Abstract
The Gal4-UAS regulatory system of yeast is widely used to modulate gene expression in Drosophila; however, there are limitations to its usefulness in transgenic zebrafish, owing to progressive methylation and silencing of the CpG-rich multicopy upstream activation sequence. Although a modified, less repetitive UAS construct may overcome this problem, it is highly desirable to have additional transcriptional regulatory systems that can be applied independently or in combination with the Gal4/UAS system for intersectional gene expression. The Q transcriptional regulatory system of Neurospora crassa functions similarly to Gal4/UAS. QF is a transcriptional activator that binds to the QUAS upstream regulatory sequence to drive reporter gene expression. Unlike Gal4, the QF binding site does not contain essential CpG dinucleotide sequences that are subject to DNA methylation. The QS protein is a repressor of QF mediated transcriptional activation akin to Gal80. The functionality of the Q system has been demonstrated in Drosophila and C. elegans and we now report its successful application to a vertebrate model, the zebrafish, Danio rerio. Several tissue-specific promoters were used to drive QF expression in stable transgenic lines, as assessed by activation of a QUAS:GFP transgene. The QS repressor was found to dramatically reduce QF activity in injected zebrafish embryos; however, a similar repression has not yet been achieved in transgenic animals expressing QS under the control of ubiquitous promoters. A dual reporter construct containing both QUAS and UAS, each upstream of different fluorescent proteins was also generated and tested in transient assays, demonstrating that the two systems can work in parallel within the same cell. The adoption of the Q system should greatly increase the versatility and power of transgenic approaches for regulating gene expression in zebrafish.
Keywords: Qa locus, QF activator, QS repressor, Gal4, transcriptional activation
1. Introduction
Transgenic tools for manipulation of gene expression are invaluable for labeling and tracking cell populations and for assessing genetic and cellular functions in developmental, physiological and behavioral studies. In zebrafish, transgenic approaches have been used extensively to test putative tissue-specific enhancers, monitor cell fates, modify or measure neural responses and to ablate cells for functional studies. In particular, two component or binary systems, such as the transcription factor Gal4 and the upstream activating sequence (UAS) to which it binds, produce high levels of gene expression and permit precise spatial and temporal regulation in vivo. The Gal4/UAS system has become the mainstay of genetic analyses in Drosophila and, coupled with other strategies (i.e., Gal80 repressor, LexA/lexO, Cre/loxP, Flp/frt), provides a powerful and versatile set of genetic tools for control of transcriptional activation (refer to [1]).
Adoption of the Gal4-UAS system has met with mixed success in the zebrafish model. The system works exceptionally well in transient assays of newly injected embryos, where high levels of gene expression can be attained by transcriptional activation from multicopy UAS sites [2]. However, in stable transgenic lines generated by Tol2-mediated transposition, CpG residues in the commonly used 10 or 14 copy upstream activator sequences (10XUAS or 14XUAS) are progressively methylated in each generation, eventually resulting in transcriptional silencing [3]. Depending on the chromosomal position of transgene integration, evidence of silencing can be found as early as in the F1 population [4]. Considerable effort has been expended on developing UAS-regulated transgenes for zebrafish by many laboratories because of the utility and flexibility of these reagents. However, transcriptional silencing of recovered transgenic lines has been a disappointing outcome and a frustrating investment in time and resources. Although it is possible to modify the copy number and repetitive nature of the UAS, for example with 4 non-repetitive upstream activator sequences 4XnrUAS, [4] expression levels from the recovered transgenes are usually not as high. For this reason, we sought to adapt another binary transcriptional regulatory system to the zebrafish.
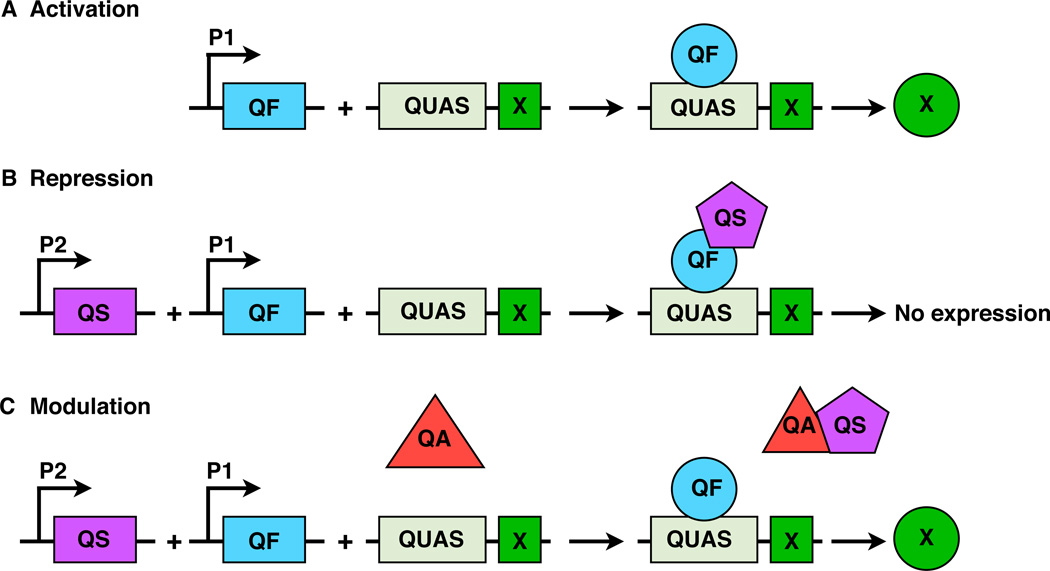
In the filamentous fungus Neurospora crassa, the quinic acid gene cluster (Qa) allows quinic acid to be used as a carbon source under conditions where glucose is limiting [5]. Components of the Qa locus include a gene encoding the transcription factor QF, a QF binding site known as QUAS that is found upstream of QF regulated genes, and a gene encoding QS, a repressor that inhibits QF from activating QUAS. The inhibitory interaction between QS and QF can also be blocked through the addition of quinic acid (Fig. 1). The so-called Q system was successfully applied to regulate gene expression in Drosophila, in cultured mammalian cells [6] and more recently in C. elegans [7].
Fig. 1. Components of the Q regulatory system.
(A) The transcription factor QF activates QUAS-regulated genes to produce reporter or effector proteins (indicated by X). (B) QF activity is blocked by the repressor QS. (C) QS repression of the QF/QUAS interaction is alleviated by addition of quinic acid (QA). P1 and P2 represent promoter/enhancer sequences for QF and QS transcription.
