53 tumor suppressor protein. Curr Opin Cell Biol. 2001;13:332–337. doi: 10.1016/s0955-0674(00)00216-7. [DOI] [PubMed] [Google Scholar]
- 9.Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 10.Gebel TW. Arsenic and drinking water contamination. Science. 1999;283:1458–1459. doi: 10.1126/science.283.5407.1455e. [DOI] [PubMed] [Google Scholar]
- 11.Soignet SL, Maslak P, Wang ZG, Jhanwar S, Calleja E, Dardashti LJ, et al. Complete remission after treatment of acute promyelocytic leukemia with arsenic trioxide. N Engl J Med. 1998;339:1341–1348. doi: 10.1056/NEJM199811053391901. [DOI] [PubMed] [Google Scholar]
- 12.Zhang XW, Yan XJ, Zhou ZR, Yang FF, Wu ZY, Sun HB, et al. Arsenic trioxide controls the fate of the PML-RARalpha oncoprotein by directly binding PML. Science. 2010;328:240–243. doi: 10.1126/science.1183424. [DOI] [PubMed] [Google Scholar]
- 13.Yih LH, Lee TC. Arsenite induces p53 accumulation through an ATM-dependent pathway in human fibroblasts. Cancer Res. 2000;60:6346–6352. [PubMed] [Google Scholar]
- 14.Lenardo MJ, Baltimore D. NF-kappa B: a pleiotropic mediator of inducible and tissue-specific gene control. Cell. 1989;58:227–229. doi: 10.1016/0092-8674(89)90833-7. [DOI] [PubMed] [Google Scholar]
- 15.Yu Y, Wan Y, Huang C. The biological functions of NF-kappaB1 (p50) and its potential as an anti-cancer target. Curr Cancer Drug Targets. 2009;9:566–571. doi: 10.2174/156800909788486759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 17.Wang YJ, Wang JT, Fan QX, Geng JG. Andrographolide inhibits NF-kappaBeta activation and attenuates neointimal hyperplasia in arterial restenosis. Cell Res. 2007b;17:933–941. doi: 10.1038/cr.2007.89. [DOI] [PubMed] [Google Scholar]
- 18.Song L, Li J, Zhang D, Liu ZG, Ye J, Zhan Q, et al. IKKbeta programs to turn on the GADD45alpha-MKK4-JNK apoptotic cascade specifically via p50 NF-kappaB in arsenite response. J Cell Biol. 2006;175:607–617. doi: 10.1083/jcb.200602149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maillet A, Pervaiz S. Redox regulation of p53, redox effectors regulated by p53: a subtle balance. Antioxid Redox Signal. 2012;16:1285–1294. doi: 10.1089/ars.2011.4434. [DOI] [PubMed] [Google Scholar]
- 20.Xu Y. Regulation of p53 responses by post-translational modifications. Cell Death Differ. 2003;10:400–403. doi: 10.1038/sj.cdd.4401182. [DOI] [PubMed] [Google Scholar]
- 21.Fiaschi T, Chiarugi P. Oxidative stress, tumor microenvironment, and metabolic reprogramming: a diabolic liaison. Int J Cell Biol. 2012;2012:8. doi: 10.1155/2012/762825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bellodi C, Kopmar N, Ruggero D. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. EMBO J. 2010;29:1865–1876. doi: 10.1038/emboj.2010.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karawajew L, Rhein P, Czerwony G, Ludwig W-D. Stress-induced activation of the p53 tumor suppressor in leukemia cells and normal lymphocytes requires mitochondrial activity and reactive oxygen species. Blood. 2005;105:4767–4775. doi: 10.1182/blood-2004-09-3428. [DOI] [PubMed] [Google Scholar]
- 24.Ljungman M. Dial 9-1-1 for p53: mechanisms of p53 activation by cellular stress. Neoplasia. 2000;2:208–225. doi: 10.1038/sj.neo.7900073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stahnke K, Fulda S, Friesen C, Strauß G, Debatin K-M. Activation of apoptosis pathways in peripheral blood lymphocytes by in vivo chemotherapy. Blood. 2001;98:3066–3073. doi: 10.1182/blood.v98.10.3066. [DOI] [PubMed] [Google Scholar]
- 26.Yang DQ, Halaby MJ, Zhang Y. The identification of an internal ribosomal entry site in the 5[prime]-untranslated region of p53 mRNA provides a novel mechanism for the regulation of its translation following DNA damage. Oncogene. 2006;25:4613–4619. doi: 10.1038/sj.onc.1209483. [DOI] [PubMed] [Google Scholar]
- 27.Peterson RT, Schreiber SL. Translation control: connecting mitogens and the ribosome. Curr Biol. 1998;8:R248–R250. doi: 10.1016/s0960-9822(98)70152-6. [DOI] [PubMed] [Google Scholar]
- 28.Hutchinson JA, Shanware NP, Chang H, Tibbetts RS. Regulation of ribosomal protein S6 phosphorylation by casein kinase 1 and protein phosphatase 1. J Biol Chem. 2011;286:8688–8696. doi: 10.1074/jbc.M110.141754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo W, Yang Z, Xia Q, Liu J, Yu Y, Li J, et al. Arsenite stabilizes HIF-1alpha protein through p85alpha-mediated up-regulation of inducible Hsp70 protein expression. Cell Mol Life Sci. 2011;68:475–488. doi: 10.1007/s00018-010-0459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torres J, Pulido R. The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. J Biol Chem. 2001;276:993–998. doi: 10.1074/jbc.M009134200. [DOI] [PubMed] [Google Scholar]
- 31.Vazquez F, Ramaswamy S, Nakamura N, Sellers WR. Phosphorylation of the PTEN tail regulates protein stability and function. Mol Cell Biol. 2000;20:5010–5018. doi: 10.1128/mcb.20.14.5010-5018.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell. 2005;18:13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 33.Quinn JE, Kennedy RD, Mullan PB, Gilmore PM, Carty M, Johnston PG, et al. BRCA1 functions as a differential modulator of chemotherapy-induced apoptosis. Cancer Res. 2003;63:6221–6228. [PubMed] [Google Scholar]
- 34.Hellen CUT, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 35.Rossman TG, Klein CB. Genetic and epigenetic effects of environmental arsenicals. Metallomics. 2011;3:1135–1141. doi: 10.1039/c1mt00074h. [DOI] [PubMed] [Google Scholar]
- 36.Li J, Tang M-s, Liu B, Shi X, Huang C. A critical role of PI-3K//Akt//JNKs pathway in benzo[lsqb]a[rsqb]pyrene diol-epoxide (B[lsqb]a[rsqb]PDE)-induced AP-1 transactivation in mouse epidermal Cl41 cells. Oncogene. 2004;23:3932–3944. doi: 10.1038/sj.onc.1207501. [DOI] [PubMed] [Google Scholar]
- 37.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 38.Beezhold K, Liu J, Kan H, Meighan T, Castranova V, Shi X, et al. miR-190-mediated downregulation of PHLPP contributes to arsenic-induced Akt activation and carcinogenesis. Toxicol Sci. 2011;123:411–420. doi: 10.1093/toxsci/kfr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen A, Cao EH, Zhang TC, Qin JF. Arsenite-induced reactive oxygen species and the repression of alpha-tocopherol in the MGC-803 cells. Eur J Pharmacol. 2002;448:11–18. doi: 10.1016/s0014-2999(02)01901-5. [DOI] [PubMed] [Google Scholar]
- 40.Ogawara Y, Kishishita S, Obata T, Isazawa Y, Suzuki T, Tanaka K, et al. Akt enhances Mdm2-mediated ubiquitination and degradation of p53. J Biol Chem. 2002;277:21843–21850. doi: 10.1074/jbc.M109745200. [DOI] [PubMed] [Google Scholar]