In this article, we describe the generation of Q reagents for the production of transgenic zebrafish by Tol2 transposition. We demonstrate that QF robustly activates QUAS-driven fluorescent reporter genes in transient assays of injected embryos and in progeny from matings between stably recovered transgenic driver and reporter lines. Constitutive, low-level reporter expression was observed from QUAS-regulated reporters introduced maternally; however, fluorescent labeling was not detected in embryos generated by fertilization with QUAS:GFP transgenic sperm. Under the control of a number of different tissue-specific promoters, QF activated QUAS reporters in the expected cell types in stable transgenic lines and, after 3 generations, we have not observed significant transcriptional silencing of a QUAS-regulated gene. QS significantly represses QF activity in transient assays; however, effective QF repression from an integrated QS transgene has yet to be achieved. Strategies for using the Gal4/UAS and QF/QUAS systems in parallel or in intersectional approaches have also been initiated. The presented results indicate that the Q system of Neurospora is a promising alternative to other binary approaches for transcriptional regulation in transgenic zebrafish. The adoption of these tools, and their use in conjunction with existing methods for spatial and temporal control of gene expression, will expand the repertoire and versatility of techniques for manipulating gene activity in developing and adult zebrafish.
2. QF activation of QUAS-regulated gene expression in zebrafish
2.1. Generation of QF driver constructs to activate reporter gene expression
To determine whether the QF transcription factor would function appropriately to activate expression of QUAS-regulated genes in zebrafish, we first subcloned the same QF and QUAS sequences that had been used for Drosophila into vectors modified for Tol2 transposition. When injected with RNA encoding the Tol2 transposase, plasmids containing 5' and 3' Tol2 recognition sequences (i.e., Tol2 arms) integrate into the zebrafish genome with a high efficiency and show an increased frequency of germ line transmission [8].
Initially, we sought to drive QF activity widely using the elongation factor 1 alpha (EF1) promoter from Xenopus [9], which has frequently been employed in studies where ubiquitous gene expression is desired in the early zebrafish embryo [10, 11]. A fragment containing the QF coding sequence and SV40 termination sequence (3.19 kb) was excised from the pattB-QF plasmid [6] and cloned into the Tol2 plasmid pT2KXIG [12] directly downstream of the EF1 promoter. We refer to this QF driver plasmid as pT2KEF1 :QF.
During the course of this work, another strong, ubiquitous promoter was identified from the zebrafish ubiquitin B (ubb) gene that drives expression as early as 4 hours post fertilization (hpf) [13]. A pBT2ubb:QF plasmid was produced by PCR amplifying the ubb promoter from pENTR5’_ubi (L4-R1) [13] and inserting the fragment (3.48 kb) into the pBT2 plasmid upstream of QF. This plasmid also contains a reporter cassette with the promoter from the Xenopus laevis gamma-crystallin (gcry1) gene driving the blue fluorescent protein (BFP) gene. BFP labeling of the lens of the eye permits identification of transgenic founders by screening embryos under a Pacific Blue 31037 filter set (Chroma Technology Corp). To generate the pBT2ubb:QF; gcry1:BFP plasmid, a fragment containing gcry1:BFP (1.26 kb) was excised from pKTol2gC-TagBFP (a gift from Karl Clark) and cloned into the SalI site of pBT2ubb:QF.
2.2. Generation of QUAS reporter constructs
For generation of QUAS driven transgenes, we adopted the QUAS construct successfully used in Drosophila and mammalian cells [6]. To produce a Tol2 QUAS:GFP reporter plasmid, a fragment (451bp) containing five 16 bp QUAS (5XQUAS) QF binding sites upstream of the green fluorescent protein (GFP) gene was digested from plasmid pQUAST [6] using MluI and XhoI. This fragment was subcloned in reverse orientation into a MluI restriction site of the Tol2 plasmid pBT2 (gift of Shannon Fisher) upstream of the carp β-actin minimal promoter [12] and the GFP coding sequence. We refer to this 5XQUAS reporter plasmid as pBT2QUAS:GFP. As described below, transcriptional activation was not impeded by reversal of the 5XQUAS relative to the transcriptional start site of GFP.
2.3. QF activation of QUAS:GFP in transient assays of injected embryos
An advantage of the zebrafish system is that constructs can be readily assayed by injecting plasmids into the yolks of newly fertilized embryos, optimally at the 1–2 cell stage, and examining gene expression a day later. For such experiments, wild-type (WT) embryos were obtained from group matings between adults of the Oregon AB strain [14]. To test QF activation of QUAS, the pT2KEF1 QF and pBT2QUAS:GFP plasmids were co-injected into 1–2 cell embryos at varying concentrations (1, 5, 10, 15 or 25 ng/ l) in the presence of Tol2 transposase RNA (25 ng/ l), and 0.2% phenol red to gauge the quality of the injection. After 1 day post-fertilization (dpf), all embryos injected with 25 ng of both plasmids were GFP positive (Fig. 2A), although survival was poor at 57% (n=84). Survival greatly improved as the plasmid concentrations were decreased (15 ng resulted in a survival rate of 74% (n=118), 10 ng in 88% (n=97), 5 ng in 95% (n=90) and 1 ng in 91% (n=57)), with all embryos showing robust GFP labeling after 1 dpf. These experiments demonstrated that, at the lower plasmid concentrations, ubiquitous QF reproducibly activated a QUAS reporter in developing zebrafish embryos with minimal toxicity.
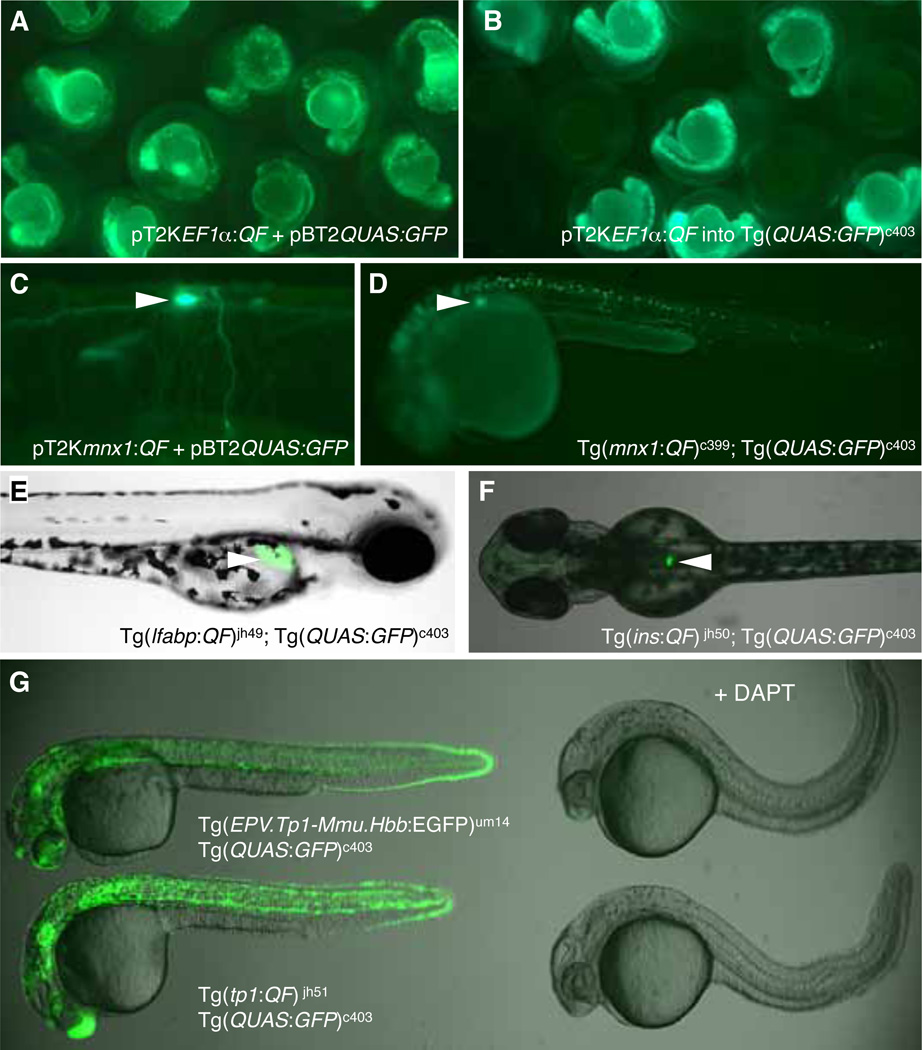
Fig. 2. QF activity in transient assays of zebrafish embryos and in stable transgenic lines.
(A) Embryos co-injected with pTK2EF1 :QF and pBT2QUAS:GFP plasmid show strong GFP labeling at 1 dpf. (B) Ubiquitous GFP labeling at 1 dpf following injection of pTK2EF1 :QF plasmid into embryos from the Tg(QUAS:GFP)c403 stable line. (C) Mosaic labeling of motor neurons at 4 dpf following co-injection of pT2Kmnx1:QF and pBT2QUAS:GFP. When mated to the Tg(QUAS:GFP)c403 reporter line, the recovered QF driver lines (D) Tg(mnx1:QF)c399 (E) Tg(lfabp:QF) and (F) Tg(insulin:QF), respectively, show tissue-specific labeling of motor neurons (white arrowhead, 1 dpf) and the presumptive pancreas (white arrowhead, 2 dpf) liver hepatocytes (white arrowhead, 2.5 dpf) and pancreatic β-cells (white arrowhead, 2 dpf). (G) Robust labeling is observed at 30 hpf with the Tg(tp1:QF) driver, when compared to the Notch-responsive line Tg(EPV.Tp1-Mmu.Hbb:EGFP)um14 (renamed from Tg(Tp1bglob:eGFP)um14 [26]). Exposure to DAPT (50 M) blocks Notch signaling and GFP labeling.
3. QF activation of QUAS:GFP from stably integrated transgenes
3.1. Production of QUAS:GFP transgenic reporters
For generation of a stable transgenic reporter line, pBT2QUAS:GFP was injected into 1–2 cell stage AB embryos (1 nl of 25 ng/ l) together with RNA encoding Tol2 transposase (25 ng/ l) and phenol red (0.05%). Of the injected embryos (F0), 90% (n=98) survived and were raised to adulthood. To identify transgenic founders, the F0 adults were mated with AB fish and their progeny injected with pT2KEF1 :QF at the 1 cell stage (Fig. 2B). Out of 34 adults screened, 5 generated embryos that showed robust GFP labeling at 1 dpf in the presence of the EF1 :QF driver. One female (c403) that produced a higher frequency of GFP-labeled embryos (19%; n=84) was mated with AB males and the embryos raised to establish the transgenic line Tg(QUAS:GFP)c403. Heterozygous F1 adults were identified by matings with stable QF driver lines that are described below.
3.2. Effect of parent of origin on expression of QUAS-regulated transgenes
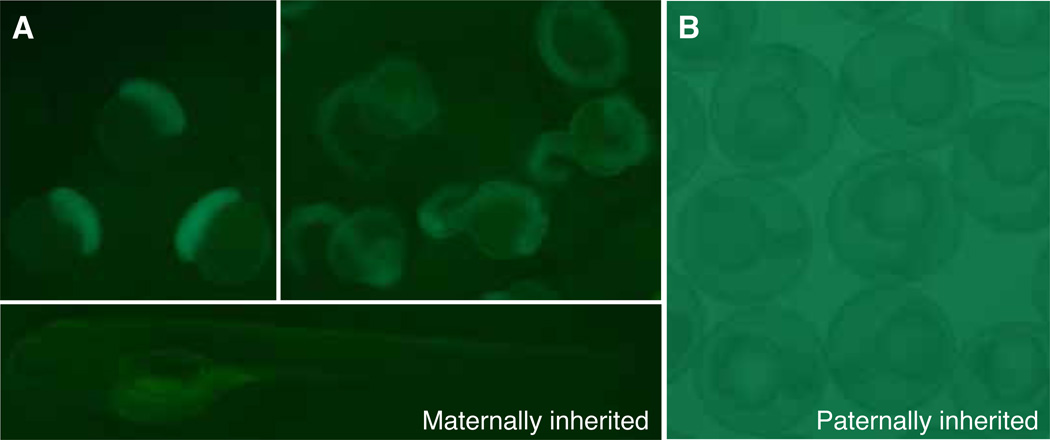
In the course of identifying QUAS:GFP transgenic founders, we observed low levels of fluorescence in some embryos being screened, even in the complete absence of QF activity. Upon examining the progeny of F1 adults, we discovered that when the reporter transgene was derived from the female genome, transgenic offspring showed homogeneous, faint labeling as early as the 1–2 cell stage (Fig. 3A). This basal GFP expression was no longer detected by 4 dpf. In contrast, when the QUAS:GFP transgene was introduced from the paternal genome, fluorescence was not observed in fertilized embryos (Fig. 3B). Thus, because of low level labeling presumably derived from basal expression in the female germ line and maternally deposited GFP, paternal transmission of QUAS-regulated transgenes is recommended.
Fig. 3. Parent of origin influences expression of QUAS:GFP transgene.
(A) In the absence of QF, embryos derived from Tg(QUAS:GFP)c403 mothers show ubiquitous GFP labeling from 2.5 hpf to 1 dpf, which gradually diminishes by 4 dpf. This is likely due to activation of the QUAS in oocytes and maternal deposition of GFP. (B) GFP labeling is not detected in the progeny of Tg(QUAS:GFP)c403 fathers (shown is 1 dpf).