- 41.Xu D, Wilson TJ, Chan D, De Luca E, Zhou J, Hertzog PJ, et al. Ets1 is required for p53 transcriptional activity in UV-induced apoptosis in embryonic stem cells. EMBO J. 2002;21:4081–4093. doi: 10.1093/emboj/cdf413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan W, Zhang Y, Zhang J, Liu S, Cho SJ, Chen X. Mutant p53 protein is targeted by arsenic for degradation and plays a role in arsenic-mediated growth suppression. J Biol Chem. 2011;286:17478–17486. doi: 10.1074/jbc.M111.231639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choudhuri T, Pal S, Agwarwal ML, Das T, Sa G. Curcumin induces apoptosis in human breast cancer cells through p53-dependent Bax induction. FEBS Lett. 2002;512:334–340. doi: 10.1016/s0014-5793(02)02292-5. [DOI] [PubMed] [Google Scholar]
- 44.Schuler M, Maurer U, Goldstein JC, Breitenbucher F, Hoffarth S, Waterhouse NJ, et al. P53 triggers apoptosis in oncogene-expressing fibroblasts by the induction of Noxa and mitochondrial Bax translocation. Cell Death Differ. 2003;10:451–460. doi: 10.1038/sj.cdd.4401180. [DOI] [PubMed] [Google Scholar]
- 45.Yu J, Wang Z, Kinzler KW, Vogelstein B, Zhang L. PUMA mediates the apoptotic response to p53 in colorectal cancer cells. Proc Natl Acad Sci USA. 2003;100:1931–1936. doi: 10.1073/pnas.2627984100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El-Deiry WS. The role of p53 in chemosensitivity and radiosensitivity. Oncogene. 2003;22:7486–7495. doi: 10.1038/sj.onc.1206949. [DOI] [PubMed] [Google Scholar]
- 47.Hinata N, Shirakawa T, Zhang Z, Matsumoto A, Fujisawa M, Okada H, et al. Radiation induces p53-dependent cell apoptosis in bladder cancer cells with wild-type-p53 but not in p53-mutated bladder cancer cells. Urol Res. 2003;31:387–396. doi: 10.1007/s00240-003-0355-9. [DOI] [PubMed] [Google Scholar]
- 48.Liu J, Weiss HL, Rychahou P, Jackson LN, Evers BM, Gao T. Loss of PHLPP expression in colon cancer: role in proliferation and tumorigenesis. Oncogene. 2009;28:994–1004. doi: 10.1038/onc.2008.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ying SY, Lin SL. Intron-derived microRNAs—fine tuning of gene functions. Gene. 2004;342:25–28. doi: 10.1016/j.gene.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 50.Zheng H, Chu J, Zeng Y, Loh HH, Law PY. Yin Yang 1 phosphorylation contributes to the differential effects of mu-opioid receptor agonists on microRNA-190 expression. J Biol Chem. 2010;285:21994–22002. doi: 10.1074/jbc.M110.112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang H, Hertlein E, Bakkar N, Sun H, Acharyya S, Wang J, et al. NF-kappaB regulation of YY1 inhibits skeletal myogenesis through transcriptional silencing of myofibrillar genes. Mol Cell Biol. 2007a;27:4374–4387. doi: 10.1128/MCB.02020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bellacosa A, Chan OT, Ahmed NN, Datta K, Malstrom S, Stokoe D, et al. Akt activation by growth factors is a multiple-step process: the role of the PH domain. Oncogene. 1998;17:313–325. doi: 10.1038/sj.onc.1201947. [DOI] [PubMed] [Google Scholar]
- 53.Jiang B-H, Aoki M, Zheng JZ, Li J, Vogt PK. Myogenic signaling of phosphatidylinositol 3-kinase requires the serine-threonine kinase Akt/protein kinase B. Proc Natl Acad Sci. 1999;96:2077–2081. doi: 10.1073/pnas.96.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]