3.3. Production of tissue-specific QF driver lines
Following the demonstration that QF could activate transcription from an integrated QUAS:GFP transgene, we proceeded to generate stable QF driver lines using known, tissue-specific zebrafish promoters. The mnx1 (formerly Hb9) promoter, originally identified in mouse [15], had been isolated from zebrafish and found to drive expression in spinal motor neurons, the epithalamus and pancreatic beta cells [16, 17]. A fragment (3.07 kb) encoding the promoter region was PCR amplified from pmnx1:GFP (provided by M. Granato) and ligated simultaneously with the QF coding and SV40 termination sequence into pT2KXIG [12] to produce pT2Kmnx1:QF.
After confirming that co-injection of pT2Kmnx1:QF (1 nl of 12.5 ng/ l) and pBT2QUAS:GFP (1 nl of 25 ng/ l) along with RNA encoding Tol2 transposase (25 ng/ l) led to robust GFP labeling in primary motor neurons at 4 dpf as well as in muscle fibers (Fig. 2C), we raised embryos injected with both plasmids as well as those injected only with the pT2Kmnx1:QF construct. To identify founders, the F0 adult fish were mated with Tg(QUAS:GFP)c403 heterozygotes. Out of 26 adults screened, 3 generated embryos that showed GFP labeling in motor neurons, the pineal anlage and the presumptive pancreas at 1 dpf (Fig. 2D). Embryos from two of the Tg(mnx1:QF) founders also showed extensive labeling of muscle fibers. To establish and propagate the Tg(mnx1:QF)c399 line, we used the third founder, which yielded embryos with fewer labeled muscle cells and strong fluorescence in the spinal cord and the pineal gland. This labeling persists in the nervous system of adult fish (data not shown).
To further validate the specificity of QF drivers, we generated three constructs; each designed to express QF in a discrete population of cells, namely, the hepatocytes of the liver, the insulin producing β-cells of the pancreas and cells undergoing Notch signaling. We used the Tol2 plasmid pT2KXIG [12] to introduce the QF coding sequence along with the SV40 polyA signal and a cassette consisting of the promoter/enhancer of beta B1 crystallin (crybb1) driving the enhanced cyan fluorescent protein (CFP) gene fused to the PEST destabilization domain [18]. Regulatory sequences for the liver fatty acid binding protein (lfabp) gene [19, 20] or the insulin (ins) gene [21] were PCR amplified and cloned upstream of the QF coding sequence using the In-Fusion® PCR cloning system (Clontech). To express QF in cells undergoing Notch signaling, we utilized a known reporter of Notch pathway activation. Following cleavage, the Notch intracellular domain (NICD) translocates to the nucleus, where it associates with the RBP-Jκ transcription factor and activates transcription of target genes. A six copy concatemer of the enhancer from the viral terminal protein 1 (tp1) gene, which provides a total of 12 Rbp-Jκ binding sites [22, 23], was placed upstream of the minimal promoter of rabbit β-globin [24, 25] and the QF coding sequence. The three Tol2 constructs were injected separately with Tol2 transposase mRNA [8] and germ line transmission was monitored by the expression of CFP in the larval lens at 4 dpf.
Tissue-specific QF activity was gauged by mating the QF driver lines to the Tg(QUAS:GFP)c403 reporter and observing GFP labeling in the resultant progeny. As expected, in Tg(lfabp:QF; crybb1:eCFP-PEST)jh49 (abbreviated to Tg(lfabp:QF)jh49) larvae, GFP expression was hepatocyte-specific (Fig. 2E) and was restricted to pancreatic β-cells in Tg(ins:QF; crybb1:eCFP-PEST)jh50 (abbreviated to Tg(ins:QF)jh50) individuals (Fig. 2F).
The Notch-responsive activity of Tg(tp1:QF; crybb1:eCFP-PEST)jh51 (abbreviated to Tg(tp1:QF)jh51) was demonstrated by mating to Tg(QUAS:GFP)c403 fish and incubating embryos in 50 µM of the γ-secretase inhibitor N-[N-(3, 5-Difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT; Sigma, D5942) from 3 to 24 h of development. Preventing γ-secretase activity blocks endogenous Notch signaling and, therefore, should inhibit Notch-dependent transcriptional activation of QF [25]. The tp1:QF transgene behaved in the expected Notch-responsive fashion (Fig. 2G) and, in the absence of DAPT, GFP labeling was enhanced relative to embryos from a previously established Notch-responsive line [26].
4. Repression and modulation of QF activity
4.1. QS blocks QF-driven expression in transient assays
The third component of the Q-system, the repressor QS, inhibits QF activation of QUAS-regulated genes. QS expressed under the control of the Drosophila tubulin promoter or the C. elegans DA neuron-specific unc-4c promoter blocked QF-mediated transcriptional activation in fly and worm embryos, respectively [6, 7]. A high QS:QF ratio was required for transcriptional repression of QUAS-regulated gfp expression in Drosophila [6].
To test whether the QS repressor restricts QF transcriptional activation in zebrafish, we expressed QS under the control of the ubiquitous EF1 and ubb promoters [13]. A fragment containing the QS coding sequence and SV40 polyA tail (3.65 kb) was PCR amplified from pCasper4-tubulin-QS [6] and cloned downstream of the EF1 promoter within the pBT2 backbone. To facilitate the identification of transgenic founders, as above, the gcry1:BFP reporter was inserted in the pBT2EF1 :QS plasmid to produce pBT2EF1 :QS; gcry1:BFP. The pBT2ubb:QS; gcry1:BFP plasmid was generated by replacing EF1 with a zebrafish ubb promoter fragment PCR amplified from pENTR5’_ubi (L4-R1) [13]
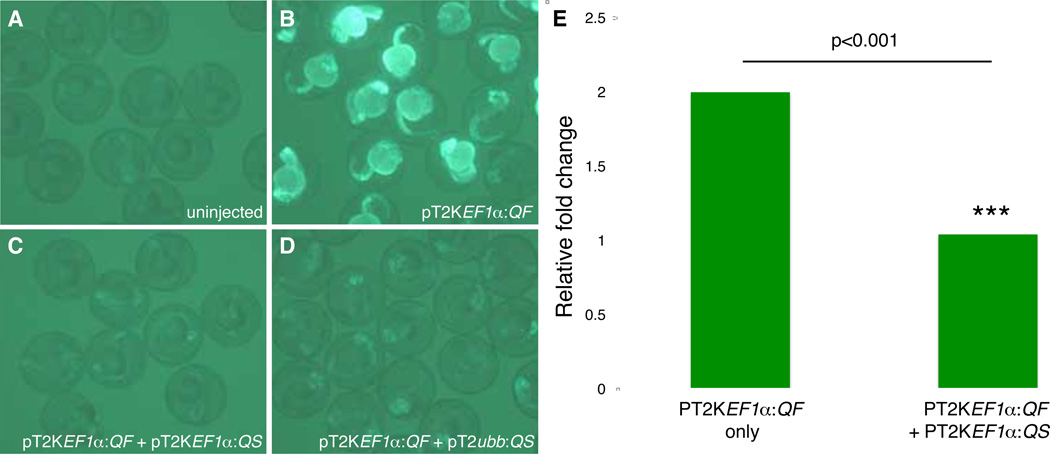
To assess QS repression of QF, we performed a transient assay by co-injecting pT2KEF1 :QF (3.2 ng/ l) with either pT2KEF1 :QS (50 ng/ l) or pT2Kubb:QS (50 ng/ l) into Tg(QUAS:GFP)c403 heterozygous embryos at the 1-cell stage. QF-dependent GFP labeling was significantly reduced in the presence of either QS construct (Fig. 4A–D). To quantify QS suppression more objectively, we imaged GFP positive embryos at 1 dpf using a SPOT Xplorer 1.4 monochrome camera mounted on a Leica MZFLIII fluorescent stereomicroscope. Fluorescence intensity measurements (i.e., the mean gray value) were obtained for individual embryos using ImageJ 1.42q software (rsbweb.nih.gov/ij/) and compared by one–way ANOVA followed by Turkey’s post-hoc comparison using R v2.15.2 software (R Core Team, 2012; http://www.R-project.org/). The average intensity of GFP labeling was reduced by approximately 50% in Tg(QUAS:GFP)c403 embryos co-injected with pT2KEF1 :QF and pBT2EF1 :QS compared to those injected with pT2KEF1 :QF alone (n=38; Fig. 4E). No difference in fluorescence intensity was found between pT2KEF1 :QS injected and uninjected embryos. Moreover, the presence of QS does not suppress fluorescent reporter expression from QF independent transgenic lines (data not shown).
Fig. 4. QS represses QF activity.
(A) Tg(QUAS:GFP)c403 embryos were uninjected, (B) injected with the QF driver construct pT2KEF1 QF, co-injected with (C) pT2KEF1 QF and the QS repressor plasmid pT2KEF1 QS or with (D) pT2KEF1 QF and the QS repressor plasmid pT2Kubb QS. (A–D) are 1 dpf. (E) Quantification of GFP intensity in Tg(QUAS:GFP)c403 embryos that received QF driver plasmid alone (T2KEF1 QF, averaged from n=29) or in combination with pT2KEF1 QS (averaged from n=25) relative to baseline levels (uninjected controls, n=38). GFP labeling was significantly reduced in the presence of the QS plasmid (Student’s T test, p<0.001).
To produce a QS transgenic line, pBT2EF1 :QS; gcry1:BFP (12.5 ng/ l) was co-injected with mRNA encoding Tol2 transposase (25 ng/ l) in phenol red (0.2%) into 1-cell stage embryos, which were raised to adulthood. F0 adults were mated with AB and their progeny screened for BFP labeling in the lens using a Leica MZFLIII fluorescent stereomicroscope. Out of 55 adults screened, one F0 male (c430) produced a single BFP-labeled embryo. This embryo was raised and mated with AB adult fish to establish the transgenic line Tg (EF1 :QS; gcry1:BFP)c430.
To test whether transgenic F2 embryos produce sufficient levels of QS expression to repress QF-mediated activation of QUAS:GFP, we mated F1 adults with doubly transgenic fish carrying Tg(mnx1:QF)c399 and Tg(QUAS:GFP)403. No difference was detected in the intensity of GFP-labeled motor neurons in embryos that had inherited the QS transgene (as indicated by BFP expression in the lens) compared to siblings that had not. Additional experiments are required to determine the number of transgenic insertions individual embryos carry and to calibrate the amount of transgene-derived QS that is needed for QF repression in vivo.
4.2. Evaluating the effect of quinic acid exposure on embryonic zebrafish
In Drosophila and C. elegans, quinic acid blocks QS repression of QF thereby permitting transcription of QUAS-regulated genes [6, 7]. Because quinic acid seems to function in an opposite manner in mammalian cultured cells and, surprisingly, enhances QF repression [6], we do not have a clear idea of how it might influence the Q system in zebrafish cells. Rigorous testing of the action of quinic acid in zebrafish embryos, will require the establishment of invariable conditions for QS repression of QF, which is difficult to achieve in transient assays. However, as a first step toward this goal, we have assessed the effects of prolonged exposure to quinic acid on zebrafish embryos.
We found that doses of quinic acid greater than 0.5 mg/ml in fish water are toxic whether provided at 2.5 hpf or at 1.5 dpf. A dose of 0.5 mg/ml is not lethal but results in developmental abnormalities. Morphologically normal embryos and larvae are obtained with concentrations of 0.3 mg/ml or lower for up to 5 dpf of treatment. There was no difference when quinic acid was added to system water or water containing 1-Phenyl-2-Thiourea (PTU) (0.003%) to inhibit melanin formation [27]. These initial tests of toxicity should serve as a useful guideline for determining whether quinic acid application can alleviate or enhance QS-mediated suppression of QF once optimal conditions for this interaction are achieved using stable transgenic lines.
5. Dual reporter system for activation by QF and Gal4
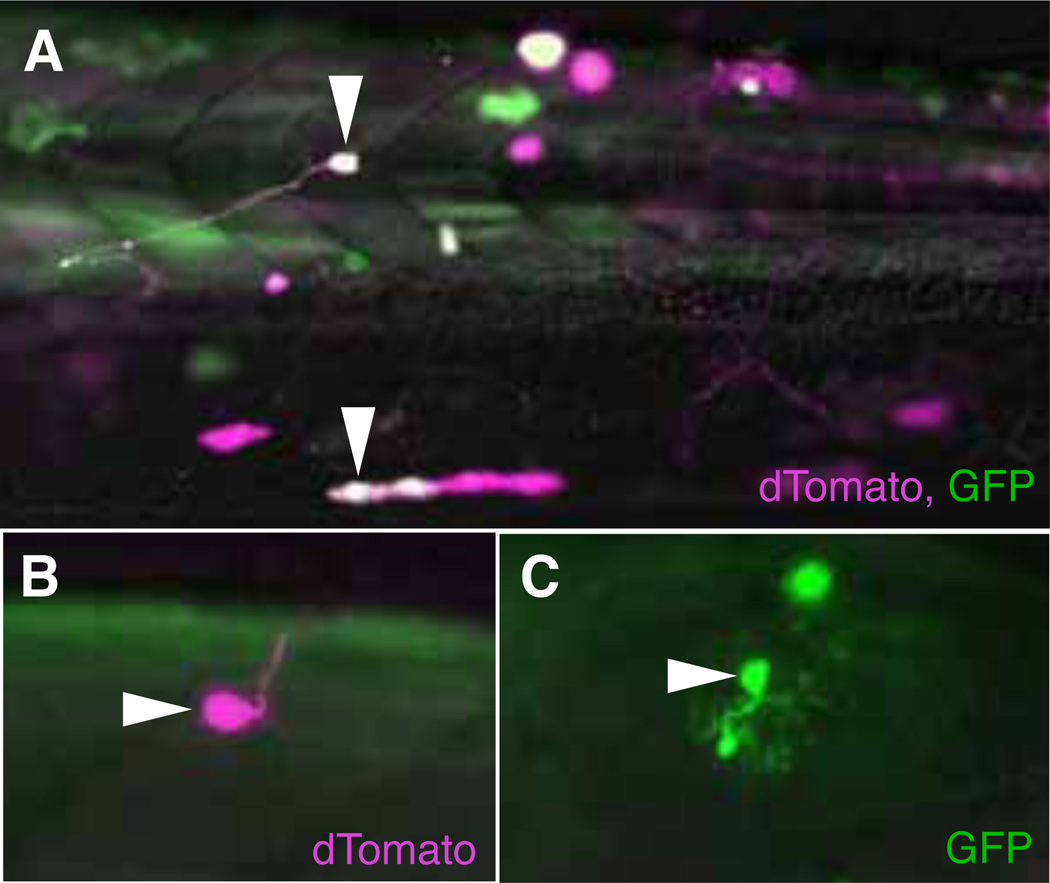
One drawback to the zebrafish model is the length of time it takes to introduce several transgenes into a single animal, particularly if homozygosity is beneficial for expression. To simplify intersectional strategies, we tested whether it would be possible to use a single construct or “dual reporter”, in which UAS controls expression of one reporter gene and QUAS regulates another. In the pBT2QUAS:dTomato-4XnrUAS:GFP plasmid, the two transcription units were placed in reverse orientation separated by plasmid sequence (160 bp) or by the crybb1:CFP cassette [18]. Injection of the dual reporter plasmid (12.5 ng/ l) together with ubb:QF and EF1 :Gal4 plasmids yielded embryos that showed the expected mosaic expression, with dTomato and GFP labeling of small numbers of cells (Fig. 5A). Infrequently, both fluorescent proteins were expressed in the same cell, indicating the presence of all three plasmids.
Fig. 5. QF and Gal4 activation from a dual reporter construct.
(A) Co-injection of the pBT2QUAS:dtomato-4xnrUAS:GFP dual reporter with QF (pT2Kubb QF) and Gal4 (pT2KEF1 Gal4) driver constructs results in mosaic labeling, with some cells expressing dTomato, GFP, or both fluorescent proteins (white arrowheads, 2 dpf, lateral view). (B) dTomato labeled motoneuron (white arrowhead) and (C) GFP labeled hindbrain neuron (white arrowhead) in the same 3 dpf larva, following injection of the dual reporter plasmid into a Tg(mnx1:QF)/+; Tg(ptf1:Gal4)/+ doubly transgenic embryo.
To confirm that tissue-specific labeling could be obtained from the dual reporter plasmid, we mated Tg(mnx1:QF)c399 fish to the Gal4 driver line Tg(ptf1a:Gal4-VP16)jh16 that expresses Gal4-VP16 in hindbrain neurons and in the pancreas [26]. Injection of the pBT2QUAS:dTomato-4XnrUAS:GFP dual reporter into 1-cell stage embryos resulted in dTomato labeled motor neurons and GFP labeled hindbrain neurons (Fig. 5B,C).
We co-injected pBT2QUAS:dTomato-4XnrUAS:GFP (12.5 ng/ l) and mRNA encoding Tol2 transposase (25 ng/ l) in phenol red (0.2%) into 1-cell stage embryos to generate a stable transgenic line. Of 12 adults mated to AB fish, 3 were determined to be F0 founders by a fraction of their progeny showing CFP labeling in the lens at 2 dpf. When F1 embryos were raised to adulthood, mated, and their progeny co-injected with pBT2ubb:QF (5 ng/ l) and pBT2EF1 :Gal4 (5 ng/ l) plasmids, dTomato positive, GFP positive and doubly-labeled cells were obtained (data not shown).
6. Generation of Tol2 Gateway compatible reagents for the Q system
To facilitate the rapid production of Q-based reagents in backbones with Tol2 arms, we subcloned the Q components into entry vectors for the Gateway Cloning Technology system (Invitrogen, Life Technologies).
The 5XQUAS was inserted into a 5' entry clone (p5E) to enable transcriptional control of downstream reporters. p5E-QUAS (L4-R1) contains a 563 bp fragment that includes the 5XQUAS and the carp -actin minimal promoter assembled through Gateway BP recombination into the donor vector pDONR P4-P1R (Invitrogen, Life Technologies). The QF and QS genes were introduced as middle entry clones (pME), enabling their regulation by any promoter of choice present in a 5' entry clone. QF and QS coding sequences along with the SV40 termination signal (3.18 and 3.66 kb) were PCR amplified from pattB-QF and pCasper4-tubulin-QS [6], respectively, and recombined into donor vector pDONR 221 (Invitrogen) to create the pME-QF and pME-QS middle entry clones.
To assist in the identification of transgenic founders, we also produced a p3E-crybb1:CFP 3' entry clone by PCR amplification of a fragment (1.52 kb) from pBT2-crybb1:CFP. The final recombination reactions were performed using the tripartite destination vector pDestTol2pA2 that had been previously modified for Tol2 transposition [28].
The Gateway reagents were tested by assembling pENTR5’_ubi (L4-R1) [13] with pME-QF (L1-L2) and p3'- crybb1:CFP (R2-L3) into pDestTol2pA using the LR clonase II enzyme mix (Invitrogen, Life Technologies). The recombined vector (25 ng) along with phenol red (0.2%) was injected into Tg(QUAS:GFP)c403 heterozygous embryos at the 1-cell stage. Broad GFP labeling was observed in at 1 dpf (data not shown), indicating that the QF Gateway construct was functional.
7. Discussion
Reliable and effective tools to modulate gene expression in vivo are an important priority for zebrafish researchers. Modifications of the yeast Gal4/UAS system that promote high levels of activity have been valuable in transient assays of newly injected embryos, but more problematic for generating and maintaining high-expressing, stable transgenic lines. With the advent of Tol2 transposon-mediated transgenesis, it was thought that single copy integrations of UAS-regulated transgenes might resolve these issues; however, many groups have observed gradual silencing of transgenic lines (refer to [29–31] in part due to CpG methylation at multicopy UAS binding sites for Gal4 [3, 4]. Efforts have also been made to modify bacterial regulators for use in transgenic zebrafish, such as an inducible LexA repressor-progesterone receptor fusion protein [18] or a tetracycline-inducible transcriptional activator (TetA) [32]. These approaches have not yet been widely adopted by the field, perhaps on account of the requirement for chemical ligands to induce expression, or the paucity of reagents currently available for tissue-specific regulation. Similar reagents have shown evidence of leaky expression and lower levels of activation in Drosophila compared to Gal4 (refer to [33, 34]).
We turned to the Q transcriptional regulatory system of Neurospora crassa because it had been successfully applied to modulate gene expression in Drosophila and mammalian cells [6], and more recently in C. elegans [7]. Moreover, in cultured mammalian cells, the QF/QUAS system achieved a much higher level of activation of reporter gene expression compared to Gal4/UAS-mediated induction [6]. The Gal4/UAS and QF/QUAS systems also function independently and do not influence one another, which is important for devising intersectional strategies [1, 6].
Our initial results on the application of the Q system in zebrafish are promising and will likely stimulate further validation and the generation of additional resources. QF driven by known tissue-specific promoters induces robust expression of a QUAS reporter in the appropriate temporal and spatial patterns in stable transgenic zebrafish embryos and larvae. Importantly, after 4 generations, we have not found evidence of diminished expression, an obvious advantage over multicopy UAS-regulated genes activated by Gal4. Preliminary data from DNA bisulfite sequencing also suggested that CpG methylation of the QUAS region was minimal in larvae from the F3 generation (M. Goll, unpublished observations).
Despite these encouraging findings, some issues remain to be addressed for optimization of the Q system in zebrafish. First, in the complete absence of QF, low levels of constitutive activity are observed from QUAS-regulated transgenes that are introduced from the maternal genome. Embryos that inherited the QUAS:GFP transgene from their mothers showed ubiquitous, albeit dim, fluorescent labeling that persists for several days. Mutating nucleotides that are not essential for QF binding might eliminate constitutive promoter/enhancer activity of the multicopy QUAS element. However, a simpler option is to introduce QUAS-regulated transgenes from the paternal genome, since GFP labeling was undetectable in the Tg(QUAS:GFP)/+ heterozygous progeny of males bearing the transgene. We are also currently testing for parent of origin expression of transgenes driven by the 5X QUAS maintained in the orientation originally used in Drosophila [6]. Although we have not found evidence for loss of expression of QUAS-regulated transgenes in the F4 generation, elimination of nonessential CpG dinucleotides would reduce the potential for progressive silencing of transgenes by DNA methylation. Thus, a systematic mutagenesis approach to identify a modified QUAS that is only activated in the presence of QF, is an ideal target for QF binding, and refractory to transcriptional silencing would be of benefit.
Another potential concern is the toxicity of QF. High levels of Gal4-VP16 activity are known to be toxic to the early zebrafish embryo [2, 35] and our transient assays of QF under control of ubiquitous promoters also indicate that, at high concentration, QF can be detrimental to normal development. The recovery of multiple, transgenic lines in which tissue-specific promoters drive full-length QF, that in turn induces robust expression of a QUAS:GFP transgene, suggests that toxicity may only be a problem when QF is provided throughout the early embryo rather than in restricted cell types. Toxicity had been previously noted in Drosophila as well, when QF was under the regulation of certain promoters [6]. To remedy this, new, truncated versions of QF have been generated that are fully active and less toxic in Drosophila (C. Potter, unpublished observations), but have yet to be tested in zebrafish.
Although we were able to achieve strong repression of QF activity by QS in zebrafish embryos co-injected with plasmids, this has not been accomplished using stable transgenes. In equimolar amounts, QS does not adequately suppress QF [6]. In zebrafish, it may therefore be necessary to increase QS transgene copy number and expression levels [33, 36] or select for low expressing QF driver lines, in order to modulate the ratios of the two proteins and determine the optimal range for QF activation as well as repression in the presence of QS. Until such conditions are established, it is difficult to assess with accuracy the ability of quinic acid to alleviate the QS block in developing zebrafish. However, we were able to determine the maximum concentration of quinic acid that is compatible with embryonic growth and survival. An additional point to consider in future studies is the timing of reactivation of QUAS-regulated transgenes after the addition of quinic acid. Recovery of QF activity appears to be faster in transgenic worms than in flies exposed to the same quinic acid concentration [7].
For any transcriptional regulatory system to be feasible and widely accepted, the generation of a large number of useful reagents is needed. Similar to Gal4, QF can be used to isolate and assemble a collection of transgenic driver lines for activation of QUAS reporter/effectors in specific cells or tissues of interest. QF Tol2 constructs have been adapted for enhancer trapping (M. Macurak and M.E. Halpern, unpublished observations), to recover numerous, diverse cell type-specific QF driver lines efficiently.
Having another well-developed gene regulatory system for the zebrafish will expand the combinatorial strategies that can be used for increasingly more refined control of gene expression and cell labeling. Because there does not appear to be any interaction between Gal4/UAS and QF/QUAS in Drosophila [6] or in the zebrafish, the two systems can be used in parallel in the same cells. Two binary, repressible systems allow, for example, the Gal4 repressor Gal80 to be regulated by QF, QS to be regulated by Gal4 or QF expression to require Gal4, which, respectively, produces intersectional expression where Gal4 is inhibited in a subset of QF-expressing cells, QF is inhibited in a subset of Gal4-expressing cells, or there is overlapping expression in cells that produce both Gal4 and QF [34]. The number of generations that are required to introduce multiple transgenes into a single animal and the length of generation time, however, make such strategies seem daunting for the zebrafish. Our initial results injecting a dual QUAS and UAS reporter plasmid into embryos obtained from matings between Gal4 and QF driver lines suggest that it may be possible to accelerate the process. However, although transgenic founders were identified as carrying the dual reporter, mating of these fish to QF or Gal4 driver lines has not resulted in fluorescent labeling of cells. Thus, in the context of a zebrafish chromosome, the dual reporter might be refractory to transcriptional activation that can only be overcome through injection of a high concentration of plasmids bearing the same drivers. Additional work is required to determine the most effective configuration, distance and termination sequences for QUAS- and UAS-mediated transcription within the same transgenic region. Coupling the Q system with recombination based approaches, such as Cre or Flp recombinases, or employing split QF reagents [7] should further expand the repertoire of techniques for selective regulation of gene expression in zebrafish.
Finally, it is essential for the widespread adoption of a new gene regulatory system to develop reagents that can be readily modified by other researchers are compatible with currently employed methods. Gateway recombination cloning is an efficient strategy for the production of reporter or effector Tol2 constructs under the control of tissue-specific promoters [28, 37] or transcriptional regulators, and has become favored by many zebrafish researchers for the preparation of transgenic constructs. For ready distribution and ease of use, we have placed the QF, QUAS and QS components into the same three-insert Gateway system that was the basis of the Tol2 kit initially produced by the Chien laboratory [28]. With the Gateway cloning system, it will be straightforward to generate new transgenic vectors for targeted cell ablation, multicolor fluorescent labeling and optogenetic approaches in the zebrafish, under the control of the Q system.
Highlights.
Q transcriptional regulatory system of Neurospora crassa functions in zebrafish
Tissue-specific QF driver lines activate a QUAS:GFP transgenic reporter
Silencing of QUAS-regulated transgenes not observed in F4 generation
Q reagents cloned into Tol2 Gateway vectors for ease of use and distribution
Acknowledgments
We thank Brittany Hay, Marlow Minor and Gregory Moore for zebrafish care, Mahmud Siddiqi for assistance with fluorescence intensity measurements and Steven D. Leach for his valuable input on this study. This work was supported by grants from the NIH (R01DK080730) and the Juvenile Diabetes Research Foundation to MJP and by a joint NIH grant (1R01HD058530) to MJP and MEH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.del Valle Rodriguez A, Didiano D, Desplan C. Nat Methods. 2012;9:47–55. doi: 10.1038/nmeth.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koster RW, Fraser SE. Dev Biol. 2001;233:329–346. doi: 10.1006/dbio.2001.0242. [DOI] [PubMed] [Google Scholar]
- 3.Goll MG, Anderson R, Stainier DY, Spradling AC, Halpern ME. Genetics. 2009;182:747–755. doi: 10.1534/genetics.109.102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akitake CM, Macurak M, Halpern ME, Goll MG. Dev Biol. 2011;352:191–201. doi: 10.1016/j.ydbio.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giles NH, Geever RF, Asch DK, Avalos J, Case ME. J Hered. 1991;82:1–7. doi: 10.1093/jhered/82.1.1. [DOI] [PubMed] [Google Scholar]
- 6.Potter CJ, Tasic B, Russler EV, Liang L, Luo L. Cell. 2010;141:536–548. doi: 10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei X, Potter CJ, Luo L, Shen K. Nat Methods. 2012;9:391–395. doi: 10.1038/nmeth.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawakami K, Shima A, Kawakami N. Proc Natl Acad Sci U S A. 2000;97:11403–11408. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson AD, Krieg PA. Gene. 1994;147:223–226. doi: 10.1016/0378-1119(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 10.Amsterdam A, Lin S, Hopkins N. Dev Biol. 1995;171:123–129. doi: 10.1006/dbio.1995.1265. [DOI] [PubMed] [Google Scholar]
- 11.Linney E, Hardison NL, Lonze BE, Lyons S, DiNapoli L. Dev Biol. 1999;213:207–216. doi: 10.1006/dbio.1999.9376. [DOI] [PubMed] [Google Scholar]
- 12.Urasaki A, Morvan G, Kawakami K. Genetics. 2006;174:639–649. doi: 10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mosimann C, Kaufman CK, Li P, Pugach EK, Tamplin OJ, Zon LI. Development. 2011;138:169–177. doi: 10.1242/dev.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker C. Methods Cell Biol. 1999;60:43–70. doi: 10.1016/s0091-679x(08)61893-2. [DOI] [PubMed] [Google Scholar]
- 15.Nakano T, Windrem M, Zappavigna V, Goldman SA. Dev Biol. 2005;283:474–485. doi: 10.1016/j.ydbio.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 16.Zelenchuk TA, Bruses JL. Genesis. 2011;49:546–554. doi: 10.1002/dvg.20766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arkhipova V, Wendik B, Devos N, Ek O, Peers B, Meyer D. Dev Biol. 2012;365:290–302. doi: 10.1016/j.ydbio.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emelyanov A, Parinov S. Dev Biol. 2008;320:113–121. doi: 10.1016/j.ydbio.2008.04.042. [DOI] [PubMed] [Google Scholar]
- 19.Farooq M, Sulochana KN, Pan X, To J, Sheng D, Gong Z, Ge R. Dev Biol. 2008;317:336–353. doi: 10.1016/j.ydbio.2008.02.034. [DOI] [PubMed] [Google Scholar]
- 20.Her GM, Chiang CC, Chen WY, Wu JL. FEBS Lett. 2003;538:125–133. doi: 10.1016/s0014-5793(03)00157-1. [DOI] [PubMed] [Google Scholar]
- 21.Pisharath H, Rhee JM, Swanson MA, Leach SD, Parsons MJ. Mech Dev. 2007;124:218–229. doi: 10.1016/j.mod.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grossman SR, Johannsen E, Tong X, Yalamanchili R, Kieff E. Proc Natl Acad Sci U S A. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henkel T, Ling PD, Hayward SD, Peterson MG. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 24.Minoguchi S, Taniguchi Y, Kato H, Okazaki T, Strobl LJ, Zimber-Strobl U, Bornkamm GW, Honjo T. Mol Cell Biol. 1997;17:2679–2687. doi: 10.1128/mcb.17.5.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pisharath H, Parsons MJ. Methods Mol Biol. 2009;546:133–143. doi: 10.1007/978-1-60327-977-2_9. [DOI] [PubMed] [Google Scholar]
- 26.Parsons MJ, Pisharath H, Yusuff S, Moore JC, Siekmann AF, Lawson N, Leach SD. Mech Dev. 2009;126:898–912. doi: 10.1016/j.mod.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karlsson J, von Hofsten J, Olsson PE. Mar Biotechnol (NY) 2001;3:522–527. doi: 10.1007/s1012601-0053-4. [DOI] [PubMed] [Google Scholar]
- 28.Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 29.Halpern ME, Rhee J, Goll MG, Akitake CM, Parsons M, Leach SD. Zebrafish. 2008;5:97–110. doi: 10.1089/zeb.2008.0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asakawa K, Kawakami K. Dev Growth Differ. 2008;50:391–399. doi: 10.1111/j.1440-169X.2008.01044.x. [DOI] [PubMed] [Google Scholar]
- 31.Distel M, Wullimann MF, Koster RW. Proc Natl Acad Sci U S A. 2009;106:13365–13370. doi: 10.1073/pnas.0903060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knopf F, Schnabel K, Haase C, Pfeifer K, Anastassiadis K, Weidinger G. Proc Natl Acad Sci U S A. 2010;107:19933–19938. doi: 10.1073/pnas.1007799107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfeiffer BD, Ngo TT, Hibbard KL, Murphy C, Jenett A, Truman JW, Rubin GM. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Potter CJ, Luo L. Nat Protoc. 2011;6:1105–1120. doi: 10.1038/nprot.2011.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogura E, Okuda Y, Kondoh H, Kamachi Y. Dev Dyn. 2009;238:641–655. doi: 10.1002/dvdy.21863. [DOI] [PubMed] [Google Scholar]
- 36.Pfeiffer BD, Truman JW, Rubin GM. Proc Natl Acad Sci U S A. 2012;109:6626–6631. doi: 10.1073/pnas.1204520109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Villefranc JA, Amigo J, Lawson ND. Dev Dyn. 2007;236:3077–3087. doi: 10.1002/dvdy.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]